Abstract
The TWINKLE protein is a hexameric DNA helicase required for replication of mitochondrial DNA. TWINKLE displays striking sequence similarity to the bacteriophage T7 gene 4 protein (gp4), which is a bi-functional primase-helicase required at the phage DNA replication fork. The N-terminal domain of human TWINKLE contains some of the characteristic sequence motifs found in the N-terminal primase domain of the T7 gp4, but other important motifs are missing. TWINKLE is not an active primase in vitro and the functional role of the N-terminal region has remained elusive. In this report, we demonstrate that the N-terminal part of TWINKLE is required for efficient binding to single-stranded DNA. Truncations of this region reduce DNA helicase activity and mitochondrial DNA replisome processivity. We also find that the gp4 and TWINKLE are functionally distinct. In contrast to the phage protein, TWINKLE binds to double-stranded DNA. Moreover, TWINKLE forms stable hexamers even in the absence of Mg2+ or NTPs, which suggests that an accessory protein, a helicase loader, is needed for loading of TWINKLE onto the circular mtDNA genome.
INTRODUCTION
DNA helicases catalyze unwinding of double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA), a process energetically coupled to nucleotide triphosphate hydrolysis. This group of enzymes is involved in processes assuring genome stability such as DNA replication, repair and recombination. Pathogenic mutations disturbing helicase function can affect these basic cellular mechanisms and lead to human disease, including cancer and premature aging (1). TWINKLE is the only known DNA helicase involved in replication of human mitochondrial DNA (mtDNA) and mutations in the TWINKLE gene can cause autosomal dominant progressive external opthalmoplegia (adPEO), a neuromuscular disorder associated with multiple deletions in mtDNA (2). TWINKLE is a hexameric helicase that can unwind short stretches (<20 bp) of dsDNA in the 5′ to 3′ direction (3). Overexpression of TWINKLE variants containing amino acid substitutions in the helicase active site leads to mtDNA depletion in both human and drosophila cell lines (4,5). Together with the mtDNA polymerase gamma (POLγ) and the mitochondrial ssDNA-binding protein (mtSSB), TWINKLE forms a processive replication apparatus, a replisome that can synthesize ssDNA molecules of up to 16 kb, roughly corresponding to the size of the human mtDNA molecule (6). Homology searches revealed a striking sequence similarity between TWINKLE and the bacteriophage T7 gene 4 protein (gp4), a bi-functional primase-helicase required at the replication fork of bacteriophage T7 DNA (2).
Electron microscopy and X-ray crystallography studies have demonstrated that gp4 assembles as both ring-shaped hexamers and heptamers (7–9). ssDNA binds within the central hole of the gp4 hexamer and contributes to high processivity of translocation. The gp4 can be divided functionally and structurally into two distinct regions. The N-terminal part of the protein harbors a primase domain (residues 1–245) and the C-terminal part possesses DNA helicase activity (residues 272–566). A 26-amino acid linker region required for hexamer formation separates the two functional domains (10). The C-terminal helicase domain includes the conserved sequence motifs characteristic of the RecA/DnaB family of replicative helicases (11) and is responsible for DNA-dependent dTTP hydrolysis, translocation along DNA and DNA unwinding (12). Moreover, 17 amino acids at the very C-terminus are involved in physical interactions between the helicase and T7 DNA polymerase (13,14). The N-terminal primase domain of gp4 comprises a functional Zn2+-finger and an RNA polymerase domain that act together to synthesize ribonucleotide primers required for initiation of DNA synthesis. The primase domain also stimulates primer utilization by the T7 DNA polymerase (15).
Similar to the gp4 protein and related ring helicases, TWINKLE forms hexameric complexes in solution (2). TWINKLE can also be divided into three regions, an N-terminal primase-like domain, a short linker region and a C-terminal DNA helicase domain. Bioinformatic analyses suggest that TWINKLE is an active DNA primase in most eukaryotes, but that this activity has been lost in metazoan cells (16). In support of this notion, we have not been able to identify a primase activity in purified human TWINKLE (unpublished data). Human TWINKLE contains primase motifs II and III, but has no discernible Zn2+-finger motif (motif I), which in gp4 is involved in nucleotide sequence recognition as well as in transfer of the primer to the polymerase (2,16). Also primase motifs IV–VI are well conserved in human TWINKLE, with the notable exception of essential amino acids involved in binding to Mg2+ ions. Therefore, even in the absence of a primase activity, the N-terminal region still contains many conserved primase motifs and others have suggested that this region may play an important role in mtDNA replication, e.g. by stimulating primer utilization (16).
In the present article, we have investigated the role of the N-terminal domain of human TWINKLE using deletion mutagenesis. We also addressed the role of TWINKY, a splice variant of TWINKLE lacking a portion of the C-terminal helicase domain. Our results revealed that the N-terminal domain is required for efficient binding to ssDNA and for the DNA unwinding activity of TWINKLE. The N-terminal domain is not absolutely required for mtDNA replisome function, but is needed for the formation of long DNA products.
MATERIALS AND METHODS
Recombinant proteins
TWINKLE, mtSSB, POLγA and POLγB were expressed and purified as described previously (6). A 6× His-tagged version of wild-type (wt) TWINKLE cloned into pBacPAK9 was used to generate the truncated forms of TWINKLE by PCR. Plasmids containing the Δ1–121, Δ1–314 and TWINKY truncated genes were sequenced and used to prepare recombinant Autographa californica nuclear polyhedrosis virus as described in the BacPAK manual (Clontech). The truncated versions of TWINKLE were purified following the same protocol used for the wt protein, with the exception that TWINKY did not bind to the heparin and SP columns, but eluted in the flowthrough fractions from these two columns. The Δ372–684 truncated gene was cloned in pET-20b (Stratagene) and expressed in Escherichia coli strain BL21(DE3)pLysS. The bacterial culture was grown a 37°C to an A600 of ∼ 0.8 in LB medium. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mM and the cells were cultured for three additional hours at 30°C and harvested by centrifugation. They were frozen in liquid nitrogen, thawed in lysis buffer [50 mM NaH2PO4 (pH 8), 300 mM NaCl, 10 mM imidazole] and incubated on ice for 30 min in the presence of 1 mg/ml lysozyme. The cells were then disrupted by sonication (6 × 20 s) and centrifuged at 10 000 g for 30 min. The supernatant was mixed with 2 ml of Ni2+-NTA matrix superflow (Qiagen) equilibrated with buffer A [25 mM Tris–HCl (pH 8.0), 10% glycerol, protease inhibitors, 10 mM β-mercaptoethanol, 0.4 M NaCl, 10 mM imidazole) and rotated for 60 min at 4°C. The Ni2+-NTA matrix was collected by centrifugation (1500 g for 10 min), resuspended in buffer A (10 mM imidazole), poured into a column, and washed with 10 column volumes of buffer A. Protein was then eluted with buffer A containing 250 mM imidazole and peak fractions containing protein were combined. The Δ372–684 protein was purified further on heparin and SP columns (GE Healthcare), following the same protocol used for wt TWINKLE. The purity of the proteins was estimated (by SDS–PAGE and Coomassie blue staining) to be >95%. Co-expression of wt TWINKLE and TWINKY was performed as described earlier (17). The co-expressed proteins were partially purified on TALON™ metal affinity resin (BD Biosciences Clontech) (6).
Gel filtration
Oligomerization studies were performed by gel-filtration chromatography using a Superose 12 PC 32/30 column (GE Healthcare) equilibrated in running buffer [25 mM Tris–HCl (pH 8.0), 10% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA and 0.4 M NaCl unless otherwise stated in the figure legends]. The different versions of TWINKLE (30 ng) eluted from the TALON™ metal affinity resin were loaded onto the Superose 12 PC 32/30 column in a total volume of 200 μl and at a flow rate of 0.04 ml/min at 4°C. Fractions of 0.05 ml were collected, and the protein content (15 μl of the indicated fractions) was analyzed by SDS–PAGE and Coomassie brilliant blue staining. A calibration curve was prepared, following the instructions of the manufacturer of the column, by running thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.35 kDa), all from BioRad. Elution of marker proteins was monitored by UV photometry (280 nm). The logarithm of molecular weight was plotted against Kav, which was calculated for each protein as follows: Kav = (Ve–Vo)/(Vt–Vo), where Ve = elution volume for the protein, Vo = column void volume (0.85 ml) and Vt = total bed volume (2.4 ml for Superose 12 PC 32/30).
Gas-phase electrophoretic mobility macromolecule analysis (GEMMA)
The TWINKLE proteins were buffer exchanged by Sephadex G-25 chromatography to a buffer containing 0.8 M ammonium acetate pH 7, 1 mM DTT and 0.005% Tween-20. The proteins were diluted to appropriate conditions for GEMMA (figure legends) and analyzed as described previously (18).
Electrophoresis mobility shift assay
Unspecific ssDNA and dsDNA-binding affinity of TWINKLE was assayed in an electrophoresis mobility shift assay (EMSA) using single-stranded or double-stranded probes. The single-stranded probes consisted of poly-d(T) of different lengths (15-, 30- and 45-nt) labeled at the 5′-end with γ-32P ATP using T4 polynucleotide kinase. To prepare the dsDNA probes, the following oligonucleotides were labeled at the 5′-end: ds15, 5′-CTCTAGACTCGACCG-3′; ds30, 5′-TGGCACGACAGGTTTCCCGACTGGAAAGCG-3′; ds45, 5′TGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTCAGCTCTAG-3′ and annealed with their complementary oligonucleotide. Reactions were carried out in 15 μl volumes containing 10 fmol DNA template, 20 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 10% glycerol and TWINKLE proteins indicated in the figure legends. Where indicated in the figure legends, 10 mM MgCl2, 2 mM ATP, 2 mM ATPγS or 2 mM ADP were added to the reaction. The reactions were incubated at RT for 10 min and run immediately on a 6% native polyacrylamide gel in 1× TBE for 15 min at 150 V.
ATPase activity
ATP hydrolysis by wt and truncated TWINKLE proteins was determined in a 20 μl reaction mixture containing 20 mM Tris—HCl (pH 7.5), 4.5 mM MgCl2, 10% glycerol, 0.3 mg/ml bovine serum albumin, 0.7 mM ATP, 300 nCi of [γ-32P] ATP (GE Healthcare) and 250 fmol of TWINKLE hexamer, in the presence or absence of 188 fmol M13mp18 ssDNA. Reactions were incubated for 50 min at 32°C and terminated by the addition of 400 μl of a Norit A suspension (12% in 0.1 M HCl, 10 mM KH2PO4). The mixture was vortexed briefly and then centrifuged for 10 min at 7000g. Two hundred microliter of the supernatant was mixed with 3 ml of scintillation mixture (Ready Safe; Beckman-Coulter), and the radioactivity was measured in a liquid scintillation counter. The enzyme-dependent release of phosphate was calculated by subtracting the release of phosphate in samples without protein.
Helicase assay
The DNA substrate was prepared by annealing a 60-nt oligonucleotide (5′-ACATGATAAGATACATGGATGAGTTTGGACAAACCACAACGTAAAACGACGGCCAGTGCC-3′) to M13mp18 ssDNA. The 20 3′-bases are complementary to M13mp18 ssDNA, whereas the 40 5′-bases form a 5′ single-stranded tail. The reaction mixture (15 μl) contained 20 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 4 mM DTT, 100 μg/ml BSA, 3 mM ATP, 10% glycerol, 5 fmol of DNA substrate and TWINKLE proteins as indicated in the figure legends. The reactions were incubated at 32°C for 45 min and stopped by the addition of 3 μl stop solution [90 mM EDTA (pH 8.0), 6% SDS, 30% glycerol, 0.025% bromophenol and 0.025% xylene cyanol). The reaction products were separated by electrophoresis through a 12.5% non-denaturing polyacrylamide gel. The gel was dried onto DE81 paper (Whatman) and autoradiographed overnight at −80°C with an intensifying screen.
In vitro DNA replication
The mini-circle template for rolling-circle DNA replication was generated as previously described (19). The mini-circle template (12.5 fmol) was added to a 20 μl reaction mixture containing 25 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 4 mM ATP, 10% glycerol, 100 μM dATP, 100 μM dTTP, 100 μM dGTP, 10 μM dCTP, 2 μCi [α-32P] dCTP and the indicated amounts of the different replication factors. The reaction was incubated at 37°C for 90 min and terminated by addition of 200 μl stop buffer [10 mM Tris–HCl (pH 8.0), 0.2 M NaCl, 1 mM EDTA and 0.1 mg/ml glycogen). The samples were treated with 0.5% SDS and 100 μg/ml proteinase K for 45 min at 42°C, and precipitated by addition of 0.6 ml ice-cold 95% ethanol. The pellets were dissolved in 10 μl gel-loading buffer (98% formamide, 10 mM EDTA (pH 8.0), 0.025% xylene cyanol FF and 0.025% bromophenol blue). The samples were heated at 95°C for 5 min and separated on a 0.8% denaturing agarose gel as described previously (6). The gels were dried onto DE81 paper (Whatman) and the replication products were visualized by autoradiography after overnight exposure at −80°C with an intensifying screen.
RESULTS
Deletion analysis of human TWINKLE
To determine the functional role of the N-terminal primase-like domain of TWINKLE, we expressed and purified a series of deleted versions of the protein (Figure 1A). Wt TWINKLE lacked the mitochondrial targeting sequence (amino acids 1–42), whereas the smaller splice variant TWINKY also lacked residues 579–684 and contained four unique amino acids at the C-terminus (2). The different truncated versions of TWINKLE were based on published predictions of the protein structure (16). A region of TWINKLE corresponding to the Zn-binding domain in gp4 was deleted in the Δ1–121 construct. The Δ1–314 construct lacked nearly the entire primase-related region, but the protein still contained all amino acids positions associated with adPEO causing mutations in affected patients. In Δ372–684, we deleted the entire C-terminal part of the protein, and only expressed the N-terminal primase-related domain together with the linker region. The proteins were expressed in insect cells (WT, Δ1–121, Δ1–314 and TWINKY) or in bacteria (Δ372–684), and purified as described for the full-length protein. The purity of the proteins is shown in Figure 1B. The truncated proteins were expressed at about the same levels as wt TWINKLE and remained in solution during purification.
Figure 1.
Deletion mutagenesis of TWINKLE. (A) Schematic representation of the WT and mutant TWINKLE proteins used in this study. TWINKLE consists of a primase-related N-terminal domain (white) and a helicase domain (black), connected by a linker region (gray). (B) Purified recombinant wt and mutant versions of TWINKLE (0.2 μg) were separated by SDS–PAGE (10%) and stained with Coomassie blue. Lanes 1 and 7, size marker; lane 2, wt TWINKLE; lane 3, Δ1–121; lane 4, Δ1–314; lane 5, TWINKY; lane 6, Δ372–684.
The hexameric structure of TWINKLE is stable and nucleotide-independent
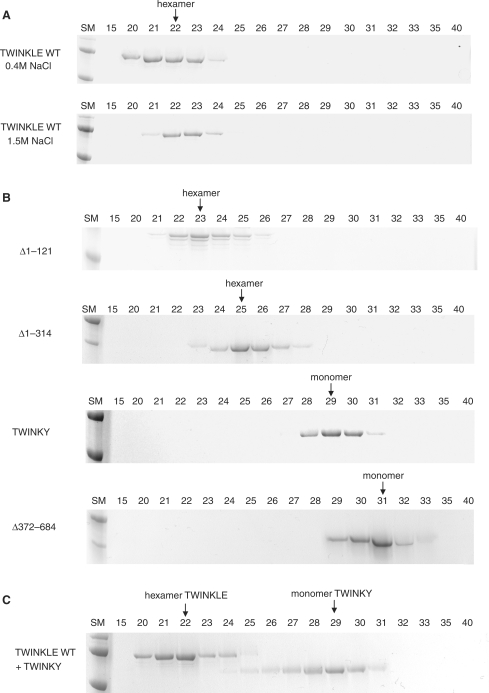
We used gel-filtration chromatography to determine the oligomeric structure of TWINKLE and noted that the protein forms hexamers even in the absence of Mg2+ or ATP, and at high ionic strength (Figure 2A). This finding contrasts with many other hexameric helicases, e.g. the T7 gp4 protein, which requires Mg2+ or nucleotide cofactors for stable hexamer formation (20). The finding that TWINKLE forms stable hexamers in solution has also recently been reported by Kaguni and coworkers (21).
Figure 2.
Hexamerization of the TWINKLE proteins. (A) Wt TWINKLE was dialyzed for 4 h in a buffer containing 0.4 M or 1.5 M NaCl and its oligomeric state was determined using a Superose 12 PC 32/30 gel-filtration column, as described in Material and Methods section, equilibrated with buffer containing either 0.4 or 1.5 M NaCl. (B) Mutant TWINKLE proteins were examined for their ability to hexamerize as described in Material and Methods section. (C) Co-expressed wt TWINKLE and TWINKY were analyzed on a Superose 12 gel-filtration column as described in Material and Methods section. The positions of the different versions of TWINKLE, as hexamers or monomers, are indicated with arrows at the tops of the gels. Fraction numbers are indicated above the gels.
The C-terminal domain of TWINKLE is needed for hexamer formation
To investigate the role of the different TWINKLE domains in hexamer formation, we analyzed the truncated derivatives by gel filtration chromatography (Figure 2B and Supplementary Data). Both Δ1–121 and Δ1–314 could form a larger complex with an apparent molecular weight corresponding to a hexameric conformation. The stability of the Δ1–314 complex was, however, somewhat reduced since the protein peak was broader and somewhat less defined than for the wt TWINKLE protein. In contrast, TWINKY and Δ372–684 migrated with an apparent molecular weight corresponding to a monomeric conformation. We verified that our gel-filtration analysis could correctly distinguish between the different oligomeric forms of TWINKLE by using GEMMA to estimate the molecular weight of Δ1–121 and Δ372–684. In this analysis, the observed molecular weight for Δ1–121 was ∼369 kDa, in nice agreement with the predicted molecular weight of 382 kDa for a Δ1–121 protein hexamer (Figure 3). The observed molecular weight for Δ372–684 was 38 kDa, in close correspondence to the predicted molecular weight of 36.9 kDa for a Δ372–684 protein monomer. Our data therefore suggest that even if the C-terminal domain is essential for oligomerization, the N-terminal domain also contributes to proper hexamerization. Since TWINKY could not hexamerize, we investigated whether it could form mixed hexamers with TWINKLE. We co-expressed the two proteins in insect cells and loaded them onto the gel-filtration column. The greatest proportion of TWINKLE eluted as a stable hexamer (Figure 2C). The majority of TWINKY migrated as a monomer, but a very small fraction of the protein appeared to eluate together with the wt hexamer. We conclude that TWINKY is a monomer in solution and unable to interact and form higher order stoichiometric complexes with the wt TWINKLE protein.
Figure 3.
GEMMA analysis of TWINKLE proteins. (A) Analysis of the Δ1–121 mutant (0.05 mg/ml). (B) Analysis of the Δ372–684 mutant (0.025 mg/ml). The buffer used for the analyses consisted of 0.4 M ammonium acetate pH 7, 0.5 mM DTT and 0.0075% Tween-20. The abscissa shows the diameter of the particles, which is correlated to the molecular weight shown on top of each peak.
Interaction of TWINKLE with single-stranded and double-stranded DNAs
We used EMSAs to study the interaction between TWINKLE and ssDNA and dsDNA. Nucleotides, e.g. dNMP, dNDP or dNTP, have been shown to stimulate DNA binding by other hexameric helicases (22). We investigated the effects of MgCl2, ATP, ATPγS and ADP on the DNA-binding capacity of TWINKLE. To monitor ssDNA binding, we used a 30-nt long poly-dT oligonucleotide (Figure 4A). TWINKLE interacted with ssDNA only in the presence of ATP or ATPγS, but not in the presence of ADP. In contrast, the binding of TWINKLE to a 30-bp dsDNA fragment was not affected by the presence of MgCl2 or nucleotides.
Figure 4.
Interactions of TWINKLE proteins with single-stranded and double-stranded DNAs. (A) DNA binding of wt TWINKLE was determined by EMSA using a 32P-labeled 30-mer poly-dT ssDNA (lanes 1–6) or a 30-bp non-specific dsDNA (lanes 7–12) as described in Material and Methods section. Lanes 3–6 and 9–12, 10 mM MgCl2; lanes 4 and 10, 2 mM ATP; lanes 5 and 11, 2 mM ATPγS; lanes 6 and 12, 2 mM ADP (B) Ten femtomole of 32P-labeled 30-mer ssDNA (lanes 1–6) or 30-bp dsDNA (lanes 7–12) were incubated with a saturating amount of wt TWINKLE (200 fmol), 10 mM MgCl2, 2 mM ATP and increasing amounts of unlabeled competitor DNA as indicated (0, 5, 10, 50 and 100 fmol). The protein–DNA complexes were incubated for 10 min at RT before being resolved by native 6% PAGE. (C) Binding reactions between wt TWINKLE and ssDNA (left) or dsDNA (right) of different lengths (15, 30 and 45 nucleotides) were performed as described in Material and Methods section in the presence of 10 mM MgCl2 and 2 mM ATP. The percentage of bound DNA was quantified by phosphor imaging and plotted against the concentration of TWINKLE hexamer. The results presented are the average obtained from two independent experiments. (D) Binding of wt and mutant TWINKLE proteins to 30-mer ssDNA (left) and 30-bp dsDNA (right) were performed and quantified as in (C).
We investigated the minimal length of DNA required for TWINKLE binding (Figure 4C). EMSAs were performed with ssDNA and dsDNA probes of different lengths (15, 30 and 45-nt) and titrations were conduced with increasing amounts of protein. TWINKLE could not interact with a short 15-nt ssDNA probe, but efficiently interacted with a dsDNA probe of similar size. TWINKLE efficiently shifted longer ssDNA and dsDNA probes (Figure 4C). The apparent Kd was ∼5 nM for the interactions observed with either a 30-nt ssDNA probe or a 30-bp dsDNA probe.
To investigate TWINKLE interactions with ssDNA and dsDNA further, we performed competitive DNA-binding experiments. To stimulate ssDNA binding, all experiments were performed in the presence of ATP. We used a constant protein concentration and increased the amount of unlabeled competitor DNA (Figure 4B). Surprisingly, the ssDNA probe bound to TWINKLE was competed off by increasing amounts of dsDNA (Figure 4B, lanes 7–12). In contrast, the binding of TWINKLE to the dsDNA probe was unaffected by the presence of increasing amounts of the ssDNA (Figure 4B, lanes 1–6). This contrasts with the behavior of the gp4 protein, which binds tightly and preferably to ssDNA (22).
Finally, we monitored ssDNA and dsDNA-binding activities for our truncated TWINKLE variants (Figure 4D). TWINKY and Δ372–684 did not display any ssDNA or dsDNA-binding activity. The lack of DNA-binding activity is most likely explained by the monomeric conformation of these two proteins. Other hexameric helicases are also unable to bind ssDNA in their monomeric state (23). The Δ1–314 protein showed reduced binding to both ssDNA and dsDNA, but the effect on ssDNA was much more severe. The Δ1–121 protein bound to dsDNA with the same efficiency as wt TWINKLE, but we observed a decrease in ssDNA binding. Our experiments suggest that the C-terminal helicase domain is responsible for DNA binding, but that the N-terminal domain specifically contributes to the ssDNA-binding activity.
ATP hydrolysis activity of the wt and truncated TWINKLE proteins
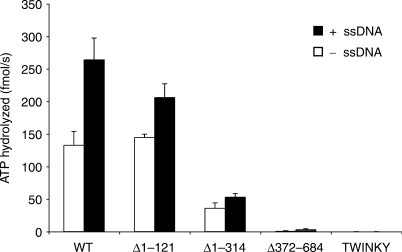
Measurements of ATP hydrolysis were performed both in the absence and presence of single-stranded M13 DNA. The DNA-independent ATPase activity provides information about the intrinsic ability of the protein to hydrolyze ATP, whereas measurement of ATPase activity in the presence of ssDNA reflects the DNA-stimulated ATPase activity (23). TWINKLE has a high intrinsic ATPase activity, which was stimulated about 2-fold in the presence of ssDNA (Figure 5). Addition of linear dsDNA did not stimulate the TWINKLE associated ATPase activity (data not shown). In the absence of DNA, the Δ1–121 protein hydrolyzed ATP at a rate very similar to that of the WT protein, Δ1–314 showed a marked decrease in activity, whereas TWINKY and Δ372–684 were completely inactive. TWINKY and Δ372–684 remained inactive in the presence of ssDNA, whereas Δ1–121 and especially Δ1–314 displayed a reduced ssDNA-dependent stimulation (Figure 5). The N-terminal region of TWINKLE is apparently not absolutely required for ATP hydrolysis, but deletions in this region have a strong negative effect on the levels of ATPase activity and also decrease ssDNA-dependent stimulation. This finding is consistent with the observation that both Δ1–121 and Δ1–314 have a reduced affinity for ssDNA (Figure 4D).
Figure 5.
ATP hydrolysis by the WT and mutant TWINKLE proteins. Wt TWINKLE and mutant TWINKLE proteins (250 fmol) were analyzed for their ability to hydrolyze ATP in the absence (white bars) and in the presence (black bars) of M13mp18 ssDNA (188 fmol). The average of three independent experiments is given. Each reaction (20 μl) was incubated at 32°C for 50 min and analyzed as described in Material and Methods section.
Helicase activity
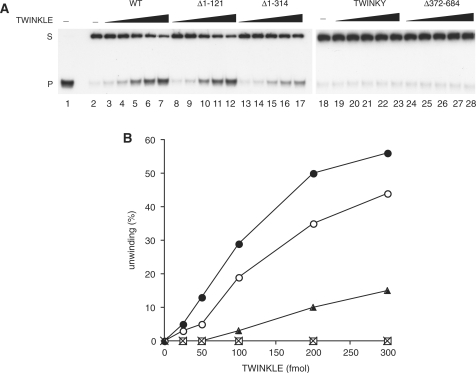
Even if the Δ1–121 protein, and to lesser extent the Δ1–314 protein, were still able to hydrolyze ATP, these truncated proteins might still be defective in their ability to use the energy of ATP hydrolysis to translocate along ssDNA and unwind duplex DNA. Thus, we examined helicase activity of the truncated TWINKLE proteins (Figure 6). To prepare the substrate used in this assay, a 60-nt oligonucleotide was annealed to M13ssDNA, to form a 20-bp duplex and a 40-nt 5′-tail. We found that the relative unwinding activity displayed by the wt and truncated proteins was essentially proportional to the activities observed in the ssDNA binding (Figure 4D) and in the DNA-dependent ATP hydrolysis reactions (Figure 5). The Δ1–121 protein was able to unwind DNA nearly as efficiently as the wt TWINKLE, whereas the Δ1–314 protein showed a significantly reduced helicase activity. TWINKY and the Δ372–684 versions were completely inactive in the DNA helicase assay.
Figure 6.
Helicase activity of the wt and mutant TWINKLE proteins. (A) The DNA unwinding activities of wt and mutant TWINKLE were measured by monitoring the amount of unwound ssDNA product (P) from the duplex DNA substrate (S) on a 12.5% non-denaturing polyacrylamide gel. The reaction was performed as described in Material and Methods section, in the presence of increasing amounts of the TWINKLE proteins indicated (25, 50, 100, 200 and 300 fmol). After a 45-min incubation at 32°C, the reaction products were run on a 12.5% non-denaturating polyacrylamide gel. Lane 1, substrate heated to 100°C before loading; lane 2, untreated substrate; S, double-stranded substrate; P, single-stranded product. (B) The helicase assay was performed exactly as in panel A and then analyzed with phosphor imaging, and the progress of unwinding was estimated from the ratio of the ssDNA product to the substrate. The data presented here are the average of two independent experiments. WT (filled circle), Δ1–121 (open circle), Δ1–314 (filled triangle), TWINKY (cross-mark) and Δ372–684 (open square).
Stimulation of POLγ activity on a dsDNA template
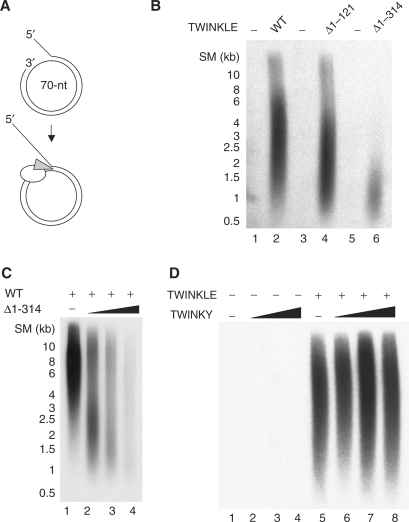
We have shown previously that TWINKLE alone is unable to unwind long stretches of dsDNA. We decided to investigate the role of the N-terminal domain for DNA synthesis on a duplex DNA template. To this end, we formed a template for DNA replication by annealing a 90-nt oligonucleotide to a 70-nt ssDNA mini-circle. The template contained a replication fork for loading the replication machinery, a 50-bp dsDNA region and a free 3′-hydroxyl terminus that could act as a primer for DNA synthesis (Figure 7A). Once initiated, leading-strand DNA synthesis coupled to continuous unwinding of the double-stranded template could in principle progress indefinitely. We have previously reported that POLγ can use the 3′-hydroxyl terminus on this mini-circle template and initiate DNA synthesis, but that the enzyme fails to elongate through double-stranded regions and only forms a 110-nt product. To elongate through the dsDNA region, the POLγ requires the DNA strand unwinding activity of the TWINKLE helicase (6). In agreement with our previous observations, there was no DNA synthesis in the absence of TWINKLE (Figure 7B, lanes 1, 3 and 5). The addition of wt TWINKLE to the reaction led to a marked increase in DNA synthesis, and allowed the formation of ssDNA products of up to 10 kb (Figure 7B, lane 2). The Δ1–121 protein was able to support DNA synthesis at almost wt levels (Figure 7B, lane 4). In contrast, Δ1–314 could only support synthesis of shorter DNA products of ∼1 kb. Addition of higher concentrations of Δ1–314 led to an increase in DNA synthesis, but had no effect on the length of the DNA products (data not shown). Addition of Δ1–314 could also severely reduce mtDNA replication in the presence wt TWINKLE, arguing for Δ1–314 being correctly folded and able to compete with the wt protein for binding to the replication fork (Figure 7C). Our data therefore demonstrate that the N-terminal domain of TWINKLE is required for full mtDNA replisome processivity. Reduced processivity could explain why overexpression of a related mutant in cell lines (Δ70–343) leads to a reduction of mtDNA copy number and stalling of the mtDNA replication fork (5).
Figure 7.
Rolling-circle DNA synthesis in the presence of the different TWINKLE mutants. (A) The mini-circle template was generated as previously described (19). The template can be efficiently replicated by the POLγ holoenzyme (white) and TWINKLE helicase (gray). (B) The mini-circle template (12.5 fmol) was incubated with constant amounts of POLγA (100 fmol), POLγB (200 fmol), mtSSB (5 pmol) for 90 min at 37°C with or without 100 fmol of the indicated TWINKLE protein. The reaction products were analyzed on a 0.8% denaturating agarose gel. (C) The reaction was performed as in (B). Increasing amounts of Δ1–314 were added in presence of 100 fmol wt TWINKLE. Lane 1, 0 fmol Δ1–314; lane 2, 50 fmol Δ1–314; lane 3, 100 fmol Δ1–314; lane 4, 200 fmolΔ1–314. (D) The reaction was performed as in (B). Increasing amounts of TWINKY were added in the absence (lanes 1–4) or presence (lanes 5–8) of 100 fmol wt TWINKLE. Lanes 1 and 5, 0 fmol TWINKY; lanes 2 and 6, 50 fmol TWINKY; lanes 3 and 7, 100 fmol TWINKY; lanes 4 and 8, 200 fmol TWINKY.
Finally, we investigated whether TWINKY could affect mtDNA replisome function in vitro. As expected from our enzymatic analysis of TWINKY, the protein could not support mtDNA replisome function (Figure 7D, lanes 1–4). It was possible that TWINKY might affect mtDNA replisome function in the presence of wt TWINKLE. We therefore added increasing amounts of TWINKY to replication reactions containing wt TWINKLE. We could not detect any effect of TWINKY on the mtDNA replisome activity, which argues against a functional role for this protein isoform at the mtDNA replication fork (Figure 7D, lanes 5–8). In support of this notion, overexpression of TWINKY in cell lines does not compromise the activity of the endogenous Twinkle protein (5).
DISCUSSION
We have attempted to identify a primase activity associated with the TWINKLE protein (data not shown). Using many different approaches, analyzing TWINKLE both in isolation and in combination with other mtDNA replication and transcription factors, we have so far failed to see any such enzymatic activities. In addition, bioinformatic analysis of mammalian TWINKLE sequences suggests that the protein has lost important protein motifs required for primase activity (16,24). Based on these observations, we decided to investigate whether the N-terminal region of TWINKLE may have other functional roles in mtDNA replisome function that are not related to primer synthesis.
TWINKLE binds to both ssDNA and dsDNA, but the two activities appear to be functionally distinct. Whereas C-terminal truncations affected both ssDNA and dsDNA binding, N-terminal truncations mainly affected binding to ssDNA. This finding leads us to speculate that ssDNA binding requires a larger protein surface, involving both the N-terminal and the C-terminal part of TWINKLE, whereas dsDNA binding is localized to the C-terminal helicase domain. In support of this idea, TWINKLE failed to interact with shorter pieces of ssDNA (15 nt), but could efficiently interact with dsDNA of similar length. In contrast, TWINKLE binds with similar efficiency to longer stretches of ssDNA and dsDNA (>30 nt or base pairs).
The existence of an ssDNA-binding region in the N-terminal region of TWINKLE has gained support from studies of structurally related proteins. Analyses of prokaryotic primases have identified catalytically important basic residues that cluster around a shallow cleft located within the catalytic core (25,26). These residues form an elongated region with significant electropositive potential, which is required for binding to ssDNA. The presence of an ssDNA-binding region in the N-terminal region of TWINKLE may also explain why binding to ssDNA, in contrast to dsDNA binding, is dependent on nucleotide cofactors. In the X-ray structure of the gp4 protein, ATP binds at the interface between two monomers. This interface is created by amino acids from the linker region and the N-terminal part of the helicase domain (27). In turn, the linker region interacts directly with amino acids in the ssDNA-binding region. This intricate web of interactions between the ATP and ssDNA-binding regions explains why nucleotide cofactors are required for gp4 binding to ssDNA. Conversely, this structural organization also explains why truncations in the N-terminal region, may affect ATPase and DNA helicase activity in the gp4 protein (12). It is likely that a similar arrangement exists in TWINKLE. In support of this notion, molecular modeling of TWINKLE suggests that the positively charged ssDNA-binding surface is conserved (unpublished data).
The N-terminal region of TWINKLE is not absolutely required for DNA helicase activity. The Δ1–314 mutant can still hydrolyze ATP and unwind dsDNA, albeit at a slower rate. In fact, the mutant is capable of supporting POLγ-dependent DNA synthesis on a duplex DNA substrate. The products are shorter than those observed for wt TWINKLE, which could be explained by the reduced helicase activity. Interestingly, a heterologous helicase was found to be unable to support POLγ-dependent DNA synthesis on the same template (6), which led to the suggestion that POLγ may interact specifically with TWINKLE at the mtDNA replication fork. In this respect, our observation that the Δ1–314 mutant can support POLγ function is intriguing. It may suggest that if direct TWINKLE interactions are required for POLγ function, then these interactions cannot be absolutely dependent on the primase-like domain of the protein.
Our analysis has also revealed efficient binding of TWINKLE to dsDNA. Interestingly, the bacterial ring-shaped helicase DnaB can accommodate both ssDNA and dsDNA in its central channel. It has been shown that DnaB requires a replication fork substrate in order to unwind DNA in the 5′–3′ direction. However, in the absence of a 3′-tail, the DnaB protein translocates from the 5′-ssDNA tail onto the dsDNA, encircles both strands and continues to translocate actively on the duplex (28). A similar mechanism has been shown for Mcm4,6,7, the putative eukaryotic replication fork helicase (29). The translocation on the duplex DNA has been proposed to accomplish several functions, including driving branch migration of a Holliday junction (30). This suggests that the DnaB protein may be directly involved in DNA recombination in vivo. We have shown previously that similarly to DnaB and Mcm 4,6,7, TWINKLE requires a fork substrate for helicase activity (3). Furthermore, we have demonstrated in this article that TWINKLE can bind dsDNA in addition to ssDNA. To date, dsDNA translocation by TWINKLE has not been reported. However, if TWINKLE is able to translocate actively along the duplex in the 5′–3′ direction, this could confer additional functions to the helicase, for example in DNA recombination or repair. In future experiments, we will address these questions to clarify the molecular function of the dsDNA-binding capacity of TWINKLE.
Finally, we have shown that TWINKLE forms stable hexamers, even in the absence of Mg2+ or NTP. This stable ring structure raises the intriguing question of how the TWINKLE protein loads onto the DNA under normal cellular conditions, when free ends of DNA are not available. Two different models may explain how DNA can enter the central channel of the preformed ring-shaped hexamer. According to the ring-opening model, the preformed hexameric ring must open to load onto the ssDNA, whereas in the disassembly model, helicases must disassemble and reassemble around the ssDNA. These mechanisms often require the presence of accessory proteins, termed helicase loaders (31). A ring-opening mechanism has been proposed for T7 gp4 (32). In this model, the N-terminal primase domain of T7 gp4 initially makes contact with DNA and acts as a helicase loader by holding the helicase next to the DNA before the DNA is transferred into the central channel of the hexamer. Given the similarities that exist between TWINKLE and T7 gp4, it will be of interest to investigate a possible role for the N-terminal primase related domain in the loading of TWINKLE onto DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants to M.F. from the Swedish Research Council, Åke Wiberg foundation, The Swedish Society of Medicine, and the Emil and Wera Cornell's foundation. Funding to pay the Open Access publication charges for this article was provided by the Karolinska Institutet.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 3.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 4.Matsushima Y, Kaguni LS. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in Drosophila mitochondrial DNA helicase expressed in Schneider cells. J. Biol. Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu X, Hingorani MM, Patel SS, Egelman EH. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 1996;3:740–743. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 8.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 9.Crampton DJ, Ohi M, Qimron U, Walz T, Richardson CC. Oligomeric states of bacteriophage T7 gene 4 primase/helicase. J. Mol. Biol. 2006;360:667–677. doi: 10.1016/j.jmb.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Tabor S, Richardson CC. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J. Biol. Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 12.Bird LE, Hakansson K, Pan H, Wigley DB. Characterization and crystallization of the helicase domain of bacteriophage T7 gene 4 protein. Nucleic Acids Res. 1997;25:2620–2626. doi: 10.1093/nar/25.13.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 14.Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J. Biol. Chem. 2006;281:25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- 15.Kusakabe T, Hine AV, Hyberts SG, Richardson CC. The Cys4 zinc finger of bacteriophage T7 primase in sequence-specific single-stranded DNA recognition. Proc. Natl Acad. Sci. USA. 1999;96:4295–4300. doi: 10.1073/pnas.96.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shutt TE, Gray MW. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006;62:588–599. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- 17.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 18.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J. Biol. Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 19.Falkenberg M, Lehman IR, Elias P. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc. Natl Acad. Sci. USA. 2000;97:3896–3900. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picha KM, Patel SS. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 21.Ziebarth TD, Farr CL, Kaguni LS. Modular architecture of the hexameric human mitochondrial DNA helicase. J. Mol. Biol. 2007;367:1382–1391. doi: 10.1016/j.jmb.2007.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hingorani MM, Patel SS. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–12487. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 23.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- 25.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA polymerase domain of E. coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 26.Lee SJ, Richardson CC. Interaction of adjacent primase domains within the hexameric gene 4 helicase-primase of bacteriophage T7. Proc. Natl Acad. Sci. USA. 2002;99:12703–12708. doi: 10.1073/pnas.202471499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan DL. The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan DL, Davey MJ, O’Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003;278:49171–49182. doi: 10.1074/jbc.M308074200. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan DL, O’Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 31.Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- 32.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19:3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]