Abstract
A new method for measuring distances between points in the AraC–DNA complex was developed and applied. It utilizes variable lengths of single-stranded DNA that connect double-stranded regions containing the two half-site binding sequences of AraC. These distances plus the protein interdomain linker distances are compatible with two classes of structure for the dimeric AraC gene regulatory protein. In one class, the N-terminal regulatory arm of one dimerization domain is capable of interacting with the DNA-binding domain on the same polypeptide chain for a cis interaction. In the other class, the possible arm-DNA-binding domain interaction is trans, where it adds to the dimerization interface.
INTRODUCTION
Much is known about the extensively studied Escherichia coli l-arabinose operon regulatory protein, AraC (1). The structure of its N-terminal dimerization domain including most of its 18 residue N-terminal arm has been determined both with and without arabinose bound (2,3), and has been related to function of the protein in a number of genetic and physical studies (4–8). Although the structure of the C-terminal, DNA-binding domain of AraC has not been directly determined, its approximate structure can be inferred because it is homologous to the MarA and Rob proteins, whose structures when bound to DNA have been determined (9,10). The approximate position and relative orientation of the two DNA-binding domains with respect to the dimerization domains in the arabinose-free state is also unknown. The objective of this work has been to determine constraints on the possible positions and orientations of these domains.
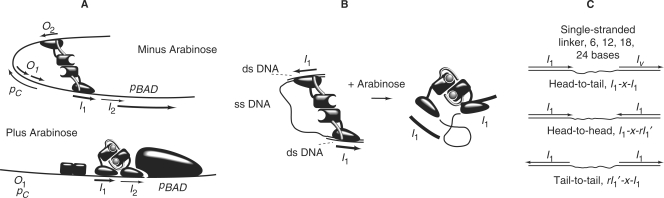
Multiple studies (7,11–15), have shown that in vivo, in the absence of arabinose, AraC protein binds principally to the O2 and I1 half-sites (Figure 1A) that are separated by several hundred base pairs rather than binding to the adjacent I1 and I2 half-sites. Such distal binding forms a DNA loop that represses the nearby ara pBAD and pC promoters. Binding of arabinose, deletion of the arm or most mutations in the arm cause AraC to shift from binding O2 and I1 to instead to bind to I1 and I2 (14,15). Binding in this mode activates transcription of the ara pBAD promoter. In vitro, the addition of arabinose increases the affinity of AraC protein for binding to adjacent DNA half-sites including direct repeat half-sites, inverted repeat half-sites and direct repeat half-sites separated by an additional 11 base pairs (12). Apparently, in the absence of arabinose, the N-terminal arms play an essential role in holding the DNA-binding domains in positions and orientations that favor looping of DNA by binding to well separated DNA half-sites and disfavor binding to half-sites located near one another, (Figure 1A). Addition of arabinose frees the DNA-binding domains and allows them to reorient and reposition. This allows the domains to bind easily to adjacent direct repeat half-sites and reduces the protein's propensity for looping.
Figure 1.
(A) Schematic of the arabinose operon in the absence of arabinose when AraC protein represses expression from pBAD by forming a loop between the araO2 and araI1 half-sites. The arm–DBD interaction is represented here as cis although for convenience in drawing, the structural details are highly stylized. The araO1 site and the pC promoter regulate expression of the AraC gene itself. In this representation in the presence of arabinose, the N-terminal arms of AraC bind over arabinose, thereby freeing the DNA-binding domains to bind to the adjacent araI1 and araI2 half-sites. Induction of the pBAD promoter results from this binding. (B) Illustration of how DNA with a flexible connector between two I1 half-sites can bind DNA in the presence or absence of arabinose with similar affinities. (C) Schematic structures of the head-to-tail, head-to-head and tail-to-tail DNA constructs that are used to determine the corresponding distances in AraC.
Several additional facts are consistent with the above picture. First, connecting the two AraC DNA-binding domains by a peptide linker (16), or dimerizing them with a leucine zipper (17), yields proteins that bind adjacent half-sites and induce the ara pBAD promoter but do not loop and repress. Second, arabinose affects the affinity of AraC for binding to two adjacent half-sites, but has little effect when the half-sites are connected by a flexible linker of sufficient length (16), (Figure 1B). That is, arabinose does not alter the intrinsic DNA-binding affinity of the individual DNA-binding domains of AraC, whereas it does alter their collective affinity for binding to two rigidly connected and adjacent half-sites.
The importance and involvement of the N-terminal arms of AraC in the arabinose response is demonstrated by the existence of arm deletions and arm mutations that reduce or eliminate the DNA looping (4,8,11). It is also demonstrated by the fact that arabinose has no significant effect on the core dimerization domain structure (2,3), leaving changes in the N-terminal arm as the most probable direct response to the binding of arabinose. Overall then, it appears that the principal factor determining the DNA-binding behavior of AraC protein is the positioning of the DNA-binding domains with respect to the dimerization domains and that the arms play a major role in positioning the DNA-binding domains in the absence of arabinose. Therefore, before the molecular and physical details of arm action on positioning the DNA-binding domains can be studied and understood, it is necessary to determine where the DNA-binding domains are positioned with respect to the dimerization domains.
In the work reported here, we develop and apply a method involving special DNA substrates for the measurement of distances between points in the AraC protein–DNA complex. We used the method to estimate values for three distances that separate parts of the protein–DNA complex when the complex is in the minus arabinose state. The acceptable ranges of the determined distances, the length of the linker connecting the dimerization and DNA-binding domains and the requirement that the N-terminal arms be able to contact the DNA-binding domains, restrict the possible positions and orientations of the DNA-binding domains with respect to each other and lead to two basic potential structures. In one of these, the DNA-binding domains are positioned such that the N-terminal arm on one dimerization domain can contact the DNA-binding domain of the same subunit in what is called a cis interaction. In the other, the arm on one dimerization domain contacts the DNA-binding domain of the other subunit for a trans interaction. In both cases, the DNA-binding domains lie on the same side of the protein's core consisting of the two dimerization domains.
MATERIALS AND METHODS
Tape measure DNA
DNA oligonucleotides were from Integrated DNA Technologies, Coralville, Iowa, USA. For the head-to-tail DNA, I1-6-I1, the top strand was, 5′-ACCCTAGCATTTTTATCCATAAGAC
CCATAGACCCTAGCATTTTTATCCATAAGACC. To each end of this oligonucleotide were hybridized two 25-base oligonucleotides, leaving in the middle a stretch of six bases of single-stranded DNA. For convenience in describing other sequences, consider the three parts: first, the double-stranded region with a top-strand sequence of 5′-ACCCTAGCATTTTTATCCATAAGAC, next, the single-stranded spacer region, 5′-CCATAG, and finally, the second double-stranded region with a top-strand sequence of 5′-ACCCTAGCATTTTTATCCATAAG ACC. The underlined region indicates the bases of the I1 site that are contacted by AraC protein. The double-stranded regions were made by annealing slightly less than stoichiometric amounts of complementary sequence, 5′-GTCTTATGGATAAAAATGCTAGGGT containing the fluorophore Cy5 on the 5′ end. The I1-12-I1, I1-18-I1 and I1-24-I1 DNA molecules were similar, but with single-strand spacers of sequence 5′-CTACTGGCATAG, 5′-CTACTGGTTCATGCATAG and 5′-CTACTGGTACCGTCTCATGCATAG, respectively. The sequences of the single-stranded regions were chosen to possess no secondary structure at the temperatures and salt concentrations used in the experiments.
In the head-to-head DNA, I1-6-rI1′, the second double-stranded region was changed to 5′-CAGCTATGGACATAATTGCTGACAGC by altering the sequence of the full-length oligonucleotide, and by annealing an oligonucleotide of complementary sequence as well as the fluorescent-labeled oligonucleotide complementary to the first double-stranded region. If the sequence of the first double-stranded region were merely inverted to form the sequence of the second double-stranded region on this DNA, the long oligonucleotide would possess self-complementary 25-base regions and the two shorter oligonucleotides would be self-complementary as well, both of which could greatly interfere with the formation of the desired DNA tape measure molecule. Therefore, in the 17-base region of DNA that is contacted by AraC, four bases that contribute little to the binding energy of AraC were altered (18), changing the sequence of the contacted region from 5′-TAGCATTTTTATCCATA to 5′-CAGCAATTATGTCCATA, where the underlined bases have been altered. Additionally, five bases outside the contacted region were also altered. Finally, the resulting 25-base sequence was inverted, yielding the sequence presented above. The 6, 12, 18 and 24-base single-stranded regions in this series possessed the same sequences as the single-stranded regions in the I1-x-I1 series.
In the tail-to-tail DNA molecules, it was the first double-stranded region that was inverted. Its sequence and that of the single-stranded spacer regions were as described above.
Unlabeled competitor DNA was a modification of the native I1-I2 AraC-binding site in which the I2 half-site has been changed to the tighter binding I1 half-site (11). The sequence of the top strand of this DNA is 5′-GCCATAGCATTTTTATCCATAAGATTAGCATTTTTATCCATACCT, where the two I1 half-sites have been underlined.
AraC protein
Partially purified, >90% pure, AraC for these experiments was prepared as described (19), dialyzed to remove arabinose, and stored at 4°C.
DNA tape measurements
Dissociation rates were measured in 70 μl of binding buffer, 10 mM Tris–acetate pH 7.4, 1 mM EDTA, 1 mM DTT, 5% glycerol, 0 or 50 Mm arabinose, and KCl was between 75 and 175 mM depending on the linker length. Salt concentration had no affect on the presence or absence of an arabinose effect on dissociation rates. Salt concentration was adjusted to place dissociation rates in convenient ranges for experimental measurement. 5′Cy5-labeled DNA was present at 2–5 × 10−9 M, and AraC at 1.5 × 10−7 M. After 5 min for equilibration, all further binding of AraC was blocked by the addition of the unlabeled I1-I1 competitor DNA to a concentration of 1.5 × 10−6 M. After intervals similar to those indicated, (Figure 2), 10 μl volumes were removed and added to 30 μl of 10 mM Tris–acetate pH 7.4, 1 mM EDTA, 25 mM L-arabinose. From this, 10 μl were immediately loaded onto 2″ × 3″ 6% acrylamide 0.1% methylene-bis-acrylamide horizontal gels containing 10 mM Tris–acetate pH 7.4, 1 mM EDTA and subjected to electrophoresis at 50 v for a total of 45 min. Three minutes after loading, circulation of buffer from one reservoir to the other of the two-chamber horizontal gel apparatus was begun to prevent electrode reactions from altering the pH. Following separation, DNA bands were imaged on a GE Typhoon 9410 fluorescence scanner and quantitated with ImageJ software (20,21). For measurement of the dissociation rates from the various DNAs, the relative amounts of protein–DNA complex at the different time points were determined for at least four time points. Scatter in these points yielded dissociation rates with a standard error of <10%. First-order exponential decay constants were determined with KaleidaGraph 4 software.
Figure 2.
Representative data showing arabinose-dependent dissociation of AraC from rI1′-6-I1, performed in 150 mM KCl, and arabinose-independent dissociation from rI1′-24-I1 DNA performed in 125 mM KCl. The labeled lanes indicate the times allowed for dissociation.
Model construction
PDB file 2ARC was used for the structure of the dimerization domain and 1BL0 was used as an analog for the DNA-binding domain plus bound DNA. Using the molecular mechanics program CHARMM (22), a DBD with bound DNA was randomly positioned and oriented anywhere within 100 Å of the dimerization domain. Its symmetry-related partner was generated. Restraining potentials of the following form were established, E(R) = 0.5K × (R − Rmin)2 for R < Rmin, E(R) = 0 for Rmin < R < Rmax and E(R) = 0.5K × (R − Rmax)2 for Rmax < R, where R is the distance between the two atoms in angstroms. K was 30 kcal/mol Å2. To hold the N-terminus of the DBDs near the C-terminus of the dimerization domain, the two constrained atoms were the alpha carbon of residue 168 of the dimerization domain and the alpha carbon of residue 9 of the MarA protein, corresponding to residue 175 of AraC, and Rmin = 5 and Rmax = 15 for each subunit. For the head-to-head distances, the two constrained atoms were O′4 of residue 401 on DNA strand A and O′4 of residue 401 on DNA strand C with Rmin = 45 and Rmax = 60. For the tail-to-tail distances, the two constrained atoms were O′4 of residue 423 of DNA strand A and O′4 of residue 423 of DNA strand C with Rmin = 55 and Rmax = 70. For the head-to-tail distances, the two constrained atoms were O′4 of residue 401 of DNA strand A and O′4 of residue 423 of DNA strand C and between O′4 of residue 401 of strand C and O′4 of residue 423 of strand A with Rmin = 55 and Rmax = 70. The energies of trial structures were taken as the sum of the inter-atomic interaction energies as calculated by the normal CHARMM potentials plus that produced by the restraining forces as defined above.
RESULTS
Measuring distances with a DNA tape measure
As discussed in the introduction, arabinose shifts the ability of AraC to bind to two DNA half-sites by altering the rigidity with which the DNA-binding domains are held with respect to each other. This mechanism is in contrast to shifting the DNA-binding affinity by changing the intrinsic DNA-binding affinity of the individual domains without changing their relative positions. In the absence of arabinose, the DNA-binding domains of AraC are held such that the protein prefers DNA looping. Hence, when looping is not possible and AraC binds to adjacent half-sites in double-stranded DNA in the absence of arabinose, the DNA-binding domains must overcome these positional constraints. The energy for this bending or restructuring of AraC comes from the strength of the protein–DNA interactions. As a result, AraC binds to such DNA significantly less tightly than if the DNA-binding domains had not been constrained. When arabinose is present, the DNA-binding domains are less constrained, and the protein binds to the adjacent DNA half-sites more tightly.
Consider the binding of AraC to a special DNA consisting of two double-stranded half-sites connected with a long and flexible single-stranded linker. If the linker is of sufficient length to span the separation of the DNA-binding domains in the minus arabinose state, (Figure 1B), then the protein can bind to this DNA tightly both in the presence and absence of arabinose (16). That is, its binding affinity to such DNA should be largely arabinose independent. This behavior provides a means to measure the distance separating parts of the DNA in the minus arabinose state. When the single-stranded flexible linker connecting the two DNA-binding sites is too short to span the distance separating the DNA-binding domains, DNA binding by the protein will display increased arabinose dependence. On the other hand, when the linker can span the distance, a significantly smaller arabinose dependence will be seen. Using a collection of DNA samples with different lengths of single-stranded linker permits determination of the minimum separation distance.
A DNA half-site to which a DNA-binding domain of AraC binds is not symmetric, (Figure 1A). Consequently, a ‘head’ and ‘tail’ end of the DNA-binding half-site can be defined and the head-to-head, head-to-tail and tail-to-tail distances are meaningful and distinguishable. This would not be the case if the binding half-sites were symmetric. The three different end-to-end distances in the AraC–DNA complex can be measured individually by constructing DNA tape measure molecules with different lengths of single-strand connector and the appropriate half-site orientation, (Figure 1C).
To construct the head-to-tail set of DNA tape measure molecules, single-stranded regions of 6, 12, 18 and 24 bases were connected to the two double-stranded I1 half-site regions. To minimize end effects on AraC binding, in all cases, four additional base pairs were provided at each end beyond the 17 bp AraC-I1 contact region (12). For the head-to-head DNA constructs, the second I1 site was inverted. As this inversion generates a sequence and its inverse-complement on the same strand, hairpin or lollypop structures are possible. To minimize their formation, the bases outside the second AraC-binding half-site were altered. In addition, 4 bp in the middle of the AraC contact site whose identity has a minimal effect on AraC binding (18), were also altered. The changes are shown in the Materials and Methods section. The resulting altered and reversed half-site is denoted as rI1′. For tail-to-tail DNA constructs, the first half-site was reversed and the second remained unchanged. If we denote I1-x-I1 as DNA containing the AraC half-sites in the head-to-tail orientation separated by x bases of single-stranded linker, I1-x-rI1′ contains the sites in head-to-head orientation, and rI1′-x-I1 contains the sites in tail-to-tail orientation.
The DNA migration retardation assay is a convenient method for separating free DNA from AraC-bound DNA (13). Accurate determination of the amounts of free DNA and protein–DNA complex in solution requires ‘freezing’ the state of the system so that no further association or dissociation occurs while the samples are being loaded onto the gel or during their electrophoretic separation. Control experiments showed that additional association of AraC with the fluorescent-labeled DNA at the end of the incubation period could be blocked by the addition of a 10-fold excess of unlabeled I1-I1 DNA. Further dissociation of AraC from the labeled DNA at the end of the incubation period was reduced to negligible amounts by two means. First, the salt concentration was reduced by dilution with buffer that contained no salt. This is effective because the dissociation rate of AraC from DNA (19), like that of most DNA-binding proteins, is strongly dependent on the salt concentration. Second, arabinose was added, thereby reducing the dissociation rate of AraC from DNA molecules by lessening the rigidity of AraC. The increased flexibility allows both DNA-binding domains of the protein to freely contact DNA-binding sites. Following these additions, samples were immediately loaded on gels. Once samples are within the gel, little dissociation occurs (13).
The affinity of AraC for its native binding site, I1-I2, or the much tighter binding I1-I1 site, is too great to allow convenient direct measurement of the equilibrium-binding constant with fluorescent-labeled DNA. While equilibrium competition-binding experiments were possible, the variation in affinity created by the different lengths of single-strand linker separating half-sites and by changing the orientation of a half-site would have added complexity. We therefore chose to extract the needed information from binding and dissociation kinetics. Experiments showed that the association rate of AraC for I1-6-I1 and I1-24-I1 DNA, both in the presence and absence of arabinose were within a factor of two of one another (data not shown). Thus, as is seen for many DNA-binding proteins, the forward rate constant does not significantly change as the binding affinity is changed by alteration of the DNA-binding site or effector binding to the protein. We therefore measured the dissociation rates of AraC from the different DNAs. These measurements can be performed at easily detected concentrations of DNA, and because AraC could be added at significant excess, the measurements are insensitive to variations in the activity or stability of the protein.
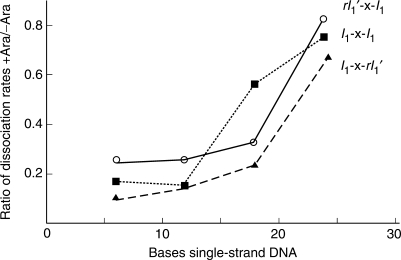
The results of typical separations used in the determination of the dissociation rate constants are shown, (Figure 2). In the data shown, it can be seen that the presence of arabinose significantly affects the dissociation rate of AraC from rI1′-6-I1, tail-to-tail DNA with a six-base flexible linker, but that arabinose does not significantly affect the dissociation rate from rI1′-24-I1 DNA. For all experiments, the ratio of bound DNA to total DNA was determined by quantitation of the fluorescent bands, and the dissociation rate constants were determined by fitting the data to a first-order kinetic decay equation. The ratio of the dissociation rates in the presence and absence of arabinose for each of the DNAs used in the experiment are shown, (Figure 3). Based on the lengths at which the arabinose dependencies begin to change, the head-to-tail and tail-to-tail distances can be estimated to be around 18 bases, and the head-to-head distance to be around 15 bases.
Figure 3.
Dissociation of AraC from the tape measure DNA containing different lengths of single-stranded linker between the two double-stranded regions containing I1 sites. Shown is the ratio of the dissociation rates in the presence and absence of arabinose as a function of the single-strand linker length connecting the two I1 half-sites.
Modeling domain locations in AraC
Construction of models that are consistent with the single-stranded lengths determined in the previous section requires converting from bases of single-stranded DNA to physical distances. Full extension of single-stranded DNA is opposed by the thermal motion of the polymer elements and by base–stacking interactions so that the theoretical relationship between force and extension is not simple. Therefore, we used experimental data derived from single molecule experiments to estimate the distance ranges. We estimate that the probable lengths of the flexible single-stranded segments employed in our experiments are 3–4 Å/base (23). Thus, we restrained the head-to-head distances to be values in the range of 45 and 60 Å (15 bases), and the tail-to-tail as well as the two head-to-tail distances to be between 55 and 70 Å (18 bases). Models possessing straight-line distances corresponding to the single-stranded regions of the test DNA molecules that were outside these ranges were increasingly penalized.
An additional restraint incorporated in the models is the length of the linker that connects each dimerization domain to its DNA-binding domain. Based on the structures of the dimerization domain of AraC (2,3), and the DBD homolog, MarA (10), this linker is about seven residues in length. Therefore, the physical distance used in the model building between the last residue, Asn168 of the dimerization domain, and the first residue of the DNA-binding domain, Met175 (residue 9 of MarA), was taken to lie in the range of 5–15 Å.
We also assumed that each of the DNA-binding domains would interact similarly with the rest of the protein. Since the dimerization domain possesses a 2-fold axis of symmetry, the DNA-binding domains and consequently, the entire modeled protein, will also possess a 2-fold axis of symmetry.
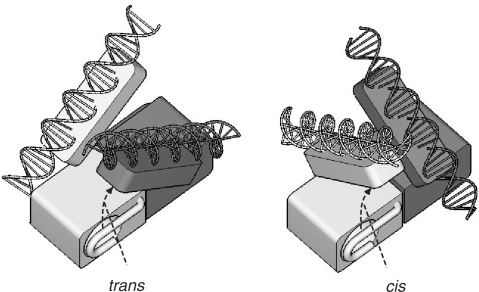
Manual attempts at positioning and orienting the DNA-binding domains so as to satisfy the four distance constraints described above proved to be awkward, innacurate and subjective. Therefore, we turned to computer construction of models. Systematic consideration of possible placements of the DNA-binding domains followed by energy minimization with the explicit inclusion of the single-stranded DNA proved to require far more computational power and sophistication than are justified by the experimental data. We therefore searched for acceptable structural models by the following method. The molecular mechanics program CHARMM (22) was programmed to work with the dimerization domain from AraC and a homolog of the DNA-binding domain of AraC, MarA bound to its 17-base binding site with four additional base pairs extending from each end. One such DNA-binding domain plus DNA was randomly placed anywhere within 100 Å of the center of the dimerization domain in a random orientation. The program then constructed the symmetry-related partner subunit, and calculated the energy of the resulting complex subject to the inter-atomic potentials of CHARMM and distance constraints corresponding to the length of single-stranded DNA between the appropriate ends of the DNA molecules. These distance constraints were implemented as potentials favoring the desired distances as described in Materials and Methods section. The lowest energy structure from 2000 trial structures was then energy minimized using a Monte Carlo search, but with separate translation and rotation steps of 1 Å and 1° and using the Metropolis criterion with a temperature of 300° for accepting proposed structures with energy higher than the current minimum. Of the 100 independent structures that were constructed and examined ∼25 positioned the DNA-binding domains such that they could be contacted by the N-terminal arms. These can be summarized as falling into two basic classes. In one of these, the possible arm–DBD interaction is within the same subunit and can be described as cis interaction, and in the other, the interaction is between the two subunits, and is a trans interaction. Representatives from the two classes are shown in Figure 4.
Figure 4.
Schematic showing the two general models for positioning the two DBDs of AraC that are consistent with the DNA tape measure distances determined for AraC. The figure on the left places the DNA-binding domain from one subunit near the N-terminal arm of the other subunit, and thus trans arm–DBD interactions are possible. The figure on the right depicts a structure compatible with cis interactions. In both figures, the tail region of the I1 half-site binds to the end of the DNA-binding domain that lies closer to the dimerization domain.
DISCUSSION
In addition to techniques like X-ray diffraction or NMR that can be used to determine protein structures, several techniques, including fluorescence resonance energy transfer, mechanical triangulation (24) and chemical cross-linking can sometimes be used for the determination of particular inter-residue distances in proteins. Here we describe the application of a new technique for determining distances between points in AraC–DNA complex. The method relies on two properties of the protein. First, the affinity of a single DNA-binding domain of AraC for an individual half-site is arabinose independent. Second and more important, the method utilizes the fact that the relative positions of the DNA-binding domains in AraC are restrained in the absence of arabinose and relatively unrestrained in the presence of arabinose (8,11,25). In this method, a segment of variable length single-stranded DNA connecting two double-stranded half-sites of the protein is used as a DNA tape measure. Consequently, if the single-stranded DNA segment is sufficiently long to span the distance between the restrained DNA-binding domains, then the DNA can bind tightly to AraC in the absence as well as in the presence of arabinose. Conversely, if the single-stranded DNA is too short to span the distance, the DNA can bind tightly only in the presence of arabinose. A collection of DNAs with different linker lengths allows approximate determination of the length of the single-stranded DNA needed to span the distance separating different ends of the DNA-binding sites in the minus arabinose state of AraC.
Conversion from the number of single-stranded bases in the DNA tape measure to a physical distance requires knowledge of the force versus extension properties of single-stranded DNA. Single molecule experiments show that single-stranded DNA begins to show substantial resistance to further extension above ∼3.5 Å/base (23). Therefore, we used a range between 3 and 4 Å/base in estimating distance ranges. A more precise conversion value depends on the stiffness of the protein and the tightness with which the protein binds to DNA. As more experience is gained with the distance measurement method, it is likely that more accurate and precise values will result.
Three distances were measured in the work described here. One was the separation of the head of one AraC-binding half-site and the head of the other AraC-binding site, i.e. the head-to-head distance. In addition, the head-to-tail and tail-to-tail distances were measured. These three distances plus the probable length of the linker connecting the dimerization and DNA-binding domains of AraC restrict the possible locations of the DNA-binding domains with respect to each other and with respect to the dimerization domains. If we assume that the interactions between the dimerization domains and the DNA-binding domains are identical for the two subunits, then the positions of the DNA-binding domains will retain the same 2-fold symmetry as is possessed by the dimerization domain. This plus the distance restraints, the fact that the dimerization and DNA-binding domains cannot overlap, and the fact that the N-terminal arms must be able to interact with the DNA-binding domains limit the possible structures to variants of the two forms shown in Figure 4. In one, the predominant arm–DBD interactions are cis. That is, they are between the arm on one dimerization domain and the DNA-binding domain of the same subunit. In the other case, the predominant interactions are trans. In both cases, the DNA-binding domains are inclined with respect to each other and both are located on the same face of the pair of dimerization domains in contrast to the 2D representation that is usually used to summarize the arm–domain interactions of the protein. In the latter, as in Figure 1A, the arm–DNA-binding domain interactions are depicted as trans for convenience in drawing. Although the magnitudes of the uncertainties in the determination of the dissociation rates from the different DNAs can be estimated as ∼10%, the uncertainties in the physical lengths of the single-stranded DNA segments cannot at this point be easily estimated. Despite this, the distance determinations seem to exclude the arrangements of the domains of AraC and the DNA as are represented in the schematic of Figure 1A, and require a 3D arrangement like those shown in Figure 4.
The data of Figure 3 suggest that the head-to-head distance in the AraC complexes is somewhat shorter than the head-to-tail or tail-to-tail distance. The same basic structures were found however, in the modeling if these distances were modeled as 45 and 55 Å, 50 and 55 Å 55 and 60 Å, or 45–55 Å and 55–65 Å, 45–55 Å and 45–55 Å or 55–65 Å and 55–65 Å. Thus, any effect of a somewhat shorter head-to-head distance is not reflected in the model building performed here. Both the cis and trans structures appear equally plausible at this point.
In summary, the experiments reported in this article provide the approximate positions and orientations of the DNA-binding domains of AraC with respect to the dimerization domains in the minus arabinose conformation of the protein. The fact that the DNA-binding domains are not planar, but instead are inclined at ∼90° with respect to each other facilitates DNA loop formation in supercoiled DNA and bears on in vitro studies of DNA looping in the ara system. Furthermore, knowledge of the approximate positioning of the DNA-binding domains with respect to the N-terminal arms that control them will facilitate study of the arm interactions.
ACKNOWLEDGEMENTS
We thank Katie Frato, Victoria Hargreaves and Jennifer Seedorf for comments on the manuscript and NIH grant GM18277 for support. This work was supported by National Institutes of Health grant GM18277 to R.S. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schleif R. The AraC protein: a love-hate relationship. BioEssays. 2003;25:274–282. doi: 10.1002/bies.10237. [DOI] [PubMed] [Google Scholar]
- 2.Soisson S, MacDougal-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- 3.Weldon JE, Rodgers ME, Larkin C, Schleif R. Structure and properties of a truely apo form of AraC dimerization domain, Proteins. 2007;66:646–654. doi: 10.1002/prot.21267. [DOI] [PubMed] [Google Scholar]
- 4.Ross J, Gryczynski U, Schleif R. Mutational analysis of residue roles in AraC function. J. Mol. Biol. 2003;328:85–93. doi: 10.1016/s0022-2836(03)00262-6. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh M, Schleif R. Biophysical evidence of arm–domain interactions in AraC. Anal. Biochem. 2001;295:107–112. doi: 10.1006/abio.2001.5213. [DOI] [PubMed] [Google Scholar]
- 6.Wu M, Schlelif R. Strengthened arm–dimerization domain interactions in AraC. J. Biol. Chem. 2001;276:2562–2564. doi: 10.1074/jbc.M008705200. [DOI] [PubMed] [Google Scholar]
- 7.Wu M, Schleif R. Mapping arm–DNA-binding domain interactions in AraC. J. Mol. Biol. 2001;307:1001–1009. doi: 10.1006/jmbi.2001.4531. [DOI] [PubMed] [Google Scholar]
- 8.Saviola B, Seabold R, Schleif R. Arm–domain interactions in AraC. J. Mol. Biol. 1998;278:539–548. doi: 10.1006/jmbi.1998.1712. [DOI] [PubMed] [Google Scholar]
- 9.Kwon HJ, Bennik MH, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 10.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl Acad. Sci. USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seabold R, Schleif R. Apo-AraC actively seeks to loop. J. Mol. Biol. 1998;278:529–538. doi: 10.1006/jmbi.1998.1713. [DOI] [PubMed] [Google Scholar]
- 12.Carra J, Schleif R. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993;12:35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson W, Schleif R. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J. Mol. Biol. 1984;174:611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- 14.Martin K, Huo L, Schleif R. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc. Natl Acad. Sci. USA. 1986;83:3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobel R, Schleif R. DNA looping and unlooping by AraC protein. Science. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- 16.Harmer T, Wu M, Schleif R. The role of rigidity in DNA looping-unlooping by AraC. Proc. Natl Acad. Sci., USA. 2001;98:427–431. doi: 10.1073/pnas.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bustos S, Schleif R. Functional domains of AraC protein. Proc. Natl Acad. Sci. USA. 1993;90:5638–5642. doi: 10.1073/pnas.90.12.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niland P, Hühne R, Müller-Hill B. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 1996;264:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 19.Martin K, Schleif R. Equilibrium DNA-binding of AraC protein, compensation for displaced ions. J. Mol. Biol. 1987;195:741–744. doi: 10.1016/0022-2836(87)90193-8. [DOI] [PubMed] [Google Scholar]
- 20.Rasband WS, Image J. Maryland, USA: US National Institutes of Health, Bethesda; http://rsb.info.nih.gov/ij/, 1997–2007. [Google Scholar]
- 21.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with Image J. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 22.Brooks BR, Bruccoleri R, Olafson B, States D, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations, J. Comput. Chem. 1983;4:178–217. [Google Scholar]
- 23.Smith S, Cui T, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1999;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 24.Dietz H, Rief M. Protein structure by mechanical triangulation. Proc. Natl Acad. Sci. USA. 2006;103:1244–1247. doi: 10.1073/pnas.0509217103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gryczynski U, Schleif R. A portable allosteric mechanism. Proteins. 2004;57:9–11. doi: 10.1002/prot.20233. [DOI] [PubMed] [Google Scholar]