Abstract
Functional analysis using the Gene Ontology (GO) is crucial for array analysis, but it is often difficult for researchers to assess the amount and quality of GO annotations associated with different sets of gene products. In many cases the source of the GO annotations and the date the GO annotations were last updated is not apparent, further complicating a researchers’ ability to assess the quality of the GO data provided. Moreover, GO biocurators need to ensure that the GO quality is maintained and optimal for the functional processes that are most relevant for their research community. We report the GO Annotation Quality (GAQ) score, a quantitative measure of GO quality that includes breadth of GO annotation, the level of detail of annotation and the type of evidence used to make the annotation. As a case study, we apply the GAQ scoring method to a set of diverse eukaryotes and demonstrate how the GAQ score can be used to track changes in GO annotations over time and to assess the quality of GO annotations available for specific biological processes. The GAQ score also allows researchers to quantitatively assess the functional data available for their experimental systems (arrays or databases).
INTRODUCTION
Elucidation of the complete human genome sequence (1,2) was a watershed event for both biology and computer science. As more genome sequence projects have been initiated, the amount of biological data and number of databases have proliferated (3,4). Methods for high-throughput, genome-wide analysis of biological systems have been developed and applied to an increasing number of organisms. Foremost among these techniques are functional genomics using microarrays and proteomics. The current challenge for functional genomics experiments is to translate large lists of genes or gene products into biologically relevant models. The Gene Ontology (GO) (5,6) was developed in part to answer this problem and has since become the de facto method for functional annotation of gene products (7).
GO annotations are provided by literature curation or by computational analysis that must be continually updated by human biocurators. For example, the European Bioinformatics Institute GO Annotation (EBI-GOA) Project (8) currently provides annotations for over 122 199 different species; GO annotations for all but 33 of these organisms have been generated by mapping functional motifs and domains to GO terms [‘inferred by electronic annotation’ (IEA) annotations] (9). These IEA annotations account for more than 90% of GO annotations and the basis for these annotations is continually reviewed so that all IEA annotations are updated on a weekly basis. Moreover, IEA annotations are generalized to apply to a diverse range of species and usually only represent very broad functions such as ‘protein binding’ and ‘enzyme binding’. In effect, this means that as functional genomics data is modeled using GO annotation, there are no curated GO annotations for many gene products and a large proportion of the remaining data describes only very broad biological concepts.
One axiom of GO is that the amount of functional information for any gene product varies from species to species, depending on the literature and databases available for different species. To assist researchers and biocurators with assessing the overall species-specific GO annotation quality of a particular dataset we developed the GO Annotation Quality (GAQ) score. The GAQ score is a quantitative measure of the GO annotation of a set of gene products (e.g. all annotated proteins in a species) based on the number of GO annotations available, the level of detail of the annotation and the types of evidence used to make these GO annotations. We demonstrate the utility of the GAQ score by comparing the current state of GO annotation in nine taxonomically diverse eukaryotes, by quantifying the improvement in GO annotation for two biomedical model species (chicken and mouse) relative to the time a dedicated GO annotation effort commenced for each species, and by demonstrating how the GAQ score can be used by biocurators to better direct GO annotation efforts and facilitate comparative functional annotation.
MATERIALS AND METHODS
The GAQ score
The overall GO annotation quality of a set of gene products is related to the coverage of gene products with GO annotation (breadth), the level of detail of GO annotation (depth), the types of evidence used to make these GO annotations (GO evidence code) and the completeness of the annotations based on how much of the current literature containing relevant information has been annotated.
We used quantitative information from breadth, depth and GO evidence code to derive a quantitative measure of GO annotation quality which we call the GAQ score. We define the GAQ score for an annotation (a) as the product of its depth in the ontology (Dd) and the evidence code rank (ECR) of the annotation:
The GAQ score for a set of gene products (S) with a total of A GO annotations is defined as:
The ‘breadth’ in this study is defined as ‘the number of annotations assigned to each of the gene products in the dataset.’ Note that, in some cases, it may be more informative to compute a separate GAQ score for each of the three GO ontologies and to consider the ‘breadth of annotation’ for each ontology. When considering the annotation, breadth of a specific gene product should be evaluated separately for each ontology.
GO annotation ‘depth’ is quantified by the depth of each GO annotation term within the ontology structure. The gene ontologies are structured as directed acyclic graphs (DAGs) where each ‘leaf’ term represents the most detailed level of information in relation to the parent level. Therefore, DAG depth from the root to an annotation term a (child node) is an indicator of the level of functional detail captured in the annotation. It has recently been argued that DAG structural levels are not good indicators of specificity for GO terms when grouping terms for functional analysis and that information theory can be used to partition GO terms into groups with similar specificity as measured by information content (10). However, this approach results in different groupings of terms for different species and would make cross-species comparisons very difficult. We have chosen to use DAG depth because we feel it gives the best overall view of the level of annotation detail, it is easily understood and because it facilitates comparison of annotation levels among different species. Since the GO ontologies are DAGs and not trees, there may be several paths from a child term to the root node. We define the GO DAG depth (Dd) of an annotation term as the length of the longest path from the term to its top-level parent in the ontology (either ‘molecular function’, ‘biological process’ or ‘cellular compartment’). We use the longest path rather than the shortest because the ‘true path rule’ used by the Gene Ontology (http://www.geneontology.org/GO.annotation.shtml#general) implies annotation to all parents on any path to the root. Note that different GO annotations will have different path lengths (which represent granularity) and that such annotations depends on the type of experiment performed, the amount of literature available for the gene product in question and the species being annotated. Therefore, a less granular GO term does not equate to a lesser annotation. We also define the Dd for an entire ontology as the sum of the Dd for each term in the ontology. Likewise, the average Dd for ontology is the Dd of all the terms divided by the number of terms in the ontology.
Each GO annotation indicates the type of evidence used to make that annotation and we initially assigned each GO term an evidence code rank (ECR) on a scale of 1 to 5 based on whether the evidence was direct or indirect (Table 1). However, like the GO itself, evidence code usage is evolving and we expect that ECRs will change over time. To test how any change in the ECR will affect the GAQ score we also used two other ranking systems to calculate GAQ (Supplementary Data). The average ECR for a species is a reflection of how much of the GO annotation is based on direct experimental evidence.
Table 1.
GO evidence codes and their corresponding rank used for this study.
| Code | Code definition | Evidence code rank |
|---|---|---|
| IDA | Inferred from Direct Assay | 5 |
| IGI | Inferred from Genetic Interaction | 5 |
| IMP | Inferred from Mutant Phenotype | 5 |
| IPI | Inferred from Physical Interaction | 5 |
| IC | Inferred by Curator | 4 |
| TAS | Traceable Author Statement | 4 |
| IEP | Inferred from Expression Pattern | 3 |
| RCA | Inferred from Reviewed Computational Analysis | 3 |
| IGC | Inferred from Genomic Context | 3 |
| ISS | Inferred from Sequence or Structural Similarity | 2 |
| IEA | Inferred from Electronic Annotation | 2 |
| NAS | Non-traceable Author Statement | 2 |
| NR | Not Recorded | 1 |
| ND | No Biological data available | 0 |
Direct experimental evidence codes (IDA, IMP, IGI and IPI) are ranked higher than indirect evidence codes. The IC and TAS evidence codes are based on expert judgment (of either the GO annotator or the researcher, respectively). The IEP, IGC and RCA codes refer to functions inferred from expression pattern, genomic context and reviewed computation analysis, respectively, and rank lower than direct functional evidence. The ISS evidence code is used for annotations made based on structural or sequence similarities. In contrast, the IEA evidence code is used for annotations that depend on automated transfer of annotations. Since some IEA annotations assigned by some groups may be of the same quality as ISS annotations assigned by other groups we assigned the same rank to both codes. NAS refers to uncited statements in reviewed articles and this data is not readily traced or the author may be referring to experiments done in a different species. The NR evidence code is a historical artifact of the GO and is used for older GO annotations made before the evidence code ontology was developed; since the evidence source is unrecorded, it must be presumed to be of lesser rank. ND is assigned where there are no biological data available. Other ranking systems used in this study are outlined in Supplementary data 1.
The breadth of annotations for a set of gene products (for example all annotated gene products for a species) can be measured in two ways. First, the total GAQ score for the set is an indication of both the number of products annotated and the quality of the annotation. In order to evaluate the breadth of annotation for each annotated gene product, we also define the meanGAQ score for a set of gene products as the GAQ score for the set divided by the total number of gene products (n) annotated:
The meanGAQ for a species is defined as the meanGAQ for all annotated gene products for that species.
Two in-house Perl scripts (DAGdepth.pl and GAQ.pl) have been implemented to determine the Dd of a given GO term and the GAQ score for a set of gene products.
GO annotation statistics for model eukaryotes
We obtained GO annotation statistics for nine species that have a dedicated GO annotation effort (Table 2). The number of GO annotations for each species, number of gene products that have annotations and percentage of GO annotations that are IEA were all obtained from EBI-GOA statistics (http://www.ebi.ac.uk/GOA/proteomes.html; 05/05/2007). A quantitative measure of the literature curated to the GO (Lc) for each species was obtained by downloading the EBI-GOA gene association file and counting the number of different literature entries for each of the species. However, none of these statistics allow a quantitative comparison of ‘how well’ a species is GO annotated. To capture this information, we computed the average Dd for each species for each ontology (Figure 1), the mean ECR for all annotations for each species (Figure 2) and the meanGAQ for the set of all annotated gene products for each of the species (Figure 3).
Table 2.
GO annotation statistics.
| Species | Number of GO annotations | Number of annotated gene products | Number of annotations per gene product | % IEA | Lc |
|---|---|---|---|---|---|
| Bt | 85 316 | 22 812 | 4 | 96 | 193 |
| Ce | 72 558 | 12 171 | 6 | 90 | 723 |
| Dm | 83 615 | 11 363 | 7 | 65 | 3546 |
| Dr | 102 202 | 31 106 | 3 | 98 | 527 |
| Gg | 56 745 | 16 230 | 3 | 96 | 123 |
| Hs | 167 889 | 34 118 | 5 | 69 | 13 361 |
| Mm | 179 696 | 34 886 | 5 | 59 | 7834 |
| Rn | 113 012 | 27 954 | 4 | 88 | 2933 |
| Sc | 64 770 | 5536 | 12 | 54 | 6123 |
Current GO statistics (as at 05/05/2007) for B. taurus (Bt), C.elegans (Ce), D. melanogaster (Dm), D. renio (Dr), G. gallus (Gg), H. sapiens (Hs), M. musculus (Mm), R. norvegicus (Rn) and S. cerevisiae (Sc). The number of GO annotations, annotations per gene products and percentage non-IEA annotations are obtained from EBI-GOA. Literature curated (Lc) figures are obtained by parsing the total number of PubMed records in the GO association files.
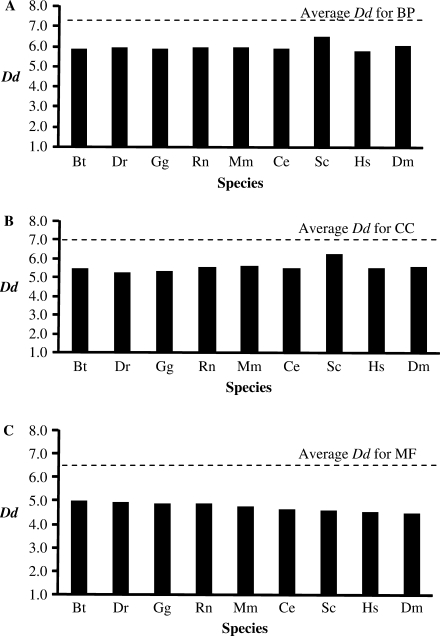
Figure 1.
The DAG depth (Dd) for each Gene Ontology. The overall average Dd (dashed line) was determined for all GO terms in each ontology (as at 05/052007). GO term Dds were compared to mean Dd of each species for (A) Biological Process (BP), (B) Cellular Component (CC) and (C) Molecular Function (MF). The species represented are B. taurus (Bt), D. renio (Dr), G. gallus (Gg), R. norvegicus (Rn), M. musculus (Mm), C. elegans (Ce), S. cerevisiae (Sc), H. sapiens (Hs) and D. melanogaster (Dm).
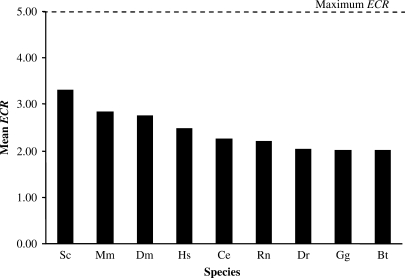
Figure 2.
The evidence code rank (ECR) for each species. GO evidence codes were ranked based on how closely they describe direct experimental evidence (Table 1) and current GO annotations were evaluated based upon these rankings. The maximum ECR, based on direct experimental evidence, is five. The species represented are S. cerevisiae (Sc), M. musculus (Mm), D. melanogaster (Dm), H. sapiens (Hs), R. norvegicus (Rn), C. elegans (Ce), B. taurus (Bt), G. gallus (Gg) and D. renio (Dr). The founder species (Sc, Mm, Dm), with a longer history of GO annotation, have the highest average ECRs. Other evidence code rankings were also used (Supplementary Data).
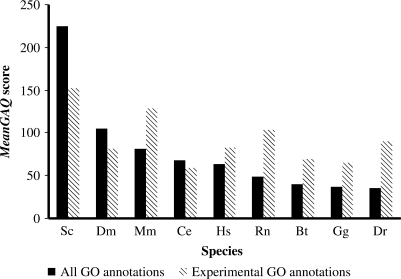
Figure 3.
Mean GO Annotation Quality (GAQ) scores for each species. To quantify GO annotation quality, we combined annotations (number of annotations per gene product), ‘depth’ (Dd) and evidence quality (ECR) to create the GO Annotation Quality (GAQ) score. The average GAQ score for S. cerevisiae (Sc), D. melanogaster (Dm), M. musculus (Mm), H. sapiens (Hs), C. elegans (Ce), R. norvegicus (Rn), B. taurus (Bt), G. gallus (Gg) and D. renio (Dr) (as at 05/05/2007) is shown. GO annotation founder species have higher overall meanGAQ scores than species with more recent GO annotation efforts. Higher scores are found in Sc, Mm, Rn and Dr, when computing meanGAQ scores from annotations made using only direct experimental evidence codes.
To compare the overall GAQ scores between species, we constructed GAQ matrices by pair-wise comparison of mean GAQ scores for all species (Table 3). Each entry in the table is the ratio of the GAQ scores of the species listed with each column divided by that of the species listed with each row.
Table 3.
The GAQ matrix obtained from pairwise comparison of meanGAQ scores for each species.
| Species | meanGAQ | Sc | Dm | Mm | Ce | Hs | Rn | Bt | Gg | Dr |
|---|---|---|---|---|---|---|---|---|---|---|
| meanGAQ(1) | 225 | 105 | 81 | 68 | 64 | 49 | 41 | 37 | 36 | |
| Sc | 225 | 1.0 | 0.5 | 0.4 | 0.3 | 0.3 | 0.2 | 0.2 | 0.2 | 0.2 |
| Dm | 105 | 2.1 | 1.0 | 0.8 | 0.6 | 0.6 | 0.5 | 0.4 | 0.4 | 0.3 |
| Mm | 81 | 2.8 | 1.3 | 1.0 | 0.8 | 0.8 | 0.6 | 0.5 | 0.5 | 0.4 |
| Ce | 68 | 3.3 | 1.5 | 1.2 | 1.0 | 0.9 | 0.7 | 0.6 | 0.5 | 0.5 |
| Hs | 64 | 3.5 | 1.6 | 1.3 | 1.1 | 1.0 | 0.8 | 0.6 | 0.6 | 0.6 |
| Rn | 49 | 4.6 | 2.1 | 1.7 | 1.4 | 1.3 | 1.0 | 0.8 | 0.8 | 0.7 |
| Bt | 41 | 5.5 | 2.6 | 2.0 | 1.7 | 1.6 | 1.2 | 1.0 | 0.9 | 0.9 |
| Gg | 37 | 6.1 | 2.8 | 2.2 | 1.8 | 1.7 | 1.3 | 1.1 | 1.0 | 1.0 |
| Dr | 36 | 6.3 | 2.9 | 2.3 | 1.9 | 1.8 | 1.4 | 1.1 | 1.0 | 1.0 |
| meanGAQ(2) | 152 | 81 | 128 | 59 | 83 | 103 | 70 | 65 | 90 | |
| Sc | 152 | 1.0 | 0.5 | 0.8 | 0.4 | 0.5 | 0.7 | 0.5 | 0.4 | 0.6 |
| Dm | 81 | 1.9 | 1.0 | 1.6 | 0.7 | 1.0 | 1.3 | 0.9 | 0.8 | 1.1 |
| Mm | 128 | 1.2 | 0.6 | 1.0 | 0.5 | 0.6 | 0.8 | 0.5 | 0.5 | 0.7 |
| Ce | 59 | 2.6 | 1.4 | 2.2 | 1.0 | 1.4 | 1.7 | 1.2 | 1.1 | 1.5 |
| Hs | 83 | 1.8 | 1.0 | 1.5 | 0.7 | 1.0 | 1.2 | 0.8 | 0.8 | 1.1 |
| Rn | 103 | 1.5 | 0.8 | 1.2 | 0.6 | 0.8 | 1.0 | 0.7 | 0.6 | 0.9 |
| Bt | 70 | 2.2 | 1.2 | 1.8 | 0.8 | 1.2 | 1.5 | 1.0 | 0.9 | 1.3 |
| Gg | 65 | 2.3 | 1.2 | 2.0 | 0.9 | 1.3 | 1.6 | 1.1 | 1.0 | 1.4 |
| Dr | 90 | 1.7 | 0.9 | 1.4 | 0.7 | 0.9 | 1.1 | 0.8 | 0.7 | 1.0 |
Species represented are S. cerevisiae (Sc), D. melanogaster (2 Dm), M. musculus (Mm), H. sapiens (Hs), C. elegans (Ce), R. norvegicus (Rn), B. taurus (Bt), G. gallus (Gg) and D. renio (Dr). The meanGAQ scores are based on number of gene products associated with the GO terms. meanGAQ(1) is based on all species' GO annotations, meanGAQ(2) is based on annotations made using only direct experimental evidence codes and in each case the meanGAQ is shown in bold at the top of each matrix. Where a species is compared to itself, the value will necessarily be one and these values are also marked in bold. A value >1 indicates that the species has higher meanGAQ score than the one it is compared against. For example, on average the meanGAQ score for the mouse gene products are two folds higher than that of chicken. Yeast consistently has the highest rates of meanGAQ scores when compared to each of the other organisms.
Measuring GAQ over time
It may be useful to know the GAQ score for a species of interest or even to compare GAQ scores between two species. Obviously, care must be taken when comparing functional annotations between species, however, because each species has its own set of literature that contains data that can be annotated directly for that species. The GAQ score is also useful for tracking how GO annotations may be improving with time (especially relative to changes in the ontology) for a given species of interest. Improving species-specific GAQ scores indicate improving functional annotation, which can be used with more confidence by researchers to model their genes or gene products to derive biological value. We used GAQ scores to measure the change in GAQ in chicken (which has only recently been actively GO annotated) and mouse (one of the GO founder species) for the first 5 years of each species’ respective GO annotation (Figure 4). Since the date of each GO annotation is recorded, we obtained annotations for each time period by parsing the chicken and mouse gene association files. The IEA annotations were excluded from this study because all IEA annotations are updated on a monthly basis and the date of these annotations changes to reflect this updating.
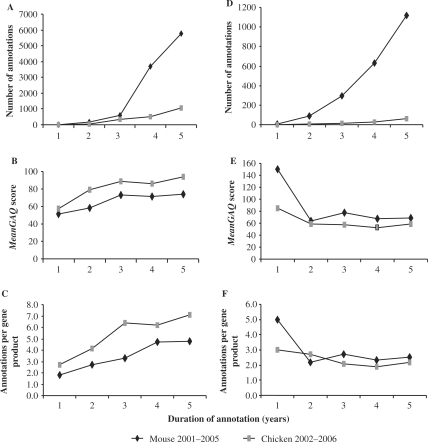
Figure 4.
Change in GO annotations and GAQ score over time. Chicken and mouse were chosen as two species with a dedicated GO annotation effort that started at different times. Number of annotations, meanGAQ scores and annotations per gene product derived from all non-IEA annotations (A, B & C) and from annotations made using only direct evidence codes (D, E & F) are shown.
Assessing GAQ scores for different areas of the GO
Since each species has its own body of functional information that can be annotated to the GO, and because some species are specifically used as model organisms for particular physiologic processes, we hypothesize that some sub-areas of the GO have more comprehensive annotation than others and that annotation cannot proceed uniformly across the entire GO. To test our hypothesis, we calculated the meanGAQ (excluding IEA annotations) for sub-areas of the chicken and mouse GO Biological Process Ontology (Table 4). We first summarized the annotations to Generic GOSlim terms using the GoSlimViewer tool at AgBase (11). Generic GOSlim terms are a subset of the GO ontologies and provide a summary level view of annotation in different major categories.
Table 4.
The 20 top-ranked chicken biological processes and the mouse GAQ score for these processes.
| Biological process | Chicken | Mouse | ||
|---|---|---|---|---|
| meanGAQ | Rank | meanGAQ | Rank | |
| Ion transport | 46 | 1 | 44 | 9 |
| DNA metabolic process | 36 | 2 | 51 | 4 |
| Response to biotic stimulus | 31 | 3 | 60 | 2 |
| Cell death | 26 | 4 | 47 | 7 |
| Anatomical structure morphogenesis | 25 | 5 | 65 | 1 |
| Multicellular organismal development | 24 | 6 | 48 | 6 |
| Lipid metabolic process | 23 | 7 | 41 | 11 |
| Nucleic acid metabolic process | 22 | 8 | 41 | 11 |
| Amino acid and derivative metabolic process | 22 | 8 | 44 | 9 |
| Cell cycle | 22 | 8 | 41 | 11 |
| Signal transduction | 22 | 8 | 44 | 9 |
| Transcription | 21 | 9 | 54 | 3 |
| Protein modification process | 21 | 9 | 44 | 9 |
| Cytoskeleton organization and biogenesis | 19 | 10 | 45 | 8 |
| Embryonic development | 19 | 10 | 33 | 18 |
| Response to stress | 19 | 10 | 25 | 25 |
| Metabolic process | 18 | 11 | 29 | 22 |
| Translation | 18 | 11 | 32 | 19 |
| Cell differentiation | 18 | 11 | 40 | 12 |
| Catabolic process | 17 | 12 | 30 | 21 |
meanGAQ scores were calculated for sub-areas of the Biological Process ontology in both chicken and mouse (excluding IEA annotations). The 20 top-ranked chicken biological processes (as summarized by the Generic GOSlim using the GoSlimViewer) are shown along with the calculated GAQ score for the chicken gene products currently described by these processes. The corresponding mouse meanGAQ score for the same sub-area and its ranking is also shown.
Assessing GAQ using available functional literature
The amount of functional literature available for curation to the GO varies for each species and estimating the amount of literature available for a species is difficult. We estimated the total PubMed entries available for a species by using that species’ scientific name, common name or taxonomy identifier. To estimate the amount of functional literature that could contain GO annotation data we used both Gene Reference Into Function (GeneRIF) (12) entries and GOPubMed (13). To determine the amount of literature curated to the GO (Lc) in each species we counted the number of unique PubMed identifiers recorded in the species’ gene association file (Table 2). The proportion of literature that contains functional data suitable for GO annotation varied significantly by species but in every case the percentage of available literature that has already been annotated using the GO is a small fraction of the functional literature available (Table 5).
Table 5.
Assessment of literature for GO annotation.
| Species | PubMed (L) | % Functional literature (Lf) | % Lc | |
|---|---|---|---|---|
| GeneRIF | GOPubMed | |||
| Bt | 301 568 | 0.49 | 4.01 | 0.06 |
| Ce | 15 920 | 7.73 | 104.22 | 4.54 |
| Dm | 61 488 | 7.81 | 27.63 | 5.77 |
| Dr | 9058 | 15.51 | 157.01 | 5.82 |
| Gg | 143 170 | 0.71 | 9.58 | 0.09 |
| Hs | 10 018 771 | 1.10 | 0.10 | 0.13 |
| Mm | 902 076 | 5.73 | 1.82 | 0.87 |
| Rn | 2 125 874 | 1.01 | 0.72 | 0.14 |
| Sc | 83 543 | 4.00 | 22.60 | 7.33 |
For consistency we searched in NCBI the total number of PubMed available for a species (L) by using the species’ scientific name, common name and/or taxonomy identifier. Species represented are B. taurus (Bt), C.elegans (Ce), D. melanogaster (Dm), D. renio (Dr), G. gallus (Gg), H. sapiens (Hs), M. musculus (Mm), R. norvegicus (Rn) and S. cerevisiae (Sc). The amount of functional literature (Lf) is from the geneRIF database and GOPubMed. GeneRIFs are often extracted directly from the document that is identified by the PubMed ID while GoPubMed is a knowledge-based search engine for biomedical texts. The amount of curated literature (Lc) is computed as the number of Pubmed IDs recorded in GO annotation (EBI-GOA; 5 May 2007). The percentage of Lf and Lc is computed based on L available for a species.
RESULTS
GO annotation statistics of the study species
While it might be expected that organisms with the longest history of active GO annotation would have the most comprehensive GO annotations, the number of GO annotations does not accurately reflect the overall GO annotation quality (GAQ) for a species. This is because so many GO annotations are based on nondirect experimental evidence (e.g. ISS and IEA). For example, zebrafish has more annotations than two of the ‘founder’ species (fruitfly, yeast), but a much smaller percentage of these annotations are based on direct experimental evidence (Table 2). Moreover, each species has its own body of direct experimental evidence that can be used for functional annotation and each group annotating to the GO have prioritized their annotation efforts based on their resources and the needs of the scientific community that they serve.
The GAQ score
The overall average Dd of Biological Process is 7.1, Cellular Component is 6.9 and Molecular Function is 6.1 (dashed line in Figure 1). In general, we found that there is very little variation for Dd between the species, although Saccharomyces cerevisiae (Sc) has a higher average Dd for both Biological Process and Cellular Component ontologies when compared to the other species. Also, the mean ECR for each species is higher in yeast, mouse and fruitfly, the founder species of GO annotation (Figure 2). This is expected because these species have the earliest dedicated, literature biocuration effort.
The meanGAQ score was calculated from all GO annotations and compared to that obtained from annotations that are only based on direct experimental evidence codes (Figure 3). Intuitively, GAQ scores should reflect the amount of dedicated GO annotation effort in each species. Yeast, fruitfly and mouse have the highest overall meanGAQ scores. This is expected because these three species (the GO founder species) have the longest effort of GO annotation. However, cow is an interesting exception to this trend as the effort to annotate bovine gene products is relatively new, yet it has slightly higher GAQ scores than chicken. We expect that this is because, as a mammalian species, cow benefits more from the transfer of GO annotations from other species such as mouse and human.
To compare the magnitude of meanGAQ scores between different species we used a GAQ matrix (Table 3). A score of 1 means that the two species compared in the pair-wise comparison have equal GAQ scores. A score >1 means that the species listed in column has better quality annotation than the one it is compared against in the corresponding row. Yeast consistently has the highest meanGAQ when compared to each of the other organisms. Although by no means completely GO annotated, yeast may be considered as the current ‘gold standard’ species for GAQ.
Measuring GAQ over time
Since the structure of the GO DAG, the available functional literature and the investment and effort in GO annotation change over time, it is desirable to be able to compare GO annotation progress over time. We compared the progression of annotation and GAQ scores in chicken and mouse (Figure 4; Supplementary Data). As we expected, based on the investment in GO annotation for these species, the number of annotations for both species increased over time (Figure 4A and D), with mouse annotations showing a rapid increase after the third year of annotation. Interestingly, although mouse has more annotations, chicken has higher overall meanGAQ scores (Figure 4B). But mouse has a higher meanGAQ score when using only annotations based on direct experimental evidence codes (Figure 4E) are used in the calculation. The meanGAQ score is directly proportional to the numbers of annotations per gene product (Figure 4C and F) rather than overall numbers of GO annotations.
Assessing GAQ scores for different areas of the GO
By using the meanGAQ score to evaluate specific regions of the Biological Process ontology, we found that some regions of the GO have more comprehensive annotation than others (Table 4). This also applies when either comparing GO annotation within a species (chicken). In general, chicken meanGAQ scores for the 20 highest-ranked regions of the Biological Process ontology are lower when compared to those of mouse. The exception is ion transport.
Assessing GAQ using available functional literature
By estimating the amount of literature available for annotation to the GO, we were able to assess what proportion of functional literature has been curated. Since it is difficult to assess how much functional literature is available, we used two different methods to estimate the amount of functional literature (Lf) that is available (Table 5). Some ‘model species’ (e.g. mouse and rat) have a low Lf while Caenorhabditis elegans and D. renio have a high Lf. However, while the Lf differs from one species to another, in all cases the percentage of literature curated (Lc) is very small. This is partially due to the amount of time and resources it takes to do literature curation but also because the amount of literature available is increasing dramatically.
DISCUSSION
Oftentimes it is difficult for researchers to assess the quality of functional annotation associated with their gene expression arrays or proteomics databases and it is often not easy to determine when they were last updated. Ideally, an overall assessment of the current GO annotation status for a genome would include the average number of GO annotations per gene. However, for many species the number of genes is not known or the number of reported genes differs significantly depending on the source used. This problem is compounded when comparing different species because it is even more difficult to find comparable information for a diverse range of species. Moreover, the number of GO annotations does not provide information about the quality of the available GO annotations. We developed the GAQ score as a quantitative measure of GO quality.
The GAQ score is derived from the number of GO annotations (breadth), DAG depth (Dd) and GO Evidence Code Rankings (ECR). In this instance, when we are discussing the ‘breadth of annotation’ we are referring to the total number of annotations assigned to each of the gene products in the dataset of interest. However, the overall GAQ score for a dataset provides little information about GO annotation for individual genes. For example, when GO annotations for mouse or chicken POLA1 and BASP1 are combined, there are 37 GO annotations for the mouse proteins and 34 GO annotations for the chicken proteins (Table 6). While this is a comparable number of GO annotations, the BASP1 mouse protein has annotations for each of the three ontologies while chicken BASP1 has no molecular function and the majority of GO annotations are to cellular component. The mouse BASP1 protein has fewer GO annotations but greater GO annotation breadth.
Table 6.
Example of breadth of GO annotations for mouse and chicken.
| Gene product | Total annotations | Number of annotations | ||
|---|---|---|---|---|
| MF | BP | CC | ||
| Mouse POLA1 | 33 | 14 | 12 | 7 |
| Chicken POLA1 | 27 | 9 | 11 | 7 |
| Mouse BASP1 | 4 | 1 | 1 | 2 |
| Chicken BASP1 | 7 | 0 | 1 | 6 |
| Mouse Total | 37 | 15 | 13 | 9 |
| Chicken Total | 34 | 9 | 12 | 13 |
Using the number of GO annotations as a measure of annotation breadth shows the overall GO annotation breadth of a dataset but does not reflect the annotation breadth of individual gene products. In this example mouse and chicken GO annotations are obtained from EBI-GOA (6 November 2007) for polymerase (DNA directed), alpha 1 (POLA1) and brain abundant, membrane attached signal protein 1 (BASP1) for each GO ontology. The three GO are molecular function (MF), biological process (BP) and cellular component (CC). Although the overall number of GO annotations is comparable for both species, the chicken BASP1 GO annotations are predominately CC annotations. When examined individually, the mouse BASP1 has better GO annotation breadth as there are annotations to all three ontologies. The UniProtKB accession numbers for the proteins are: chicken POLA1–Q59J86; mouse POLA1–P33609; chicken BASP1–P23614; and mouse BSAP1–Q91XV3.
The GO DAGs are designed so that the more detailed terms are deeper in the structure. As expected, none of the species in this study reach the average Dd for any of the three ontologies. Even comprehensively GO-annotated orthologs from different species have different Dd, reflecting the type of experiments performed in each species, the amount of species-specific literature available for that gene and inter-species variation in gene function. However, while a less granular GO term does not equate to a lesser annotation, it does mean less detailed functional information. The only way to assess the maximum granularity possible for a species is to have completed literature annotation for each of the gene products of interest; this is not possible nor is it currently possible to accurately and quantitatively assess the amount of granularity currently available in comparison to the functional detail available in current literature. Despite these practical limitations, our method still provides a quantitative measure of GO annotation that enables researchers to assess the GAQ of a specific dataset at a given time.
It is unlikely that any one species will have direct experimental evidence to be annotated to the most detailed (or deepest) GO terms across the enormous range of the GO. Detailed GO annotation relies on continued funding of new and existing annotation efforts, including support for developing the GO, maintaining existing data and database resources and updating existing GO annotations. Literature curation to the GO across a wider range of different species will provide more detailed and species-specific information in addition to informing functional annotation in closely related species.
Our ECR also reflected the importance of species-specific GO annotation. However, GO evidence code usage changes over time and the IEA and ISS evidence codes are particularly broad. To assess how the ECR may skew results we did additional analyses using different ranking systems (Supplementary Data) but the meanGAQ showed little change. We hypothesized that annotations based on direct experimental support will provide the ‘best-case scenario’ for assessing the GAQ and this is supported by our results (Figure 3). The use of GO evidence codes is evolving and that ranking GO evidence codes should be done knowledgably and to best suit the needs of specific datasets, questions and requirements.
To test the GAQ score we measured the GO annotation effort over a period of time and we also assessed GO quality for different sub-areas of the GO for both chicken and mouse. We chose chicken and mouse because they represent two species that we expected to have very different bodies of literature (based on the fact that the mouse is a purely model organism while the chicken is an agricultural species as well as a biomedical model). Moreover, the mouse and chicken GO annotation efforts started at different times and their annotation efforts employed different strategies for annotating literature; moreover, as a GO founding species, mouse annotators were heavily involved in the development of the GO during this period. By tracking GAQ score over time, we observed that for the first 5 years of GO annotation effort mouse had more annotations than chicken, but chicken had a higher average GAQ score. The mouse annotation effort focuses on biocurating the latest available literature while the biocurators for chicken gene products annotate all the literature for specific gene products, so that initially the average number of annotations per gene product is higher in chicken than that of mouse (eight compared to five). However, when only annotations based on direct experimental evidence are considered, mouse has a higher meanGAQ score, reflecting the early emphasis on literature biocuration in this species. A high GAQ score does not necessarily mean the most direct experimental knowledge has been captured for a species; it is more a general annotation coverage. Nevertheless, the improvement of the chicken GAQ over time demonstrates the effectiveness of a gene product-directed literature curation effort for newly sequenced species.
By using the GAQ score to quantitatively assess GO annotation for different sub-areas of the GO we show that GO annotation does not progress evenly across the ontology. This is in part due to differences in experimental literature available for each species and in part due to the focus of the GO annotation efforts. Analysis of sub-areas is useful as many research projects are directed at specific functional processes. By determining the quality of functional annotation available for different species, researchers may choose to target their research for experimental models that have the best-curated functional data for the processes they are studying.
The ability to assess what functional literature is available for a particular species is very difficult and it was this lack of accessibility for functional data that could be compared across species that initially drove the development of the GO (5). PubMed contains most of the published papers but one of the problems we faced is how to accurately assess the amount of literature (L) and functional literature (Lf) available for a specific species. We used GeneRIF (12) and GOPubMed (13) to estimate Lf. The GeneRIF database contains statements about the function of a gene and each geneRIF entry links to the PubMed ID and the gene name. While anyone may add GeneRIFs, National Library of Medicine (NLM) curators also add GeneRIFs and it may be this effort that skews GeneRIFs numbers to favor human, mouse and rat publications while other species are under-represented. GOPubMed is a sophisticated tool that combines PubMed searching with controlled vocabulary terms and does not have the same species as GeneRIFs. However, adding GOPubMed numbers for publications that have biological process, molecular function or cellular component terms will overestimate the number of papers that have functional literature, as many papers will be counted more than once. Neither method can effectively account for GO term synonyms, recognize variations in gene product names or account for functional data that may not be mentioned in the title and abstract of an article. Trained biocurators are essential for recognizing and curating experimental data from published literature but cannot keep up with the increasing amount of functional literature without improved tools and resources to support biocuration. However, by capturing the different direct experimental evidence for different species it is possible to extrapolate functional data to other, less well-annotated species. Given the increasing number of organisms to which functional genomics and proteomics analyses is applied, providing quality functional annotations for a diverse range of organisms is a critical research need. By developing a quantitative measure to assess GO quality, we provide a means for researchers to make the most of existing GO annotations and for biocurators to more efficiently focus their GO annotation efforts. The GAQ scripts will be freely distributed via the AgBase website (http://www.agbase.msstate.edu) and users provided with assistance in using or calculating GAQ scores to suit their specific needs.
In summary, we demonstrate the utility of the GAQ score for assessing GO annotation quality in nine different species that have varying levels of GO annotation and by assessing the improvement in GO annotation for both chicken and mouse based on time since a dedicated GO annotation effort commenced for each species. We also show how the GAQ score may be used to assess specific areas of the ontologies and this can also be applied to specific datasets (including microarrays). A quantitative assessment of GO quality will help biocurators to better direct current GO annotation efforts to specific areas that are important for their organisms’ research community and provides researchers with valuable information about their model systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Financial assistance for T.J.B. and F.M.M. is provided by the USDA NRI, MSU Office of Research, MSU Bagley College of Engineering, MSU College of Veterinary Medicine, the MSU Life Science and Biotechnology institute and the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number MISV-329140. Funding to pay the Open Access publication charges for this article was provided by the Department of Basic Sciences, College of Veterinary Medicine, Mississippi State University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Istrail S, Sutton GG, Florea L, Halpern AL, Mobarry CM, Lippert R, Walenz B, Shatkay H, Dew I, Miller JR, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl Acad. Sci. USA. 2004;101:1916–1921. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Gregory TR, Nicol JA, Tamm H, Kullman B, Kullman K, Leitch IJ, Murray BG, Kapraun DF, Greilhuber J, Bennett MD. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–D338. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liolios K, Tavernarakis N, Hugenholtz P, Kyrpides NC. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis SE. Gene ontology: looking backwards and forwards. Genome Biol. 2005;6:103. doi: 10.1186/gb-2004-6-1-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas M, O’Rourke JF, Camon E, Fraser G, Kanapin A, Karavidopoulou Y, Kersey P, Kriventseva E, Mittard V, Mulder N, et al. Applications of InterPro in protein annotation and genome analysis. Brief. Bioinform. 2002;3:285–295. doi: 10.1093/bib/3.3.285. [DOI] [PubMed] [Google Scholar]
- 10.Alterovitz G, Xiang M, Mohan M, Ramoni MF. GO PaD: the Gene Ontology Partition Database. Nucleic Acids Res. 2007;35:D322–D327. doi: 10.1093/nar/gkl799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy FM, Bridges SM, Wang N, Magee GB, Williams WP, Luthe DS, Burgess SC. AgBase: a unified resource for functional analysis in agriculture. Nucleic Acids Res. 2006;35:D599–D603. doi: 10.1093/nar/gkl936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, Cohen KB, Hunter L. Finding GeneRIFs via gene ontology annotations. Pac. Symp. Biocomput. 2006:52–63. [PMC free article] [PubMed] [Google Scholar]
- 13.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.