Abstract
Alternative splicing is thought to be one of the major sources for functional diversity in higher eukaryotes. Interestingly, when mapping splicing events onto protein structures, about half of the events affect structured and even highly conserved regions i.e. are non-trivial on the structure level. This has led to the controversial hypothesis that such splice variants result in nonsense-mediated mRNA decay or non-functional, unstructured proteins, which do not contribute to the functional diversity of an organism. Here we show in a comprehensive study on alternative splicing that proteins appear to be much more tolerant to structural deletions, insertions and replacements than previously thought. We find literature evidence that such non-trivial splicing isoforms exhibit different functional properties compared to their native counterparts and allow for interesting regulatory patterns on the protein network level. We provide examples that splicing events may represent transitions between different folds in the protein sequence–structure space and explain these links by a common genetic mechanism. Taken together, those findings hint to a more prominent role of splicing in protein structure evolution and to a different view of phenotypic plasticity of protein structures.
INTRODUCTION
Alternative splicing is one of the major sources for functional diversity in the proteomes of multicellular organisms. It refers to assembling the exons of a gene in different ways during pre-mRNA splicing such that different mRNAs and, thus, proteins are produced from the same gene. Based on EST data it is estimated that up to 74% of all human multi-exon genes are alternatively spliced (1) which drastically increases the number of proteins in the human proteome and, together with time and tissue-specific regulation of alternative splicing, largely increases the functional complexity of an organism. Alternatively spliced proteins are involved in many biological processes such as apoptosis (2) or the control of synaptic function (3) and play important roles in human diseases like cancer where the influence of alternatively spliced genes on transcription factors of signalling pathways have been described (4). The effects of alternative splicing on the function of a single protein range from changes in substrate or interaction partner specificity to the regulation of DNA-binding properties (5). In order to change the function of a protein by alternative splicing, its structure may be changed accordingly.
To date, the structures of less than 10 isoforms are available in the Protein Data Bank (PDB) (6) and known structural implications of splicing events on some of those proteins have been reviewed in Ref. 5. Those examples include a protein called Piccolo and the surprisingly drastic rearrangement of its C2-domain altered by a short insert of nine residues (7). Despite those few examples, only little is known from experimental data about how and to what extent alternative splicing affects protein structures. Due to this lack of knowledge from biological data, several recent studies (8–10) have mapped alternative splicing events onto predicted protein structures and have analysed features of the regions being affected. While they could link some structural properties like protein disorder (8) to a group of splicing events, the effects on many isoforms appear to be non-trivial. Based on this surprising complexity of alternative splicing on the proteome level, the most recent study by Tress et al. (9) even comes to the converse conclusion that ‘it seems unlikely that the spectrum of conventional enzymatic or structural functions can be substantially extended through alternative splicing’.
In this article, we present the results from comprehensively mapping all splice variants annotated in Swissprot (11) onto known protein structures from the PDB leading to almost 500 isoforms annotated to more than 350 Swissprot entries whose structures can be modelled with a very high accuracy (see Materials and Methods section). While about half of the events fall into variable regions of protein structures or affect complete domains, the effects on the other half appear to be non-trivial since they affect structured and well-conserved regions of the corresponding protein family. The large number of such non-trivial events, which are also found to be conserved among different species, can be explained in two ways:
First, non-trivial splicing events are non-functional on the mRNA or protein level leading to nonsense-mediated mRNA decay or unstructured proteins that are degraded after translation. This would indeed allow only few exons of an organism to be alternatively spliced, clearly questioning the importance of splicing on the proteome level.
Second, they may represent evidence that non-trivial splice events may produce functional isoforms where the absence of highly conserved parts of the structure might even allow for new structural and new functional properties of the isoform.
Here we provide evidence for the second hypothesis. Having mapped a large set of splicing events onto protein structures, we first explore the natural variation of the corresponding protein structure family, namely the ‘evolutionary isoforms’ of the respective protein, which allow us to explain ∼50% of the splicing events. The other half of the isoforms defines the set of non-trivial splicing events which cannot be explained by the observed amount of variation in the respective protein family and which we examined in more detail.
Based on evolutionary considerations of known fold-changing events (12), we group non-trivial events into eight different categories comprising different effects to be expected on the structure level. We then show that an extensive search of the biological literature provides clear evidence of stable protein products originating from such isoforms as well as evidence for a well-defined functional role of those proteins in the cell. The existence of such isoforms will help to sharpen our understanding of a protein's tolerance against major structural changes and, additionally, largely increases the importance of alternative splicing for generating functional and structural diversity. We will therefore review the function as well as the structural complexity of some of those isoforms.
Based on those findings, we try to explain the tolerance of such isoforms against the splicing events, which cannot be explained in their own fold, by members from different folds. We find evidence that such links between different folds in the sequence–structure space may indeed exist, and, for the first time, suggest a simple and common genetic mechanism, namely alternative splicing, for nature to explore them in vivo. Finally, we show that new experimental methods like Affymetrix exon array chips provide interesting data to prove or falsify our hypothesis in the future.
MATERIALS AND METHODS
In the following, the methods and data sources used for our study are described in detail. The methods are summarized in the Supplementary Figure S2.
Alternative splicing and literature data
The data for alternatively spliced proteins used in this work was obtained from the Swissprot protein database (September 2006), which annotates splicing events for 9135 out of 231 434 protein entries. Those 9135 entries harbour 20 845 alternative splicing events where 56.6% of the events are deletion events and the other 43.3% of the events represent replacements. In 22.2% of the replacements, the original sequence is shorter than the replacement sequence (insertions); in 27.7%, the replacement sequence is shorter than the original sequence (deletions) while in 50.1% of the cases the original sequence and the replacement sequence are of the same length. Literature assignments to different isoforms are also provided by Swissprot. We have examined them manually for evidence for the experimental proof of a stable protein product as well as experimental validation of its function.
Protein structure assignment
To obtain protein structure data we ran BLAST (13) against all proteins in the PDB (August 2006) for all Swissprot proteins with annotated splicing events. For each alternatively spliced protein, we then used free-shift alignment (14) (Pam250 matrix, gap open: 12, gap extend: 1) to compute full-length sequence–structure alignments of the alternatively spliced Swissprot entries with their respective homologues identified by BLAST. From all alternatively spliced Swissprot proteins, only those whose structure could be modelled with a very high sequence identity of at least 60% between template and target and whose sequence is covered to at least 75% by protein structure are used for further analysis.
Assignment of protein structures to families
The protein structures used to model the Swissprot proteins have been assigned to their corresponding SCOP (15) families as defined in SCOP version 1.71 (December 2006). All Swissprot entries that are modelled with structures not yet classified in the SCOP version 1.71 were assigned to their respective protein families using Vorolign (16). The final dataset contains 367 Swissprot proteins with 488 annotated isoforms, which are classified into 166 different families, 134 superfamilies and 119 folds with respect to the SCOP hierarchy.
Multiple structure alignments and evolutionary ‘isoforms’
Multiple structure alignments were computed from multiple structure superpositions with STACCATO (17), which has been shown to compute accurate alignments with respect to both, sequence and structure. In order to guarantee enough variability within the set of evolutionary related protein structures, we use proteins from the same SCOP superfamily. On this level of the SCOP hierarchy, protein structures exhibit enough structural variance to allow the definition of conserved and variable regions without overestimating structure conservation due to too similar proteins. Each set must contain at least three members and their structural similarity is measured by the TM-Score (18). Proteins in a set have to be similar enough (indicated by a pairwise TM-Score of >0.4) while still showing structural variability (TM-Score <0.8).
Given a multiple structure alignment, a ‘conserved region’ of a SCOP superfamily is defined as a block of at least 10 residues, which are conserved among all members of the superfamily. Each protein in a block may contain two gaps at most to account for some small variability within blocks. All regions outside of the conserved blocks are defined as ‘variable regions’. The proteins in the set define what we call ‘evolutionary isoforms’ which display insertions, deletions and substitutions and define the set of evolutionary events that are likely to be tolerable for a protein structure.
Alternative splicing and alternative structural models
In order to suggest alternative structures for non-trivial alternative splicing isoforms we applied the splicing event onto the structure (e.g. removed the structural parts belonging to a skipped exon). We then searched for reliable structural superpositions of the resulting structure model against all known folds (according to the SCOP classification). Such a search resulted in a number of structurally similar proteins from SCOP folds different than the proteins own fold. The soundness of such superpositions was measured by different criteria. First, as argued by Zhang and Skolnick (19) a TM-Score >0.4 is a clear evidence for a structural similarity (criterion 1). Since we believe this criterion is not strict enough to claim such remote structural similarities we also used more stringent criteria. Therefore, we filtered the superpositions to those superposing at least 80% of the spliced structure and 60% of its secondary structure elements and additionally examined the resulting superpositions manually for the conservation of core secondary structure elements with the correct connectivity and topology (criterion 2).
RESULTS
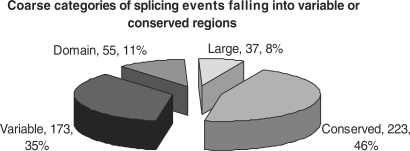
Our study is based on 367 Swissprot proteins and 488 additional splicing isoforms, which can be modelled on the structure level with a very high accuracy. The ENCODE dataset analysed in Ref. (9) contains 6 of these proteins (with 11 isoforms annotated in Swissprot) which are discussed in the Supplementary Data. As shown in Figure 1, ∼50% of the events fall into variable, often terminal, regions of the corresponding protein superfamily (evolutionary ‘isoforms’) or affect complete domains. The other half (255) of the events are harder to explain since they affect regions conserved in all superfamily members including core secondary-structure elements as well as highly conserved residues.
Figure 1.
Distribution of 488 splicing events in the four major categories. 35% of the events fall into variable regions of the corresponding superfamily while 11% affect complete domains of multi-domain proteins; 8% of the isoforms affect larger regions (more than 50% of the structure) while 46% affect conserved regions of their corresponding superfamily which are present in all superfamily members.
Different categories of non-trivial splicing events
Based on evolutionary considerations about possibly fold-changing events (12), we defined eight categories describing different types of those splicing events. Due to their hydrogen-bonding patterns, β-strands being located at the edge of a larger β-sheet are known to be more variable than internal β-strands. Therefore, two categories describe events affecting peripheral or internal β-strands. Similarly, in proteins belonging to αβ-fold classes, often αβ-secondary structure motifs tend to be affected and, accordingly, we defined two classes comprising peripheral and internal αβ-motifs. Additional categories contain conserved coil regions, conserved helices, large-scale events (affecting more than 50% of the structure) as well as events affecting repetitive protein-structure families whose repeat number is known to vary in evolution. Table 1 shows the distribution of the splicing events in the different categories.
Table 1.
The distribution of non-trivial isoforms in the eight categories defined based on evolutionary considerations with respect to different features
| Coil | α | β (p) | β (i) | αβ (p) | αβ (i) | Repeat | Large | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Conserved region affected (class) | 14 | 50 | 29 | 25 | 49 | 35 | 21 | 37 | 255 |
| Isoform confirmed (protein level) | 4 | 15 | 2 | 9 | 6 | 3 | 1 | 3 | 43 |
| Function described | 2 | 7 | 1 | 5 | 5 | 3 | 1 | 2 | 26 |
| Log Odds: Function/Class | 0.36 | 0.41 | −1.16 | 0.78 | 0 | −0.2 | −0.81 | −0.67 |
(p) and (i) indicate the position of the corresponding β-strands either at internal or peripheral positions of the sheet. The ‘Conserved region affected’ row displays the number isoforms which affect conserved regions of the corresponding superfamily. The ‘Isoform confirmed’ row displays isoforms which have been confirmed in the literature (see Supplementary Data for complete list) on the protein level, while the ‘Function described’ row references isoforms in the different categories which have been described in the literature to perform a well-defined function. The ‘Function/Class’ row contains the log odd ratios of functionally described isoforms in the different structural classes (third row) versus the background class distribution (first row). All log odd ratios of the values given for ‘Isoform confirmed’ and ‘Function described’ against the background distribution of structural classes within isoforms with ‘conserved regions affected’ can be found in the Supplementary Data, Figure S1.
Literature search for experimentally verified and functionally characterized isoforms
The biological literature annotated to the isoforms in Swissprot provides a valuable source of information. While Swissprot annotates proteins only if they have been verified experimentally, this must not be the case for their corresponding isoforms that may originate from large-scale EST or cDNA experiments. Therefore, most isoforms have only been experimentally verified on the mRNA level, while the protein products of the isoform were not investigated in the corresponding study. Nevertheless, out of the 255 non-trivial isoforms, 43 (17%) have been experimentally validated on the protein level and for 26 isoforms (10%), the function of the spliced variant has been described. Surprisingly, and in contrast to previous findings, we find literature evidence for functionally important and well-characterized isoforms in all of our eight categories (Table 1) indicating that even large-scale events may lead to functional and interesting protein products. A complete list of all literature references and isoforms is given in the Supplementary Data. In the following, we will review some interesting isoforms from different categories.
Alternative splicing of a terminal region of a protein
Many splicing events in our dataset change amino- or carboxy-terminal parts of a protein structure, which are found to be more variable and differ significantly among the members of a protein family. One of the examples, where the splicing event can be explained by the variability observed in the corresponding protein family is found in the human protein p38α (Q16539), a member of the mitogen-activated kinase (MAPK) family. Those proteins are integral parts of several signal transduction pathways and known to play important roles e.g. in the stress response of the cell. For p38α, several splice variants are annotated in public databases and among the well-studied splice variants are two proteins known as Mxi2 (Q16539-3) and Exip (Q16539-4) which both differ in large parts of their carboxy-terminal ends compared to p38α. Also, a similar splicing event is annotated for a homologous protein in mouse (P47811-2). The splicing event annotated for Exip (20) removes 46 residues from the protein structure, in addition, 52 residues differ in their sequence due to a frameshift introduced by the event (Figure 2a). It results in the loss of a well-conserved interaction domain used to interact with upstream kinases and downstream substrates. This leads to the fact that the protein is not targeted by MKK6 anymore. Expression of the isoform in the cell leads to an earlier onset of apoptosis and seems to target signal transduction pathways that are different from those targeted by p38α (20). In the example of Exip, the splicing event indeed targets a more variable part of the protein family (SCOP superfamily d.144.1). Structures, which lack the carboxy-terminal part are known to fold into stable conformations.
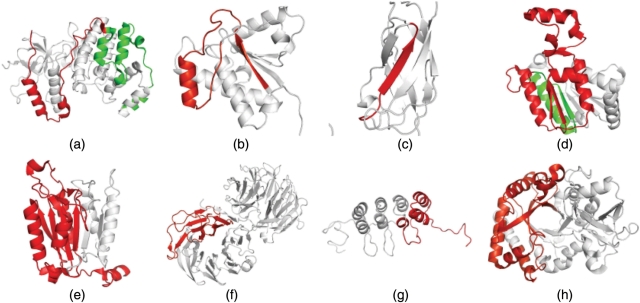
Figure 2.
Visualization of alternative splicing events on the structure level. Substitutions are coloured in green while deletions are coloured in red. All figures have been created using PyMOL (http://www.pymol.org). (a) The removal of the carboxy-terminal part from MK14_HUMAN (Q16539-4, pdb: 1zzlA), (b) the removal of one external strand and helix motif in PPAC_HUMAN (P24666-3, pdb: 5pnt), (c) the removal of an internal strand in TF65-HUMAN (Q04206-3, pdb 1nfi), (d) the removal of a large part of the protein from TIP30_HUMAN (Q9BUP3-2, pdb: 2bkaA), (e) the removal of several strands in CASP9_HUMAN (P55211-2, pdb: 1nw9B), (f) and (g) the removal of repetitive motifs from WDR1_CAEEL (Q11176-2: pdb: 1pevA) and CD2A1_HUMAN (P42771-2, pdb 2a5e) as well as (h) the removal of one half of a TIM-barrel structure in CHIA_HUMAN (Q9BZP6-3, pdb: 1vf8A). A comprehensive database of alternative splicing events mapped onto protein structures can be found at: http://www.bio.ifi.lmu.de/ProSAS/NARSupplement.html.
Alternative splicing at the edges of β-sheets
Alternative splicing events that involve β-strands naturally lead to the disruption of important hydrogen bonds in the protein structure. Nevertheless, such events are typical in the evolution of globular proteins if they affect peripheral β-strands of a larger β-sheet (12). Therefore, these events might be tolerable by a protein structure, even if they affect conserved regions of the protein's family. One such event is the removal of one peripheral αβ-motif from LMPTP (P24666), a tyrosine phosphatase. The protein is known to be expressed in three different isoforms, all of which differ in a 38-amino acid long part corresponding to one αβ-motif. The β-strand represents a peripheral strand of a β-sheet consisting of four strands in total. While the original sequence is replaced by another 38 residues in isoform 2, the corresponding part is removed in isoform 3 (LMPTP-C, P24666-3) (see also Figure 2b). Detailed analysis of LMPTP-C (21) shows that the protein is lacking phosphatase activity and can also not be phosphorylated by Lck kinase indicating that the active center has been tackled by the splicing event. When being co-expressed with isoform 2, LMPTP-C is shown to act as an antagonist to its native variant. The proposed mechanism (21) is that LMPTP-C competitively associates with the cellular substrates or regulators of its native counterparts and thereby blocks dephosphorylation of their targets. LMPTP-C represents an example how a splicing event changes a protein's function by removing the active center of the protein. While this goes hand in hand with a loss of its native function, the isoform is still able to mimic features of the native structure which allows it to act as antagonist of LMPTP.
Alternative splicing of internal strands of β-sheets
As shown above, the deletion of peripheral αβ-motif can result in a functional protein revealing an interesting mechanism for the regulation of enzyme activity. The deletion of internal strands from a β-sheet or a β-barrel appears to be more problematic since this results in the loss of hydrogen bonds on both sides of the strand and requires the formation of several new ones to retain the native-like structure of the protein. Nevertheless, there are known examples for strand deletion events that occurred in structure evolution as discussed in (12). The p65 (Q04206) subunit of the NF–κB transcriptional activator has one splice variant (Q04206-3), which exhibits such a removal event (22) as shown in Figure 2c, where nine residues, corresponding to one internal β-strand, are removed. Again, for a homologous protein in mouse (Q04207-2) the same splicing event is annotated. While the splice variant lost its capability to bind to p50, the second subunit of the NF–κB complex, it can form weak heterodimers with the native isoform of p65. Those heterodimers are found to be greatly reduced in their ability to bind DNA. This finding allows for two possible conclusions. Either, co-expression of the isoform and the native protein negatively regulates the NF–κB function, again revealing a pattern where the inactivation of a protein feature may act as an antagonist for the native protein. Or, in case that the isoform is still able to bind IκB (the inhibitor of the NF–κB complex), it may act as a regulatory ‘sink’ binding excess IκB and allowing p65 or the p65–p50 complex to enter the nucleus (22).
Alternative splicing may affect large and conserved regions of the protein structure
In the following, we give evidence for the fact that alternative splicing events may affect large and conserved regions of a protein structure and still can result in an isoform with unique functional features. CC3 (Q9BUP3) is known to be a metastasis suppressor inducing apoptosis in human cells, which is not expressed in highly metastatic lines of ‘small cell lung carcinoma’. A variant, called TC3 (Q9BUP3-2) (23), undergoes an alternative splicing event which removes 107 residues from its carboxy-terminal end and further replaces 21 residues at the new terminus which do not share any sequence similarity with the original sequence. As shown in Figure 2d, the splicing event affects more than 50% of the protein structure. It removes two peripheral β-strands from a β-sheet consisting of seven strands as well as several additional helices and strands that are not involved in the formation of the core αβα-fold. Strikingly, TC3 has, in contrast to its native variant CC3, an anti-apoptotic function that seems to be located in its unique C-terminal part. Even though the protein lost several conserved elements of its fold, it seems to be able to fold into a stable, functional isoform (see also Figure 3, rightmost column). It is shown to be short-lived due to a degradation signal located in the new carboxy-terminal end of the protein (23) that possibly represents another physiological feature. A second example that exhibits a similar splicing event that removes an even larger part from the structure is an isoform of caspase 9 (P55211-2) (Figure 2e). The isoform, named caspase 9b, is again shown to function as an endogenous apoptosis inhibitory molecule (24). The isoforms of CC3 and caspase 9 reveal a surprising tolerance of αβα-fold proteins to large-scale aberration events. This tolerance might originate in the evolutionary history of proteins of this fold class as they might have evolved by successively adding αβ-motifs to the edges of the core sheets. This might result in the fact that they can also be removed from the structure without losing capability to fold into a stable conformation.
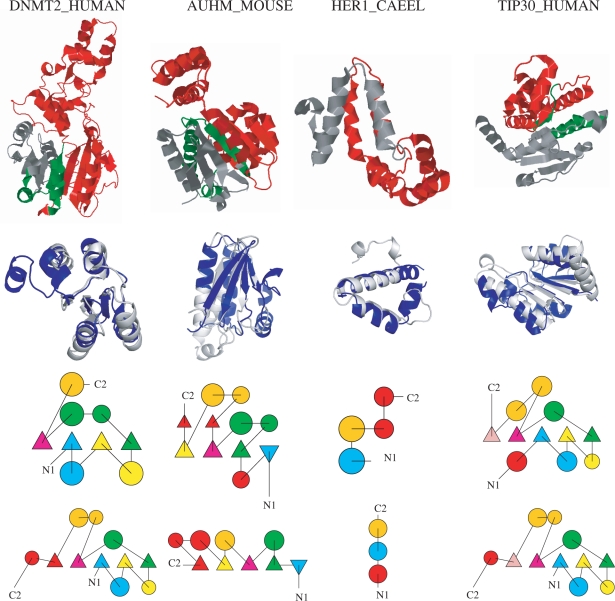
Figure 3.
This figure shows four examples for probable fold-changing events by non-trivial splicing events (i.e. those which cannot be accommodated in the native structure) identified by superposing the spliced structure to a different SCOP fold. The examples can also be explored interactively following this link http://www.bio.ifi.lmu.de/ProSAS/NARSupplement.html. Each column represents one example. In the first row, the splicing event is visualized on the native protein structure of the Swissprot protein. In the second row, the superposition of the spliced protein with the corresponding protein from a different fold is shown. Rows three and four display TOPS (34) diagrams of the spliced protein (row three) and the protein belonging to the different fold (row four). In the TOPS diagrams, corresponding secondary structure elements are coloured the same, elements missing in the other protein are coloured in red. Sometimes corresponding helices are split up which frequently results from breaks in the DSSP assignments. From left to right the following examples are shown: Column 1 DNMT2_HUMAN (O14717-6, Astral: d1g55a_, SCOP: c.66.1.26). The spliced protein superposes very well (TM-Score: 0.68) with d1gsoa2 (SCOP: c.30.1.1). Topologically the proteins are very similar, except for a very short strand (length 2) - helix (length 4) motif at the C-terminal end of d1gsoa2. Column 2 AUHM_MOUSE (Q9JLZ3-2, Astral: d1hzda_, SCOP: c.14.1.3) that superposes well (TM-Score: 0.56) with d1vc1a_ (SCOP: c.13.2.1). Topologically the proteins are similar, except for two small strands (both of length 2) and one short helix (length 3) missing in d1vc1a_. Additionally, the C-Terminal part of d1vc1a_ has an additional, short helix-strand motif. Column 3 HER1_CAEEL (P34704-2, Astral: d1szha_, SCOP: a.226.1.1) superposed with d1ni8a_ (SCOP: a.155.1.1, TM-Score 0.49). Only a small fragment (helix–turn–helix-motif) is left over by the splicing event. The two TOPS diagrams are similar with the two main helices being preserved while short helical parts are missing in either of the two proteins. Interestingly, d1ni8a_ is described to contribute to DNA binding after dimerization (35), which might also be the way how the isoform resulting from the HER1 splicing event is stabilized. Column 4 TIP30_HUMAN (Q9BUP3-2, PDB: 2bka, SCOP: c.2.1.2) that again superposes well with d1gsoa2 (see also DNMT2_HUMAN) from SCOP fold c.30.1.1 (TM-Score: 0.54). Topologically the two proteins are very similar according to TOPS except for two helices missing at the C- and N-terminal ends. The function of the isoform is discussed in the text (isoform TC3). Images have been created using Jmol (http://www.jmol.org) and PyMol (http://www.pymol.org).
Alternative splicing of highly repetitive protein structures
Internal repetition of (super-) secondary structure elements resulting from intragenic duplication and recombination events has been observed for several protein structure families. Proteins that exhibit repetitive structure are involved in many different functions in the cell while an increase of the repeat number in general affords a protein-enhanced evolutionary prospect due to an enlargement of its binding surface area (25). While repeats are found in all phyla they seem to be more common in eukaryotes which may be associated with an increasing complexity of the cellular functions that are available from assemblies of repeats. Several protein families contain repetitive elements but the major classes are β-propellers (b.67, b.68, b.69, b.70), β-trefoils (SCOP fold b.42), α-α superhelices (SCOP fold a.118), leucine-rich repeats (SCOP fold c.10) as well as the β-hairpin-α-hairpin repeats (SCOP fold d.211) (25). The fact that repeat duplication has been a successful strategy throughout protein-structure evolution that changed functional features of the proteins without losing the possibility to fold into a stable structure leads to the conclusion that such proteins should be highly tolerant against structural changes by alternative splicing. Indeed, we find that four out of the five repeat classes (all except for β-trefoils) described above harbour alternative splicing events affecting complete sets of repetitive motifs. Two examples are shown in Figure 2f and g. For most repetitive protein structures, large-scale deletion events are likely to be tolerable by the structure though, unfortunately, experimental validation and a functional categorization of the splicing variants is missing for all isoforms in our dataset. The principle of changing the number of repeats in the course of evolution in order to evolve novel functional features seems also to be used frequently to increase the functional diversity by alternative splicing as indicated by the large number of repetitive protein folds with annotated splicing events.
Alternative splicing may support hypothesis on the origin of TIM-barrels from half-barrels
For a number of protein structures and protein structure families it is well known that they resulted from ancient gene duplication and/or fusion events. Such duplication events are not always obvious from sequence data since the two subdomains have possibly already evolved to an extent where sequence similarity is random. A well-studied and recurrent motif in protein structures is the (α/β)8-barrel family (‘TIM-barrel’, SCOP fold c.1). Proteins in this family adopt a large variety of different functions and based on sequence and structure analysis it has been proposed (26) for some members of that family that they originated from a gene duplication and fusion event of two ancestral half-barrel proteins. Those ancient half-barrels probably formed a homodimer consisting of two identical half-barrels (27).
Our analysis now provides additional support for this hypothesis since it reveals two splicing isoforms (Q9BZP6-3 from CHIA HUMAN, and P27934-2 from AMY3E ORYSA) where one half of the barrel is removed by a large-scale removal event (see Figure 2h). The isoform of the human chitinase gene (Q9BZP6-3) has been described by Saito et al. (28) to be specifically expressed in lung though experimental validation of the existence of the stable protein product is lacking. In comparison to its native isoform, the protein lacks a secretory signal sequence leading to the conclusion that it might be present in the cytoplasm instead of being secreted. It also lacks the amino-terminal active site essential for chitinase activity. So far, we have no experimental validation for a functional gene product and a stable protein resulting from those splicing events. Nevertheless, based on the proposed evolutionary mechanism of fusing two half-barrels by an ancient gene duplication and fusion event, the splicing isoforms possibly form a (homo-)dimer to reconstruct the complete barrel. The possibility to express proteins of the TIM-barrel family as half-barrels might offer an increased functional variability by combining half-barrels containing different functional sites in heterodimeric complexes.
Indications for fold transitions caused by alternative splicing
For 225 non-trivial isoforms (excluding repetitive and conserved coil cases as these splice events will presumably not result in a different fold), we searched for similar structures as described in the Materials and Methods section. Applying the TM-Score criterion (TM-Score > 0.4) (19) alone we find for 139 (66%) isoform structures resulting from splicing events a similar structure from a different fold. Applying the more stringent criterion (secondary structure and isoform coverage) results in 49 isoforms (47 of which have a TM-Score >0.4). For these, we superposed the spliced structure with the target fold and visually inspected the superpositions for conservation of core secondary-structure elements as well as their connectivity, i.e. the topology of the core elements. We observe 21 (10%) highly confident superpositions, i.e. models for the spliced structures having a fold different from the fold of the non-spliced protein.
Thus, these different folds are probable structural models for the isoform, which could explain the drastic changes caused by the splicing event (four examples are shown and briefly discussed in Figure 3 and may be interactively explored at http://www.bio.ifi.lmu.de/ProSAS/NARSupplement.html). Of course, proteins resulting from splicing events might not be able to fold at all into a stable structure and often this will be the case. In other cases, the structure might be stable but will form a novel fold (so far not solved and deposited in the PDB). In rare cases, the modified structure might be similar to a known fold different from the native one. In the latter case, we would observe links between different folds by defined genetic changes (alternative splicing) transforming one stable 3D structure into a different stable 3D structure. Despite many attempts and research on structure classifications and structural descriptions and features, which led to the well-known structural resources such as SCOP and CATH (29), reliable and traceable links between fold classes are very rare. This is even more the case for evolutionary explanations of the observed similarities and events. Here we do not only observe a considerable number of such transformation events but with alternative splicing also provide a simple genetic mechanism explaining them as all the events correspond to known observed transcripts.
Validation of non-trivial splicing event using exon array data
Recently, Affymetrix released a novel type of DNA-chip which is capable of measuring most exons in human (Homo sapiens) as well as mouse (Mus musculus) and rat (Rattus norvegicus) by single probesets on the chip. The analysis of such chips allows measuring the expression of different transcripts of one gene in vivo, under different experimental conditions and in different tissues. The time and tissue-specific expression of certain transcripts may be another indication of a functional role of the corresponding gene product in the cell and will therefore help to understand functional diversity resulting from alternative splicing. Additionally, this data will be a helpful resource to validate or falsify our hypothesis that many, in particular also non-trivial splicing events may play functional roles in the cell. For this reason we have mapped all probesets provided on the human exon chip onto human exons annotated in Ensembl (30). Additionally, we have structurally modelled all human genes for which we can find reliable structural annotations in the PDB. The data can be accessed in the ProSAS database (31) at: http://services.bio.ifi.lmu.de/ProSAS. In total, we are able to cover 80.1% of all human exons with at least one Affymetrix probeset. When concentrating on high-quality structures (more than 40% sequence identity between template and human transcript) ∼35% of the human genes are covered at least partly by protein structure while 17% are completely (more than 75% coverage) modelled by a protein structure. This indicates that the combination of exon chip experiments with structural data indeed has the potential to test our hypothesis since a large number of genes (and exons) is at the same time covered by structure and measured on the chip.
DISCUSSION
Our study reveals a large number of functionally important, alternatively spliced proteins that harbour non-trivial splicing events and hints to a high degree of plasticity and a large tolerance against major rearrangements on the protein structure level. The possibility to express the antagonist of a protein as an isoform of the native variant represents an intuitive mechanism to increase the functional complexity of an organism by alternative splicing and has been discussed by several studies before. The structural explanation for this mechanism may be grounded in the removal of highly conserved parts, which are essential for the function of the native variant. If the isoform is still able to fold into a native-like structure, it can mimic native structural features and interact with native interaction partners without processing them further. Thus, alternative splicing immediately provides a mechanism for turning an activator into an effective inhibitor via a simple, possibly regulated, genetic mechanism.
The sequence–structure protein space tries to link different folds by appropriate similarities and differences but examples for fold transitions are rare and typically difficult to explain biologically. Our study provides examples for such links and explains them with a simple and common genetic mechanism. Thus, alternative splicing may be a new approach to chart the protein space and gain insights into mechanism of protein structure evolution. Future work will be on exploring the fold space as well as the changes that occur within and between folds in the context of alternative splicing in more detail. Therefore, structural analysis of alternative splicing events may help to identify common paths of protein fold evolution similar to the events discussed by (12) and to describe events that may be tolerated within protein families. This knowledge may have interesting applications in protein design.
Without experimental proof we can currently only speculate about the structures of isoforms resulting from non-trivial splicing events. Several facts indicate that at least some of those isoforms could have a well-defined structure. They perform a well-defined function in the cell and are able to mimic features of their native counterparts. Additionally, the identification of fold transitions exemplifies that they could adopt structures from different folds. Nevertheless, they might also be unstructured or fold into yet unknown conformations. Therefore, this study will provide interesting starting points for experimentalists trying to gain a deeper understanding of the non-trivial alterations of protein structure produced by alternative splicing and will lead to new insights into protein structure stability and the principles of protein fold evolution.
We also expect recently established experimental techniques like exon-level microarrays to contribute significantly to our understanding of the functional and, in the context of this study more important, the structural effects of alternative splicing. NMR-techniques (32) and mass spectrometry (33) measuring complete proteomes will contribute to validate or falsify the importance of alternative splicing for the functional diversity of complex organisms.
As, in principle, alternative splicing is a mechanism to produce a combinatorial number of transcripts, even a small percentage of stable structures implies a very large number of new protein variants. Thus, we believe that evolution makes use of alternative splicing to produce structural and functional diversity and this diversity is due to the large structural plasticity of proteins. Understanding alternative splicing on the proteome level will be one of the major challenges of computational and experimental biology in the next years.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online. Additional supplementary data, especially an interactive version of figure 3 which supports our argumentation is available at the following page: http://www.bio.ifi.lmu.de/ProSAS/NARSupplement.html
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by the Ludwig-Maximilians-Universität (LMU) München.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 2.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 5.Stetefeld J, Ruegg MA. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia J, Gerber SH, Sugita S, Sudhof TC, Rizo J. A conformational switch in the Piccolo C2A domain regulated by alternative splicing. Nat. Struct. Mol. Biol. 2004;11:45–53. doi: 10.1038/nsmb707. [DOI] [PubMed] [Google Scholar]
- 8.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PL, Albrecht M, Hegyi H, et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc. Natl Acad. Sci. USA. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grishin NV. Fold change in evolution of protein structures. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birzele F, Gewehr JE, Csaba G, Zimmer R. Vorolign – fast structural alignment using Voronoi contacts. Bioinformatics. 2007;23:e205–e211. doi: 10.1093/bioinformatics/btl294. [DOI] [PubMed] [Google Scholar]
- 17.Shatsky M, Nussinov R, Wolfson HJ. Optimization of multiple-sequence alignment based on multiple-structure alignment. Proteins. 2006;62:209–217. doi: 10.1002/prot.20665. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl Acad. Sci. USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudo T, Yagasaki Y, Hama H, Watanabe N, Osada H. Exip, a new alternative splicing variant of p38 alpha, can induce an earlier onset of apoptosis in HeLa cells. Biochem. Biophys. Res. Commun. 2002;291:838–843. doi: 10.1006/bbrc.2002.6529. [DOI] [PubMed] [Google Scholar]
- 21.Tailor P, Gilman J, Williams S, Mustelin T. A novel isoform of the low molecular weight phosphotyrosine phosphatase, LMPTP-C, arising from alternative mRNA splicing. Eur. J. Biochem./FEBS. 1999;262:277–282. doi: 10.1046/j.1432-1327.1999.00353.x. [DOI] [PubMed] [Google Scholar]
- 22.Ruben SM, Narayanan R, Klement JF, Chen CH, Rosen CA. Functional characterization of the NF-kappa B p65 transcriptional activator and an alternatively spliced derivative. Mol. Cell. Biol. 1992;12:444–454. doi: 10.1128/mcb.12.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitman S, Wang X, Shalaby R, Shtivelman E. Alternatively spliced products CC3 and TC3 have opposing effects on apoptosis. Mol. Cell. Biol. 2000;20:583–593. doi: 10.1128/mcb.20.2.583-593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasula SM, Ahmad M, Guo Y, Zhan Y, Lazebnik Y, Fernandes-Alnemri T, Alnemri ES. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res. 1999;59:999–1002. [PubMed] [Google Scholar]
- 25.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 26.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 27.Miles EW, Davies DR. Protein evolution. On the ancestry of barrels. Science. 2000;289:1490. doi: 10.1126/science.289.5484.1490. [DOI] [PubMed] [Google Scholar]
- 28.Saito A, Ozaki K, Fujiwara T, Nakamura Y, Tanigami A. Isolation and mapping of a human lung-specific gene, TSA1902, encoding a novel chitinase family member. Gene. 1999;239:325–331. doi: 10.1016/s0378-1119(99)00394-7. [DOI] [PubMed] [Google Scholar]
- 29.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birzele F, Küffner R, Meier F, Oefinger F, Potthast C, Zimmer R. ProSAS: a database for analyzing alternative splicing in the context of protein structures. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm793. , doi:10.1093/nar/gkm793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filipp FV, Sattler M. Conformational plasticity of the lipid transfer protein SCP2. Biochemistry. 2007;46:7980–7991. doi: 10.1021/bi6025616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adachi J, Kumar C, Zhang Y, Mann M. In-depth Analysis of the Adipocyte Proteome by Mass Spectrometry and Bioinformatics. Mol. Cell Proteomics. 2007;6:1257–1273. doi: 10.1074/mcp.M600476-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Michalopoulos I, Torrance GM, Gilbert DR, Westhead DR. TOPS: an enhanced database of protein structural topology. Nucleic Acids Res. 2004;32:D251–D254. doi: 10.1093/nar/gkh060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloch V, Yang Y, Margeat E, Chavanieu A, Auge MT, Robert B, Arold S, Rimsky S, Kochoyan M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.