Abstract
Friedreich ataxia is caused by an expanded (GAA•TTC)n sequence, which is unstable during intergenerational transmission and in most patient tissues, where it frequently undergoes large deletions. We investigated the effect of DSB repair on instability of the (GAA•TTC)n sequence. Linear plasmids were transformed into Escherichia coli so that each colony represented an individual DSB repair event. Repair of a DSB within the repeat resulted in a dramatic increase in deletions compared with circular templates, but DSB repair outside the repeat tract did not affect instability. Repair-mediated deletions were independent of the orientation and length of the repeat, the location of the break within the repeat or the RecA status of the strain. Repair at the center of the repeat resulted in deletion of approximately half of the repeat tract, and repair at an off-center location produced deletions that were equivalent in length to the shorter of the two repeats flanking the DSB. This is consistent with a single-strand annealing mechanism of DSB repair, and implicates erroneous DSB repair as a mechanism for genetic instability of the (GAA•TTC)n sequence. Our data contrast significantly with DSB repair within (CTG•CAG)n repeats, indicating that repair-mediated instability is dependent on the sequence of the triplet repeat.
INTRODUCTION
Friedreich ataxia (FRDA) is one of over 20 neurodegenerative diseases caused by the expansion of a triplet-repeat sequence (1–3). Whereas all other triplet-repeat diseases are caused by either expanded (CTG•CAG)n or (CCG–CGG)n sequences, FRDA is caused by an expanded (GAA•TTC)n sequence located in the first intron of the FXN gene (previously known as X25) (4). This (GAA•TTC)n sequence is polymorphic; alleles with <33 triplets do not cause disease, whereas alleles with >65 triplets are disease causing (4–6). The expanded (GAA•TTC)n sequence inhibits transcription of the FXN gene, most likely by forming a stable secondary structure such as a triplex or sticky DNA (7–10), which produces a deficiency of the mitochondrial protein frataxin (11). The levels of mature FXN transcript and frataxin protein in patient cells is inversely correlated with the length of the (GAA•TTC)n sequence (11,12), and therefore, disease severity, including the age of onset and several other clinical features, directly correlate with repeat length (13–15).
The (GAA•TTC)n sequence within the FXN gene is genetically unstable. The length of the repeat tract often changes during intergenerational transmission (15–17). The expanded repeat almost always contracts (by ∼20–30%) via paternal transmission, but shows an equal tendency for expansion or contraction during maternal transmission. The expanded (GAA•TTC)n sequence also displays somatic instability. In the dorsal root ganglia (DRG), which is the primary site of pathology in FRDA patients, there is a tendency for the (GAA•TTC)n sequence to undergo further expansion (18). Interestingly, there is an accumulation of these expansions over time, which suggests that these expansions could contribute to the development of the progressive, tissue-specific phenotype in FRDA (18). However, in contrast to DRG, all other human tissues display a marked contraction bias (18). In peripheral blood cells and in sperm, the expanded repeat tract may even revert back to the normal size range (17,19). Since FRDA is an autosomal recessive disease, the majority of the disease-causing expanded alleles are inherited via asymptomatic heterozygous carriers of expanded (GAA•TTC)n alleles. Indeed, de novo expansion from premutation alleles (with 34–65 triplets) is rare, and disease prevalence is mostly maintained via the existence of a large number of asymptomatic heterozygous carriers (5,6). Given that disease severity correlates with the length of the inherited expanded allele, and the repeat tract has a natural tendency for large contractions in most human tissues, understanding the mechanisms that cause large contractions may help in developing therapeutic strategies aimed at slowing down or preventing the progressive accumulation of large expansions in DRG.
The mechanisms responsible for triplet-repeat instability are only partially understood. Recombination (20–23), DNA repair (24–26) and epigenetic modification (27) have all been implicated. Our lab, along with others, has demonstrated that DNA replication can also mediate triplet-repeat instability. The orientation of the (CTG•CAG)n, (CGG•CCG)n and (GAA•TTC)n repeat tract relative to the origin of replication in bacteria and yeast influences instability (28–33). Furthermore, plasmid replication in transiently transfected mammalian cells in culture was a prerequisite for instability of the (CTG•CAG)n and (GAA•TTC)n repeats, with both the orientation of the repeat tract and its distance from the origin of replication acting as significant modifiers (34,35). The (GAA•TTC)n sequence has been shown to stall replication fork progression, and this occurs specifically when (GAA)n is the template for lagging strand synthesis (36–39). Stalling of replication forks may result in a double-strand break (DSB) (40–42). Others have shown that repair of a DSB near or within a (CTG•CAG)n or (CGG•CCG)n sequence results in orientation-dependent repeat instability and/or the deletion of flanking sequence. Furthermore, the instability they observed following DSB repair significantly increased in the absence of the RecA protein (43–46). We recently found that in the absence of RecA, (GAA•TTC)n instability was significantly increased specifically when GAA served as the lagging strand template (47).
Since the role of DSB repair in mediating (GAA•TTC)n instability is unknown, we investigated whether repair of a synthetic DSB within the (GAA•TTC)n sequence would influence repeat instability. We discovered that while DSB repair immediately outside of a (GAA•TTC)n sequence had no effect on repeat instability, DSB repair within the (GAA•TTC)n sequence dramatically increased instability. Indeed, almost every single repair event resulted in deletion of the repeat tract. We also found that the high level of instability was RecA-independent. Interestingly, the location of the DSB within the repeat tract significantly influenced the size of the deletion, consistent with a single-strand annealing mechanism by which DSB repair causes contractions of the (GAA•TTC)n sequence. The absence of any dependence on the length and orientation of the (GAA•TTC)n repeat, the presence/absence of RecA, and no obvious deletions involving the sequence flanking the triplet repeat, contrasts significantly with observation previously made with the (CTG•CAG)n sequence. Our results provide a potential mechanistic basis for the frequent deletions seen with the expanded (GAA•TTC)n sequence, and also highlight the dependence of the mechanism(s) of genetic instability on the sequence of the expanded triplet repeat.
MATERIALS AND METHODS
Plasmid construction
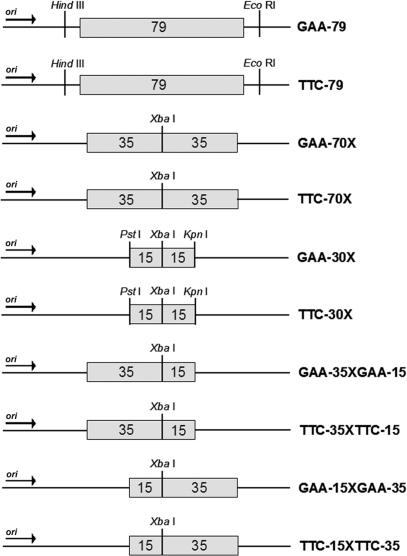
A (GAA•TTC)79 repeat sequence, isolated by PCR from the FXN locus of a human subject, was cloned into pUC19 as previously described (33). The pUC19 vector contains a portion of the ROP gene, making it a low copy number plasmid. The repeat tract plus minimal flanking sequence of intron 1 of the human FXN gene (38 bp 5′ and 35 bp 3′ of the repeat tract) was subcloned into the PstI and XbaI sites in both orientations relative to the origin of replication, such that either GAA or TTC serves as the template for lagging strand synthesis (GAA-79 and TTC-79, respectively) (Figure 1). Sequence length and purity were verified by sequencing.
Figure 1.
(GAA•TTC)n constructs used to analyze effect of DSB repair on triplet-repeat instability. pUC19 based constructs are shown containing either uninterrupted (GAA•TTC)79 or (GAA•TTC)n sequences engineered to contain a XbaI recognition sequence at specific locations within the repeat tract (see Materials and Methods section for details). Repeat tracts were cloned in both orientations relative to the origin of replication (arrow indicates direction of replication). Repeat-containing plasmids are depicted either in the ‘GAA orientation’ (e.g. GAA-79) or ‘TTC’ orientation (e.g. TTC-79), based on whether (GAA)n or (TTC)n serves as the lagging strand template, respectively. Numbers within the boxes indicate the length of the uninterrupted (GAA•TTC)n sequence.
The GAA-70X construct, which contains the (GAA•TTC)70 sequence with an XbaI site located exactly at the center of the repeat tract (Figure 1), was created using four synthetic oligonucleotides, as follows:
Oligo #1: 5′-GGCGCTCCGCTGCAGCC(GAA)35TCTAGACGCATCGCC-3′ and Oligo #2: 5′GGCGATGCGTCTAGA(TTC)35GGCTGCAGCGGAGCGCC-3′ were annealed together in 10 mM Tris, pH 8.0, and digestion buffer 3 (100 mM NaCl, 10 mM MgCl2) (New England Biolabs), followed by incubating with PstI and XbaI restriction enzymes to digest both ends of the annealed oligos. The purified insert was subcloned into the PstI and XbaI sites of pUC19, and confirmed by sequencing. Oligo #3: 5′-GGCGCTCCGTCTAGA(GAA)35CCGGTACCCGCATCGCC-3′ and Oligo #4: 5′-GGCGATGCGGGTACCGG(TTC)35TCTAGACGCATCGCC-3′were annealed in 10 mM Tris, pH 8.0 and digestion buffer 2 (50 mM NaCl, 10 mM MgCl2) (New England Biolabs), digested with XbaI and KpnI, and ligated into the similarly digested recombinant pUC19 vector containing the first (GAA•TTC)35 insert in the PstI and XbaI restriction sites (described above). The length and purity of the resulting (GAA•TTC)70 sequence with an XbaI site located at the center of the repeat tract were verified by sequencing.
The GAA-30X construct, which contains a (GAA•TTC)30 repeat sequence with an XbaI restriction site located exactly at the center of the repeat tract (Figure 1), was created using the two following synthetic oligos, which allowed the insertion of the appropriate sequence into the PstI and KpnI sites in pUC19, as described above: Oligo #5: 5′-GGCGCTCCGCTGCAGCC(GAA)15TCTAGA(GAA)15GGGGTACCCGCATCGCC-3′and Oligo #6: 5′-GGCGATGCGGGTACCCC(TTC)15TCTAGA(TTC)15GGCTGCAGCGGAGCGCC-3′. Plasmids containing the appropriate repeat insert were confirmed by sequencing.
The GAA-35XGAA-15 construct has a (GAA•TTC)35 sequence on one side of the XbaI restriction site in pUC19 (proximal to the ColE1 origin) and a (GAA•TTC)15 sequence on the other side of the XbaI site (Figure 1). Oligos #1 and #2 were annealed as described above and digested with PstI and XbaI to generate the (GAA•TTC)35 insert. Plasmid GAA-30X was digested with PstI and XbaI, releasing the intervening (GAA•TTC)15 sequence, and the gel-purified vector was ligated with the similarly digested (GAA•TTC)35 insert. The GAA-15XGAA-35 construct has a (GAA•TTC)15 sequence on one side of the XbaI restriction site (proximal to the origin) and a (GAA•TTC)35 sequence on the other side of the XbaI site (Figure 1). Oligos #3 and #4 were annealed as described previously and digested with XbaI and KpnI to generate a (GAA•TTC)35 insert. Plasmid GAA-30X was digested with XbaI and KpnI, releasing the intervening (GAA•TTC)15 sequence, and the resulting vector was ligated with the similarly digested (GAA•TTC)35 insert. Plasmids containing the appropriate repeat insert were confirmed by sequencing.
To create the corresponding TTC-70X, TTC-30X, TTC-35XTTC-15 and TTC-15XTTC-35 constructs (Figure 1), the orientation of the repeat tract within GAA-70X, GAA-30X, GAA-15XGAA-35 and GAA-35XGAA-15, respectively, was reversed. The constructs in the GAA orientation were digested with HindIII and EcoRI, which are located outside of the repeat tract. The 3′ recessed ends of the vectors and inserts were end-filled with Klenow polymerase. The vector and insert were separately gel-purified and the vector was dephosphorylated with calf intestinal phosphatase, and ligations were set-up. Colony PCR with primers: DSBTTCtest-F: 5′GCCAAGCTAATTCGAGCTCGGTACC-3′ and DSB-R (see below) was used to identify plasmids with repeat tracts in the desired TTC orientation. The sequence and orientation of the appropriate repeat inserts were verified by sequencing.
The GAA-70-spacer construct was created by inserting a synthetic sequence with the following restriction sites ‘XbaI–NcoI–BamHI–XhoI–XbaI’ into the XbaI site of GAA-70X. This effectively inserted a 28 bp spacer in between two tracts of GAA-35 (in the same orientation) that could be cut exactly in the center with BamHI. The following complementary oligonucleotides were annealed and inserted into a phosphatase-treated XbaI linearized GAA-70X vector to produce GAA-70-spacer:
5′-GGCGCTCCGTCTAGACTCGAGGATCCATGGTTCTAGACGCATCGCC-3′ and 5′-GGCGATGCGTCTAGAACCATGGATCCTCGAGTCTAGACGGAGCGCC-3′. Note: an extra ‘A’ was added between the XbaI and NcoI sites in order to make BamHI cut in the center of the 28 bp spacer.
Preparation of linear constructs
Three micrograms of each plasmid construct was linearized with the appropriate enzyme (Figure 1; see Results section) to create synthetic DSBs, and isolated from any undigested plasmid by gel purification following electrophoresis in 2% agarose gels. Linear DNA destined for use in future experiments was never exposed to ethidium bromide or UV irradiation. Southern blotting with a γ-P32 end-labeled pUC19 probe (5′-GCTGCAAGGCGATTAAGTTGG-3′) was used to verify adequacy of isolation of linear DNA from supercoiled DNA. Moreover, the transformation efficiency of linearized DNA was ∼100-fold less compared with the corresponding circular DNA.
Transformation of bacterial strains and analysis of (GAA•TTC)n instability
Isogenic Escherichia coli strains MM28 (galK2(OC) λ− IN(rrnD-rrnE)1 rpsL200(strR)) and M152 (recA3), and isogenic E. coli strains AB1157 (thr-1 araC14 leuB6(Am) DE(gpt-proA)62 lacY1 tsx-33 qsr’0 lgnV44(As) galK2(Oc) LAM− Rac-0 hisG4(Oc) rfbC1 mgl-51 rpoS396(Am) rpsL31(strR) kdgK51 xylA5 mtl-1 argE3(Oc) thi-1) and JC10287 (DE(recA-srlR)304) were obtained from the E. coli Genetic Stock Center (Yale University). The MM28, M152, AB1157 and JC10287 strains were made ‘ultra-competent’ using the Inoue method. Each transformation was performed using 5 ng of circular plasmid DNA or 25 ng of linearized plasmid DNA, followed by plating on LB-agar plates containing 100 μg/ml ampicillin. Since viability of cells transformed with linear constructs was dependent upon recircularization of the plasmid to provide ampicillin resistance, each colony therefore represented a DSB repair event. Transformation of each template was performed in triplicate and 50–100 colonies were screened from each transformation. (GAA•TTC)n instability was determined by colony PCR of transformants using primers DSB-F (5′-ACAGCTATGACCATGATTACG-3′) and DSB-R (5′-GGGTTTTCCCAGTCACGACG-3′), which are located within the flanking pUC19 sequence (40 bp upstream and 56 bp downstream from the repeat tract, respectively). Instability was defined as the number of mutation events (repeat tracts of altered length) per successful colony (DSB repair event) screened. Repeat lengths were calculated by estimating sizes of PCR products on 2.5% agarose gels relative to a DNA size standard. Since all mutation events resulted in deletion of the repeat tract, the term instability is synonymous with deletion in these experiments.
To determine whether the restriction site used to create the DSB was maintained following repair, PCR products representing deletions were digested with the restriction enzyme used to linearize the plasmid prior to transformation (HindIII and EcoRI for GAA-79, XbaI for GAA-70X, and BamHI for GAA-70-spacer). Products of digestion were run on 2.5% agarose gels alongside the original PCR product.
Statistical methods
Comparison of means was done using the independent samples t-test. Frequencies were compared using χ2 analysis. Differences were considered significant when P ≤ 0.01.
RESULTS
DSB repair within the (GAA•TTC)n sequence results in dramatically increased repeat instability
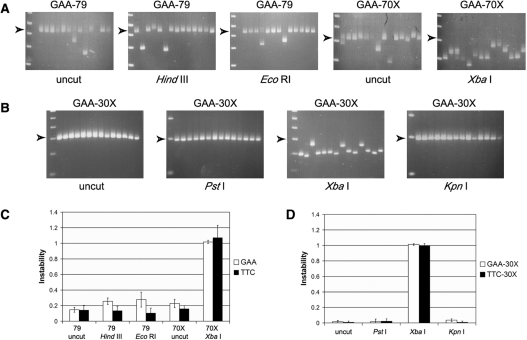
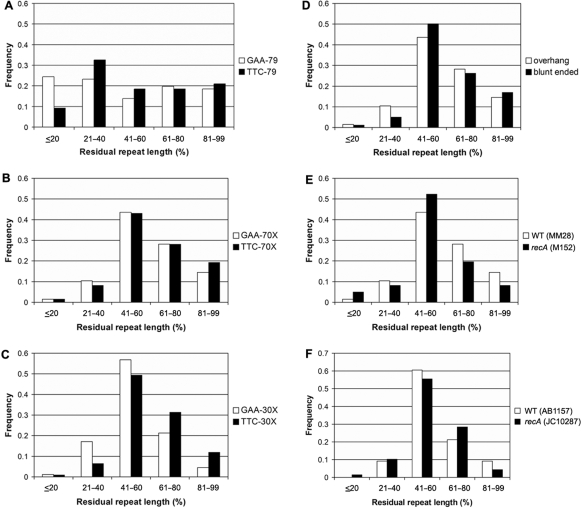
To investigate the potential effect on repeat instability of repairing a DSB within or near the (GAA•TTC)n sequence, plasmids were created that contained a pure (GAA•TTC)79 sequence, or a (GAA•TTC)70 sequence engineered to have a XbaI restriction site located exactly in the center of the repeat tract. These sequences were cloned into pUC19 in both orientations relative to the origin of replication (GAA-79, TTC-79, GAA-70X and TTC-70X; Figure 1). The GAA-79 and TTC-79 plasmids were linearized by restriction digestion with either HindIII or EcoRI to create a DSB just outside of the repeat tract (12 bp 5′ or 3′ from the repeat). The GAA-70X and TTC-70X plasmids were linearized by restriction digestion with XbaI to create a DSB exactly in the center of the repeat tract. Each of the linearized plasmids, as well as their respective circular parental plasmids, was transformed individually into E. coli MM28 (RecA+). Colony PCR was used to visualize and quantify triplet-repeat instability. The effect of DSB repair was determined by comparing the frequency of instability observed with the transformation of linearized templates, where each colony represents an individual DSB repair event, with the background instability observed with the corresponding circular plasmids that did not undergo DSB repair (see Materials and Methods section). No significant difference in the level of instability was noted between plasmids that underwent DSB repair at either the HindIII or EcoRI sites just outside of the repeat tract and the corresponding circular parental plasmid (GAA-79 and TTC-79), in either the GAA or TTC orientation, indicating that DSB repair outside of the (GAA•TTC)n sequence did not result in increased repeat instability (Figure 2A and C). However, repair of a DSB in the center of the (GAA•TTC)n sequence resulted in dramatically increased instability compared to its corresponding parental plasmid (GAA-70X and TTC-70X), in both the GAA and TTC orientations (P < 0.001; Figure 2A and C). As seen in Figure 2A, almost every DSB repair event within the repeat tract resulted in a deletion of the (GAA•TTC)n sequence. The orientation of the repeat tract with respect to the origin of replication had no significant effect on the level of instability following DSB repair within the repeat tract (P = 0.49; Figure 2C).
Figure 2.
DSB repair results in dramatically increased instability of the (GAA•TTC)n sequence when the break is located within the repeat tract. (A) Representative agarose gels with products of colony PCR showing transformants of GAA-79 [circular (uncut), or linearized with HindIII or EcoRI], and GAA-70X [circular (uncut), or linearized with XbaI]. Arrowheads indicate the position of the full-length repeat tract. DSB repair outside the repeat tract showed levels of instability that were similar to the uncut plasmids; however, DSB repair within the repeat tract resulted in a very high frequency of deletions. The first lane in each gel contains the 1 kb Plus ladder (Invitrogen) with bands from the bottom of the gel representing 0.2, 0.3, 0.4, 0.5 and 0.65 kb (note: the full-length products of GAA-79 and GAA-70X are 423 and 312 bp, respectively, due to the presence of some flanking intron 1 sequence from the human FXN gene in the former). (B) Representative agarose gels with products of colony PCR showing transformants of GAA-30X [circular (uncut), or linearized with PstI, XbaI or KpnI]. Arrowheads indicate the position of the full-length repeat tract. DSB repair outside the repeat tract showed levels of instability that were similar to the uncut plasmids; however, DSB repair within the repeat tract resulted in a very high frequency of deletions. Note that even the (GAA•TTC)30 sequence, which was otherwise extremely stable, showed a dramatic rise in the frequency of deletions. The first lane of every gel contains a DNA size marker; the markers used in the gels containing repair products of templates GAA-30X PstI and KpnI are different from all other gels. The first lane in each gel contains either the 1 kb Plus ladder (Invitrogen) (GAA-30X; PstI and KpnI with bands from the bottom of the gel representing 0.1, 0.2 and 0.3 kb, or the 50 bp DNA ladder (Invitrogen) (GAA-30X; uncut and XbaI) with bands from the bottom of the gel representing 0.1, 0.15, 0.2, 0.25, 0.3 and 0.35 kb. (C) DSB repair within a slightly unstable (GAA•TTC)70–79 sequence produced a dramatic rise in instability. Note that the (GAA•TTC)n sequence was equally unstable in the GAA and TTC orientations. (D) DSB repair within a highly stable (GAA•TTC)30 sequence produced a dramatic rise in instability. Note that the (GAA•TTC)n sequence was equally unstable in the GAA and TTC orientations. All error bars represent +/−2 SEM derived from triplicate experiments.
The sequence at the repair site was investigated by further digesting the deletion products obtained after DSB repair of the linearized templates, using the respective enzymes for the creation of the DSBs. Whereas all 63 products of the HindIII-digested GAA-79 construct, and all but one of the 61 EcoRI digested GAA-79 construct maintained their respective HindIII and EcoRI restriction sites, none of the 76 deletions from the XbaI digested GAA-70X construct had maintained the XbaI site. This indicates that the increased instability observed when DSB repair was made to occur within the (GAA•TTC)n sequence was due to the repair process itself, i.e., resulting from loss of DNA sequence at the break site. The lower level of instability observed with plasmids cut outside of the repeat tract must therefore have occurred either before the DSB was created or after the repair process itself.
To determine whether the length of the repeat tract had any influence on DSB repair mediated instability, a (GAA•TTC)30 sequence was similarly engineered with an XbaI restriction site located exactly at the center of the repeat tract, which was also cloned into pUC19 in both orientations relative to the origin of replication (GAA-30X and TTC-30X; Figure 1). The uncut GAA-30X and TTC-30X constructs were almost completely stable, as were the plasmids that underwent DSB repair just outside of the repeat tract (at PstI and KpnI; Figure 2D), and no significant difference was seen in the level of instability between the uncut and linearized plasmids in either the GAA or TTC orientations. However, DSB repair at the center of the repeat tract resulted in a significantly higher number of deletions than the uncut parental plasmid in both orientations (P < 0.001), with no significant difference in instability between the GAA and TTC orientations (P = 0.39; Figure 2D). Despite the fact that the circular GAA-30X and TTC-30X plasmids were significantly more stable than the circular GAA-70X and TTC-70X plasmids (P = 0.01 and P = 0.005 for GAA and TTC, respectively), no difference was seen in the level of instability between the 70X and 30X constructs following DSB repair at the XbaI site, in either the GAA or TTC orientation (P = 0.59 and P = 0.35 for GAA and TTC, respectively). Essentially, most of the DSB repair events within the (GAA•TTC)30 sequence resulted in deletion of the repeat tract (Figure 2B and D). Therefore, neither the length nor the orientation of the (GAA•TTC)n sequence had any influence on the frequency of instability seen when DSB repair occurred within the repeat tract.
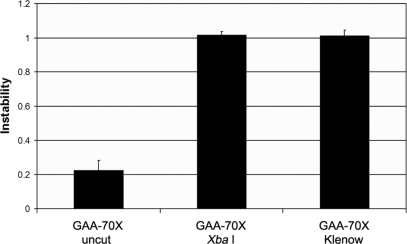
Given that DSB repair of blunt versus staggered termini may be processed differently, we compared the instability of XbaI linearized GAA-70X templates with those that were blunt-ended via end filling of the 3′ recessed XbaI ends using Klenow polymerase. As seen in Figure 3, the level of (GAA•TTC)n repeat instability was identical for the two types of termini (P = 0.78), and both were significantly more unstable than the circular GAA-70X plasmid (P < 0.001; Figure 3). These data suggest that the loss of triplets during DSB repair within the (GAA•TTC)n repeat is independent of the nature of the termini being repaired.
Figure 3.
The dramatic rise in instability of the (GAA•TTC)n sequence when DSB repair occurs within the repeat tract is independent of the nature of the termini being repaired. Repair of both the 3′ recessed XbaI termini (GAA-70X), and the same template after end filling with Klenow polymerase resulted in similar levels of instability. All error bars represent +/−2 SEM derived from triplicate experiments.
DSB repair-mediated deletions involved the repeat tract per se and there was no indication of deletions involving the plasmid (non-repeat) sequence immediately flanking the (GAA•TTC)n repeat. Sequencing of several of the deletions obtained via repair of the XbaI site at the center of the repeat in the 70X and 30X constructs revealed only a shorter but pure (GAA•TTC)n sequence with no loss of sequence outside of the repeat tract. Moreover, the location of the DSB-F and DSB-R primers close to the (GAA•TTC)n repeat would entail that even relatively small deletions of the flanking sequence would be expected to result in failure of amplification of the repeat tract. However, this was not seen, indicating that there was no significant deletion of flanking non-repeat sequence. For example, the DSB-F and DSB-R primers are located 22/26 bp upstream and 39/43 bp downstream of the (GAA•TTC)30 repeat in the GAA-30X and TTC-30X constructs, respectively, and there was no difference in the ability to amplify the repeat tract from either the uncut [522/559 (93.4%) colonies], XbaI [669/705 (94.9%) colonies] or PstI [362/373 (97%) colonies] linearized vectors (P = 0.85).
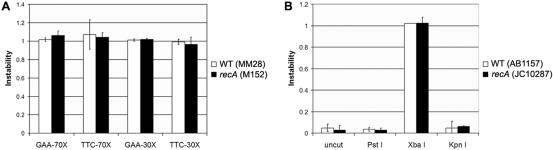
DSB repair-mediated instability of the (GAA•TTC)n repeat occurs in the absence of RecA
RecA plays an important role in DSB repair via homologous recombination. We have previously shown that the (GAA•TTC)n repeat is more unstable in recA mutants, specifically when GAA serves as the lagging strand template (47). To investigate the role of RecA in DSB repair mediated repeat instability, we examined the level of instability in plasmids GAA-70X, TTC-70X, GAA-30X and TTC-30X, that had been linearized with XbaI. Linearized templates were transformed into E. coli M152, which is isogenic to strain MM28 (RecA-proficient) used in all experiments described above, except that M152 is RecA deficient. No significant difference was noted in the level of instability in either the GAA or TTC orientation, for constructs of both lengths, in MM28 versus M152 (Figure 4A). These data suggest that the instability caused via DSB repair within the (GAA•TTC)n repeat is RecA-independent. Using a very similar experimental strategy, others have previously shown that deficiency of RecA is associated with increased instability when DSB repair occurred within a (CTG•CAG)n sequence (44). Therefore, to confirm that our result with the (GAA•TTC)n sequence was not simply due to the specific mutant strains used in our experiments, we reanalyzed our templates using the same strains tested by Hebert et al. The GAA-30X plasmid, either circular or linearized at the PstI, XbaI or KpnI restriction sites, was transformed into isogenic E. coli strains, AB1157 (RecA-proficient) and JC10287 (RecA-deficient). As expected, DSB repair within the (GAA•TTC)n sequence was associated with significantly enhanced instability compared with repair outside the repeat (Figure 4B). However, no significant difference in instability was noted for any of the four plasmid templates in AB1157 versus JC10287 (Figure 4B), further substantiating our finding that (GAA•TTC)n instability caused by DSB repair does not require RecA.
Figure 4.
DSB repair mediated instability of the (GAA•TTC)n sequence is independent of RecA. (A) Repair of linear templates obtained by XbaI digestion of GAA-70X, TTC-70X, GAA-30X and TTC-30X resulted in similar levels of instability when transformed into E. coli MM28 [wild-type (WT)] and its isogenic recA mutant strain, M152. There was no difference in the level of repeat instability between the RecA-proficient and RecA-deficient strains, regardless of repeat length or the orientation with respect to the origin of replication. (B) Instability of GAA-30X constructs either uncut or following repair at the PstI, XbaI or KpnI restriction sites in isogenic E. coli strains AB1157 (RecA-proficient) and JC10287 (RecA-deficient) showing that DSB repair-mediated instability is independent of RecA. Note that there is no difference in the level of instability with any of the four constructs in AB1157 versus JC10287. DSB repair, when specifically within the repeat tract, causes significantly increased instability in both strains. All error bars represent +/−2 SEM derived from triplicate experiments.
DSB repair at the center of the (GAA•TTC)n sequence preferentially results in deletion of approximately half of the repeat tract
In order to elucidate the possible mechanism of the DSB repair-mediated repeat instability, we analyzed the length distribution of all of the deletion products. The residual repeat length of all deletions observed after DSB repair with HindIII and EcoRI linearized GAA-79 (n = 229) and TTC-79 (n = 128) constructs, located outside of the (GAA•TTC)n sequence, was determined as a percentage of the full-length repeat tract. The length distribution of the deletions was found to be random, with an approximately equal frequency of small, medium and large deletions observed in both the GAA and TTC orientations (P = 0.57 and P = 0.2, respectively; Figure 5A). However, when DSB repair occurred at the center of the repeat tract in both (GAA•TTC)70 (n = 280 and 316 for GAA-70X and TTC-70X, respectively) and (GAA•TTC)30 (n = 728 and 652 for GAA-30X and TTC-30X, respectively), the repair process resulted in a preferential deletion of half of the repeat tract in both the GAA and TTC orientations (41–60%; Figure 5B and C). Furthermore, there were significantly more small deletions (50–99% residual repeat length) than large deletions (0–49% residual repeat length) (P < 0.001 for 70 or 30 triplets in either GAA or TTC orientations). There was no difference in the size distribution of deletions when DSB repair occurred using the 3′ recessed XbaI end or after end filling it to produce blunt-ended termini (P = 0.18; Figure 5D). We also found that RecA status did not affect the deletion spectrum, as there was no difference in the size distribution of deletions caused by repair at the XbaI site of GAA-30X in both pairs of isogenic strains; MM28 versus M152 (Figure 5E), and AB1157 versus JC10287 (Figure 5F).
Figure 5.
DSB repair at the center of the (GAA•TTC)n sequence results in the preferential deletion of approximately half (or less than half) of the total repeat length. The residual tract lengths of the (GAA•TTC)n sequence are shown (as a percentage of full-length) after transformation in E. coli MM28 of (A) EcoRI and HindIII linearized GAA-79 and TTC-79; (B) XbaI linearized GAA-70X and TTC-70X; (C) XbaI linearized GAA-30X and TTC-30X, (D) XbaI linearized (3′ recessed) and after end filling (blunt-ended) GAA-70X, (E) XbaI linearized GAA-30X in MM28 (RecA-proficient) and M152 (RecA-deficient) isogenic strains, and (F) XbaI linearized GAA-30X in AB1157 (RecA-proficient) and JC10287 (RecA-deficient) isogenic strains. Note that the size distribution of deletion products is random when DSB repair occurs outside the repeat (EcoRI, HindIII); however, approximately half of the repeat tract, or less, is preferentially deleted when DSB repair occurs at the center of the repeat tract. This is irrespective of the initial length of the repeat tract (70 or 30 triplets), the nature of the termini being repaired (staggered or blunt), or the presence/absence of RecA.
In order to determine if the length of the intervening sequence in between the two halves of the (GAA•TTC)n tract influenced the level of instability, a 28 bp spacer was inserted into the XbaI site of GAA-70X (Figure 6A; see Materials and Methods section). As previously observed for the XbaI-linearized GAA-70X, DSB repair of most of the BamHI-linearized GAA-70-spacer also underwent deletions (160 contractions from 191 independent DSB repair events), irrespective of the RecA status of the bacterial strain (Figure 6B; uncut versus BamHI, P < 0.001 for both RecA-proficient and RecA-deficient strains; no difference was noted in RecA-proficient versus RecA-deficient strains). When DSB repair occurred at the BamHI site, the repair process also resulted in a preferential deletion of approximately half of the repeat tract in both the RecA+ and RecA− strains (41–60%; Figure 6C), indicating that the length of the intervening sequence (6 bp in GAA-70X or 28 bp in GAA-70-spacer) did not significantly alter the mutational outcome. The 28 bp spacer was completely lost in almost all of the DSB repair events, as demonstrated by BamHI digestion (27 of 28 lost the site) and DNA sequencing (19 of 20 lost the entire 28 bp spacer).
Figure 6.
DSB repair-mediated instability of the (GAA•TTC)n sequence is independent of the length of the intervening sequence at the center of the repeat tract. (A) GAA-70-spacer construct containing a 28 bp spacer in the XbaI site of GAA-70X, such that BamHI would cut in the center of the spacer (indicated by the black box). (B) DSB repair at the BamHI site in GAA-70-spacer construct produced a dramatic rise in instability. Note that the (GAA•TTC)n sequence was equally unstable when transformed into E. coli MM28 [wild-type (WT)] and its isogenic recA mutant strain, M152. All error bars represent +/−2 SEM derived from triplicate experiments. (C) DSB repair at the center of the (GAA•TTC)n sequence results in the preferential deletion of approximately half of the total repeat length. The residual tract lengths of the (GAA•TTC)n sequence are shown (as a percentage of full-length) after transformation of BamHI-linearized GAA-70-spacer vector in E. coli MM28 (WT) and M152 (recA).
DSB repair located ‘off-center’ within the (GAA•TTC)n tract preferentially results in the deletion of a length equal to the shorter repeat tract flanking the DSB
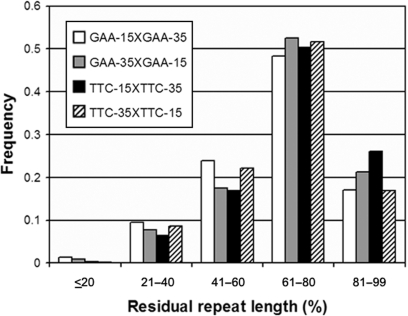
Since DSB repair at the center of the (GAA•TTC)n sequence resulted in significantly increased repeat instability with the preferential deletion of approximately half of the repeat tract, we further investigated what effect changing the location of the DSB within the (GAA•TTC)n might have on the level of instability and/or the size distribution of any resulting deletions. Two different plasmids were constructed, which contained a (GAA•TTC)50 sequence with an XbaI restriction site located off-center within the repeat tract, such that 15 and 35 repeats were located on either side of the XbaI site. As before, these off-center constructs were made in both orientations, resulting in the four following constructs: GAA-35XGAA-15, GAA-15XGAA-35, TTC-35XTTC-15 and TTC-15XTTC-35 (Figure 1). Transformation of each of these four constructs, after linearization with XbaI, was performed in E. coli MM28. Similar to DSB repair at the center of the (GAA•TTC)n sequence, nearly all repair events at the off-center location also resulted in deletions of the repeat tract (data not shown). Therefore, the position of the DSB within the (GAA•TTC)n sequence did not affect the overall frequency of instability. However, examination of the size distribution of the deletion products showed that unlike DSB repair at the center of the (GAA•TTC)n sequence, DSB repair at the off-center location resulted in deletion of less than half of the repeat tract. A preferential deletion of ∼15 repeats, i.e. equivalent to the length of the shorter of the two repeat tracts flanking the DSB, was noted for all four constructs, resulting in a residual repeat length of 61–80% of the full-length repeat tract (Figure 7). This occurred irrespective of the orientation of the repeat tract, or if the location of the shorter repeat tract was either upstream or downstream of the break site (Figure 7). Therefore, it appears that repairing a DSB anywhere within the (GAA•TTC)n results in deletion of the repeat tract; however, the location of the break within the repeat tract significantly modulates the size of the deletion resulting from DSB repair.
Figure 7.
Repair of a DSB located off-center within the (GAA•TTC)n sequence preferentially results in deletions that are approximately equivalent to the length of the side with the shorter repeat tract. A (GAA•TTC)50 sequence was linearized at an XbaI restriction site located at one of two locations off-center within the (GAA•TTC)n sequence such that there were 15 and 35 repeats on either side of the DSB. The residual repeat length of the (GAA•TTC)n sequence following DSB repair is shown as a percentage of the full-length repeat. The length distribution of the deletions show that the magnitude of the deletions were approximately equivalent to the shorter of the two sides of the repeat tract (∼15 repeats or 61–80% of full-length), regardless of the location of the DSB within the (GAA•TTC)n sequence, or the orientation of the repeat tract with respect to the origin of replication.
DISCUSSION
FRDA is exceptional among a growing number of diseases caused by triplet-repeat expansions, in being inherited as a recessive trait (4). Therefore, while most other triplet-repeat diseases are frequently the result of de novo intergenerational expansions, FRDA is most commonly caused by inheriting fully expanded (GAA•TTC)n alleles from both parents. All expanded triplet-repeat sequences are somatically unstable; however, while (CNG)n repeats (the sequence associated with most such diseases) display an expansion bias in all human tissues (48–51), the expanded (GAA•TTC)n repeat displays a strong contraction bias in almost all tissues (18,19,52).
The (GAA•TTC)n sequence has been shown to stall replication fork progression specifically when (GAA)n is the lagging strand template (39). We recently found that a deficiency of the RecA-dependent RecFOR and RecBCD pathways for restart of stalled replication forks results in orientation-dependent instability, with increased instability specifically when (GAA)n is the lagging strand template (47). These data indicate that the pathways for restart of stalled replication forks can result in stabilization of the (GAA•TTC)n sequence in the GAA orientation. However, another possible consequence of replication stalling is the generation of DSBs (40–42). Therefore, it was of interest to us to determine what effect repairing DSBs near or within the (GAA•TTC)n sequence might have on repeat instability. Here we show that while DSB repair outside of the (GAA•TTC)n sequence had no effect on the level of instability, repairing a DSB within the (GAA•TTC)n sequence resulted in a dramatic increase in frequency of deletions. Indeed, almost every repair event resulted in a partial deletion of the repeat tract. Additionally, abolition of the restriction sequence at the site of the repair indicated that the repair process itself was responsible for the loss of triplet-repeat sequence.
Our observations are in sharp contrast with those made by others using the (CTG•CAG)n sequence (44). We noted that virtually all DSB repair events within the (GAA•TTC)n sequence resulted in deletions. Moreover, our results did not depend on the length or orientation of the repeat tract, nor on the RecA status of the strain used. In contrast, others found that DSB repair within the (CTG•CAG)n sequence resulted in deletions that were dependent on the length and orientation of the repeat tract, and moreover that recA mutants showed enhanced deletion frequency (44). Furthermore, when DSB repair occurred outside of the (CTG•CAG)n sequence, both in E. coli (46) and in primate cells (43), frequent deletions of the triplet repeat along with flanking non-repeat sequence was noted. However, our data show that DSB repair outside of the (GAA•TTC)n sequence resulted in no additional instability of the repeat tract or the flanking sequence. These data indicate that the mechanism of DSB repair-mediated triplet-repeat instability is highly dependent on the sequence and/or physical properties of the repeat tract.
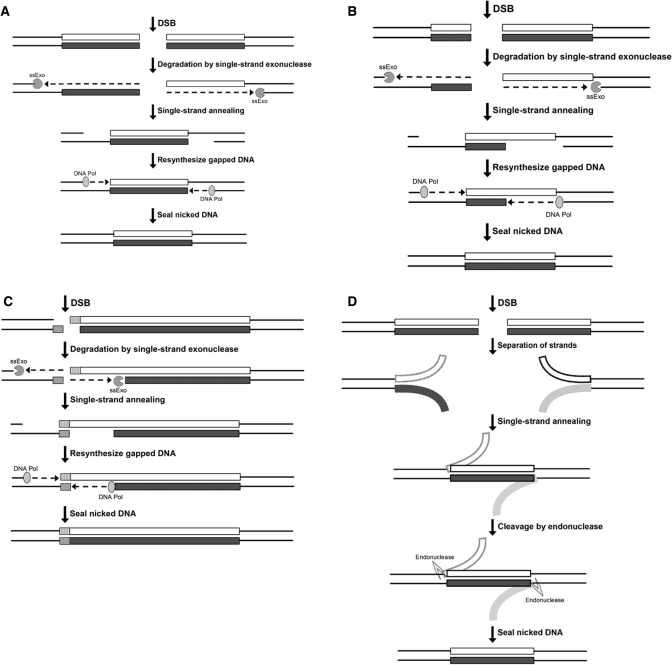
In contrast to when DSB repair occurred within the (GAA•TTC)n sequence, the restriction enzyme site used to create the DSB was always maintained when DSB repair occurred outside of the repeat sequence. These observations indicate that the presence of (GAA•TTC)n repeat sequence flanking each side of the DSB influences the repair process in such a way as to result in loss of sequence at the termini, whereas the break outside of the (GAA•TTC)n is repaired by ligation of the restriction site, with no additional loss of sequence. When repair occurred at the center of the repeat tract, it preferentially deleted approximately half of the initial repeat length, and small contractions (deletion of <50% of initial length) were more prevalent than large contractions (deletion of >50% of initial length). However, when a DSB was repaired off-center within the repeat tract, there was a shift in the length distribution of deletions, such that a length approximately equivalent to the shorter of the two repeat tracts (or less) was preferentially deleted. These data are consistent with a single-strand annealing mechanism for generating deletions via DSB repair (Figure 8). If a DSB occurs at the center of a (GAA•TTC)n sequence and a single-strand exonuclease (ssExo) degrades either 5′ to 3′ or 3′ to 5′ on both sides of the break, it would result in complementary single-stranded (GAA)n and (TTC)n sequences on opposite sides of the break. The single-stranded repeat sequences could then anneal to repair the break, with DNA polymerase filling in any resulting gaps in the DNA (Figure 8A). Due to the repetitive nature of the (GAA)n and (TTC)n sequences, these single-stranded regions could anneal at numerous locations within the repeat tract. If the two sequences were to completely overlap, it would result in deletion of half of the initial repeat tract length, which was the predominant outcome in our experiments. If the two strands were to only partially overlap, it would result in the deletion of less than half of the repeat tract, which according to our data, is significantly preferred over the deletion of more than half of the repeat tract. The same mechanism also explains the observed outcome seen when DSB repair occurred off-center within the repeat tract (Figure 8B). The number of repeats deleted during the repair process would be limited by the length of the shorter of the two repeat tracts, since the single-stranded (GAA)n and (TTC)n sequences on opposite sides of the break could only overlap by as many repeats as there are in the shorter repeat tract, or less (Figure 8B). Consistent with this model, we noted that the deletion was most likely to be equivalent to the length of the shorter of the two flanking repeat tracts. Indeed, the fact that the 28 bp spacer at the center of the repeat tract also resulted in deletion of half of the repeat tract, along with loss of the entire spacer following DSB repair, further supports this model. However, if the break were to occur outside of the repeat tract, there would be a single-stranded (GAA)n or (TTC)n sequence on one side of the break, and a single-stranded random (non-complementary) sequence on the other side. Therefore, the only way for the two single-stranded sequences to anneal would be at the site of the break, which offers a complementary restriction enzyme recognition sequence, thereby resulting in no deletion of repeat sequence (Figure 8C). Again, this is consistent with our observation that repairing a break outside of the (GAA•TTC)n did not result in repeat instability, and most repair products maintained the restriction site used to create the break. Indeed, the same outcomes would also be predicted if, instead of digestion with an ssExo, a flap endonuclease were to cleave partially annealed single strands on either side of the DSB (Figure 8D). The same length distributions would be expected as described for the single-strand exonuclease mediated mechanism shown in Figures 8A–C.
Figure 8.
Model of DSB repair within or outside of the (GAA•TTC)n sequence. (A, B) Single-strand annealing model of DSB repair is shown at the center (A) or off-center (B) within the (GAA•TTC)n sequence. If a DSB occurs within the repeat tract, a deletion could result if a single-strand exonuclease were to degrade DNA on either side of the break (only 3′ → 5′ exonuclease is depicted here). The single-stranded (GAA)n and (TTC)n sequences (white and dark gray boxes) would anneal, followed by DNA polymerase re-synthesizing the gap, and sealing of the nick by DNA ligase. Note [as seen in (A)] that DSB repair at the center of the (GAA•TTC)n sequence would result in deletion of half of the repeat tract (if the entire repeat tract on each side were digested) or less than half (if less than the entire repeat tract on each were digested), consistent with our data. Furthermore, the DSB repair at the off-center location within the repeat tract [as seen in (B)] would preferentially delete the length of the shorter of the two flanking repeats (if the entire repeat tract on each side were digested), again consistent with our observations. (C) When DSB repair occurs outside of the repeat tract, and if the same process of single-strand exonuclease degradation were to occur, the only complementary sequence at which the resulting single-stranded DNA could anneal would be at the restriction enzyme sequence used to create the initial DSB. No loss of repeat sequence, and retention of the restriction site would be predicted, which is consistent with our data. (D) Alternatively, rather than the DNA being degraded by a single-strand exonuclease, DNA endonuclease processing of partially annealed strands on either side of the DSB could occur. The resulting single-stranded (GAA)n and (TTC)n sequences could anneal on opposite sides of the DSB, and an endonuclease could remove the flaps. DNA polymerase and DNA ligase would fill in and seal the nick, resulting in a shorter (GAA•TTC)n sequence. The same outcomes would be predicted with the endonuclease-mediated processing if the DSB were located at the center, or off-center, within the (GAA•TTC)n sequence.
It should be noted that both replication stalling and repeat instability are length- and orientation-dependent when plasmids containing the (GAA•TTC)n sequence are propagated in E. coli. However, paradoxically our data show that DSB repair-mediated instability leads to the same frequency of deletions in both the GAA and TTC orientations, and this is independent of the length of the repeat tract. One way to reconcile these apparently conflicting observations is that, while a DSB in a repeat tract of any length and in any orientation is likely to result in a deletion, in nature DSBs may only be occurring in E. coli when the repeat length is in the GAA orientation and of sufficient length. It is plausible that the DSBs are a result of the replication stalling that preferentially occurs in the GAA orientation when plasmids containing repeat tracts of sufficient length are grown in E. coli, and this is why deletions are observed preferentially in the GAA orientation. It is also possible that some inherent property of the (GAA•TTC)n sequence itself may make it more prone to breakage, whether it be a consequence of the repetitive nature of the DNA sequence per se, or due to a specific secondary structure adopted by the (GAA•TTC)n sequence (7–10).
It is also interesting to note that in tissues of FRDA patients there is a significant trend for large deletions of the expanded (GAA•TTC)n sequence in the FXN gene (18,19,52). While all tissues show a significant frequency of contractions, peripheral blood cells are particularly prone to large deletions (18,19), which in some rare cases even results in complete reversion of the mutation to the non-pathogenic size. Indeed, the instability seen in blood is age dependent, with increasing mutation load as the patients get older (52). It is plausible that the proliferative nature of their precursor cells may afford multiple opportunities for DSB formation via replication fork stalling, thereby explaining the particularly large deletions in peripheral blood cells. The only other cells that show particularly large deletions, including occasional reversion to the normal size range are sperms from male FRDA patients (17). Again, human spermatozoa are generated following multiple pre-meiotic cellular divisions, thus allowing a similar opportunity to develop fork stalling, DSBs, and contractions. It is also likely, at least in FRDA patients, that the free radical-induced oxidative damage caused via mitochondrial frataxin deficiency (53) may further increase the possibility of DSBs. Both replication stalling in continuously proliferating cells and oxidative damage can potentially account for the age-dependent increase in contractions in vivo. Our data in E. coli therefore make a convincing case for the exploration of the potential role of DSB repair as a mechanism for somatic instability in mammalian cells.
ACKNOWLEDGEMENTS
This research was made possible by grants from the National Institutes of Health (NIH/NINDS), Muscular Dystrophy Association, and Friedreich Ataxia Research Alliance to S.I.B. We thank the E. coli Genetic Stock Center (Yale University) for kindly providing us with bacterial strains.
Funding to pay the Open Access publication charges for this article was provided by Muscular Dystrophy Association.
Conflict of interest statement. None declared.
REFERENCES
- 1.Di Prospero NA, Fischbeck KH. Therapeutics development for triplet repeat expansion diseases. Nat. Rev. Genet. 2005;6:756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- 2.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 3.Bowater RP, Wells RD. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 4.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, et al. Friedreich's ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 5.Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, et al. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum. Mol. Genet. 1997;6:1261–1266. doi: 10.1093/hmg/6.8.1261. [DOI] [PubMed] [Google Scholar]
- 6.Cossée M, Schmitt M, Campuzano V, Reutenauer L, Moutou C, Mandel JL, Koenig M. Evolution of the Friedreich's ataxia trinucleotide repeat expansion: founder effect and premutations. Proc. Natl Acad. Sci. USA. 1997;94:7452–7457. doi: 10.1073/pnas.94.14.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA•TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 9.Grabczyk E, Usdin K. The GAA•TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA•TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 11.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 12.Pianese L, Turano M, Lo Casale MS, De Biase I, Giacchetti M, Monticelli A, Criscuolo C, Filla A, Cocozza S. Real time PCR quantification of frataxin mRNA in the peripheral blood leucocytes of Friedreich ataxia patients and carriers. J. Neurol. Neurosurg. Psychiatry. 2004;75:1061–1063. doi: 10.1136/jnnp.2003.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dürr A, Cossee M, Agid Y, Campuzano V, Mignard C, Penet C, Mandel JL, Brice A, Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N. Engl. J. Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 14.Filla A, De Michele G, Cavalcanti F, Pianese L, Monticelli A, Campanella G, Cocozza S. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am. J. Hum. Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 15.Monros E, Molto MD, Martinez F, Canizares J, Blanca J, Vilchez JJ, Prieto F, De Frutos R, Palau F. Phenotype correlation and intergenerational dynamics of the Friedreich ataxia GAA trinucleotide repeat. Am. J. Hum. Genet. 1997;61:101–110. doi: 10.1086/513887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pianese L, Cavalcanti F, De Michele G, Filla A, Campanella G, Calabrese O, Castaldo I, Monticelli A, Cocozza S. The effect of parental gender on the GAA dynamic mutation in the FRDA gene. Am. J. Hum. Genet. 1997;60:460–463. [PMC free article] [PubMed] [Google Scholar]
- 17.De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, Cocozza S. Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich's ataxia. Hum. Mol. Genet. 1998;7:1901–1906. doi: 10.1093/hmg/7.12.1901. [DOI] [PubMed] [Google Scholar]
- 18.De Biase I, Rasmussen A, Endres D, Al-Mahdawi S, Monticelli A, Cocozza S, Pook M, Bidichandani SI. Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann. Neurol. 2007;61:55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- 19.Sharma R, Bhatti S, Gomez M, Clark RM, Murray C, Ashizawa T, Bidichandani SI. The GAA triplet-repeat sequence in Friedreich ataxia shows a high level of somatic instability in vivo with a significant predilection for large contractions. Hum. Mol. Genet. 2002;11:2175–2187. doi: 10.1093/hmg/11.18.2175. [DOI] [PubMed] [Google Scholar]
- 20.Napierala M, Parniewski P, Pluciennik A, Wells RD. Long CTG•CAG repeat sequences markedly stimulate intramolecular recombination. J. Biol. Chem. 2002;277:34087–34100. doi: 10.1074/jbc.M202128200. [DOI] [PubMed] [Google Scholar]
- 21.Pluciennik A, Iyer RR, Napierala M, Larson JE, Filutowicz M, Wells RD. Long CTG•CAG repeats from myotonic dystrophy are preferred sites for intermolecular recombination. J. Biol. Chem. 2002;277:34074–34086. doi: 10.1074/jbc.M202127200. [DOI] [PubMed] [Google Scholar]
- 22.Hashem VI, Rosche WA, Sinden RR. Genetic recombination destabilizes (CTG)n•(CAG)n repeats in E. coli. Mutat. Res. 2004;554:95–109. doi: 10.1016/j.mrfmmm.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Napierala M, Dere R, Vetcher A, Wells RD. Structure-dependent recombination hot spot activity of GAA•TTC sequences from intron 1 of the Friedreich's ataxia gene. J. Biol. Chem. 2004;279:6444–6454. doi: 10.1074/jbc.M309596200. [DOI] [PubMed] [Google Scholar]
- 24.Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, Bowater RP, Sinden RR, Wells RD. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl Acad. Sci. USA. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parniewski P, Bacolla A, Jaworski A, Wells RD. Nucleotide excision repair affects the stability of long transcribed (CTG•CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 1999;27:616–623. doi: 10.1093/nar/27.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parniewski P, Jaworski A, Wells RD, Bowater RP. Length of CTG•CAG repeats determines the influence of mismatch repair on genetic instability. J. Mol. Biol. 2000;299:865–874. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- 27.Nichol K, Pearson CE. CpG methylation modifies the genetic stability of cloned repeat sequences. Genome Res. 2002;12:1246–1256. doi: 10.1101/gr.74502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 29.Shimizu M, Gellibolian R, Oostra BA, Wells RD. Cloning, characterization and properties of plasmids containing CGG triplet repeats from the FMR-1 gene. J. Mol. Biol. 1996;258:614–626. doi: 10.1006/jmbi.1996.0273. [DOI] [PubMed] [Google Scholar]
- 30.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 31.Hirst MC, White PJ. Cloned human FMR1 trinucleotide repeats exhibit a length- and orientation-dependent instability suggestive of in vivo lagging strand secondary structure. Nucleic Acids Res. 1998;26:2353–2358. doi: 10.1093/nar/26.10.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakumaran BS, Freudenreich CH, Zakian VA. CGG/CCG repeats exhibit orientation-dependent instability and orientation-dependent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 33.Pollard LM, Sharma R, Gomez M, Shah S, Delatycki MB, Pianese L, Monticelli A, Keats BJ, Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;32:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 35.Rindler PM, Clark RM, Pollard LM, De Biase I, Bidichandani SI. Replication in mammalian cells recapitulates the locus-specific differences in somatic instability of genomic GAA triplet-repeats. Nucleic Acids Res. 2006;34:6352–6361. doi: 10.1093/nar/gkl846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, et al. GAA instability in Friedreich's ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 37.Samadashwily GM, Mirkin SM. Trapping DNA polymerases using triplex-forming oligodeoxyribonucleotides. Gene. 1994;149:127–136. doi: 10.1016/0378-1119(94)90421-9. [DOI] [PubMed] [Google Scholar]
- 38.Krasilnikov AS, Panyutin IG, Samadashwily GM, Cox R, Lazurkin YS, Mirkin SM. Mechanisms of triplex-caused polymerization arrest. Nucleic Acids Res. 1997;25:1339–1346. doi: 10.1093/nar/25.7.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michel B, Ehrlich SD, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 42.Marians KJ. Mechanisms of replication fork restart in Escherichia coli. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2004;359:71–77. doi: 10.1098/rstb.2003.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcadier JL, Pearson CE. Fidelity of primate cell repair of a double-strand break within a (CTG).(CAG) tract. Effect of slipped DNA structures. J. Biol. Chem. 2003;278:33848–33856. doi: 10.1074/jbc.M304284200. [DOI] [PubMed] [Google Scholar]
- 44.Hebert ML, Spitz LA, Wells RD. DNA double-strand breaks induce deletion of CTG.CAG repeats in an orientation-dependent manner in Escherichia coli. J. Mol. Biol. 2004;336:655–672. doi: 10.1016/j.jmb.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 45.Hebert ML, Wells RD. Roles of double-strand breaks, nicks, and gaps in stimulating deletions of CTG.CAG repeats by intramolecular DNA repair. J. Mol. Biol. 2005;353:961–979. doi: 10.1016/j.jmb.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 46.Kosmider B, Wells RD. Double-strand breaks in the myotonic dystrophy type 1 and the fragile X syndrome triplet repeat sequences induce different types of mutations in DNA flanking sequences in Escherichia coli. Nucleic Acids Res. 2006;34:5369–5382. doi: 10.1093/nar/gkl612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollard LM, Chutake Y, Rindler PM, Bidichandani SI. Deficiency of RecA-dependent RecFOR and RecBCD pathways causes increased instability of the (GAA•TTC)n sequence when GAA is the lagging strand template. Nucleic Acids Res. 2007;Oct 11 doi: 10.1093/nar/gkm810. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 49.Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 50.Wong LJ, Ashizawa T, Monckton DG, Caskey CT, Richards CS. Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am. J. Hum. Genet. 1995;56:114–122. [PMC free article] [PubMed] [Google Scholar]
- 51.Martorell L, Monckton DG, Gamez J, Johnson KJ, Gich I, Lopez de Munain A, Baiget M. Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum. Mol. Genet. 1998;7:307–312. doi: 10.1093/hmg/7.2.307. [DOI] [PubMed] [Google Scholar]
- 52.De Biase I, Rasmussen A, Monticelli A, Al-Mahdawi S, Pook M, Cocozza S, Bidichandani SI. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007;90:1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Puccio H, Koenig M. Friedreich ataxia: a paradigm for mitochondrial diseases. Curr. Opin. Genet. Dev. 2002;12:272–277. doi: 10.1016/s0959-437x(02)00298-8. [DOI] [PubMed] [Google Scholar]