Abstract
Kinetoplast DNA (kDNA) is a novel form of mitochondrial DNA consisting of thousands of interlocked minicircles and 20–30 maxicircles. The minicircles replicate free of the kDNA network but nicks and gaps in the newly synthesized strands remain at the time of reattachment to the kDNA network. We show here that the steady-state population of replicated, network-associated minicircles only becomes repaired to the point of having nicks with a 3′OH and 5′deoxyribonucleoside monophosphate during S phase. These nicks represent the origin/terminus of the strand and occur within the replication origins (oriA and oriB) located 180° apart on the minicircle. Minicircles containing a new L strand have a single nick within either oriA or oriB but not in both origins in the same molecule. The discontinuously synthesized H strand contains single nicks within both oriA and oriB in the same molecule implying that discontinuities between the H-strand Okazaki fragments become repaired except for the fragments initiated within the two origins. Nicks in L and H strands at the origins persist throughout S phase and only become ligated as a prelude to network division. The failure to ligate these nicks until just prior to network division is not due to inappropriate termini for ligation.
INTRODUCTION
Crithidia fasciculata is a protozoan parasite related to the pathogenic human parasites Trypanosoma brucei, Trypanosoma cruzi and Leishmania. These latter organisms cause devastating diseases such as African sleeping sickness, Chagas disease and leishmaniasis. These protozoan parasites diverged early in the course of eukaryotic evolution and have many unique features of their biology including antigenic variation, RNA editing, unusual genome organization, novel mechanisms of regulating gene expression and an unusual form of mitochondrial DNA termed kinetoplast DNA or kDNA (1,2). The kDNA is a large network of interlocked circular DNA molecules condensed into a disk-like structure within the mitochondrial matrix close to the flagellar basal bodies (3,4). The kDNA is physically connected to the basal bodies through a transmembrane filament system known as the tripartite attachment complex or TAC (5–7). The kDNA network consists of two types of circular DNAs, minicircles and maxicircles. The maxicircles are the equivalent of usual mitochondrial DNAs and encode ribosomal RNAs and genes for oxidative metabolism (8,9). The minicircles apparently do not code for proteins but rather encode small RNAs called guide RNAs that are involved in the editing of maxicircle transcripts by a process involving the insertion and/or deletion of U residues in the transcripts (10–12).
The kDNA in C. fasciculata consists of 5000 to 10 000 minicircles and 20–30 maxicircles. The maxicircles replicate by a theta mechanism while remaining attached to the kDNA network (13). On the other hand, the minicircles are released from the network sequentially as covalently closed circular molecules and are replicated in the mitochondrial matrix in a space between the kDNA disk and the flagellum termed the flagellar attachment zone or KFZ (14,15). Following their replication the minicircles are rejoined to the kDNA disk at antipodal sites by a DNA topoisomerase (16,17). At any given time during S phase only a few percent of the minicircles are free and undergoing replication. The newly reattached minicircles become distributed around the perimeter of the kDNA network apparently as a result of rotation of the kDNA disk relative to the antipodal attachment sites (18–20). The annular ring of newly replicated minicircles grows inward as replication proceeds and the central zone of unreplicated covalently closed minicircles shrinks in size until all minicircles have been replicated. At that point the minicircles within the double size network all contain discontinuities in the newly synthesized strands.
Crithidia minicircles have two origins of replication (oriA and oriB) located 180° apart and each origin contains three sequence elements (CSB-1, CSB-2 and CSB-3) that are conserved in kinetoplastid protozoa (21). The longest of these conserved sequences (12 nt) is CSB-3, also known as the universal minicircle sequence or UMS. Replication of the free minicircles is unidirectional with one strand (the L strand) synthesized continuously and one strand (the H strand) synthesized discontinuously (22–26). Analysis of newly replicated free minicircles in an isolated kinetoplast system showed that the minicircle L strand initiates from either of two replication origins (A and B) but not from both in the same minicircle (22,23). The 5′ terminus of the L strand is within the sequence complementary to the UMS and is usually a nick or a gap with a few ribonucleotides (23–25,27). The daughter molecule containing the new L strand has a single discontinuity at either oriA or oriB and a covalently closed circular H strand. In contrast, the nascent free minicircles containing newly synthesized H strands have a covalently closed circular L strand and have multiple discontinuities between Okazaki fragments in the H strand that all become repaired except for discontinuities at both oriA and oriB. Amazingly, the discontinuities in the minicircles newly synthesized in vivo are not repaired until all minicircles have been replicated and reattached, then only closed as a prelude to division of the network to yield two daughter networks in which all minicircles are covalently closed (18,19,28). In light of the presence of multiple DNA polymerases and DNA ligases within kinetoplasts (29,30) it is a mystery why the discontinuities in network-associated minicircles remain unrepaired until all minicircles have been replicated. To begin to address this question, we have undertaken an analysis of network-associated minicircles during kinetoplast S phase in synchronized cultures of C. fasciculata.
MATERIALS AND METHODS
Cell growth and synchronization
Crithidia fasciculata was grown in a defined medium (31) at 28°C on a rotary shaker and synchronized by release from a hydroxyurea block as described previously (32). The cell number was determined using a Z1 particle counter (Beckman-Coulter, Miami, FL, USA) and cells containing two nuclei were counted using a hemocytometer and DAPI (4′-6-diamidino-2-phenylindole) staining of the cells.
DNA pulse labeling
One milliliter of culture was removed at 30 min intervals for determining both the rate of total DNA synthesis and the rate of minicircle synthesis. Each sample was labeled by adding 155 μl of 3H-thymidine (1 mCi/ml; 80 Ci/mmol) to the cells for 5 min at 28°C. Incorporation was stopped by the addition of sodium azide to a final concentration of 0.02%. Cells were harvested by centrifugation for 1 min in a microcentrifuge and resuspended in 360 μl of 0.01 M Tris pH 8, 1 mM EDTA, 0.1 M NaCl and 0.02% sodium azide. The cells were then lysed by adding 40 μl of 10% SDS and 10 μl of proteinase K (10 mg/ml) and incubating overnight at 56°C. The lysates were extracted with 0.4 ml of Tris buffer-equilibrated phenol pH 8 and 50 μl was spotted onto DE81 filters for determining total DNA synthesis. The DE81 filters were washed three times in 250 ml of 0.3 M NH4COOH pH 7.8 for 5 min each, once for 5 min in 95% ethanol, dried and counted in a liquid scintillation counter. The remainders of the cell lysates were each ethanol precipitated and resuspended in 200 μl of 0.01 M Tris pH 8, 1 mM EDTA and 22 μl of 10 × NEB 3 buffer followed by digestion for 60 min at 37°C with Mlu I and XhoI restriction enzymes (New England Biolabs) to fragment the DNA. The digested DNAs were treated with 2 μl of RNase A (Sigma, 10 mg/ml) for 10 min at 37°C and then hybridized to nylon filters containing either DNA from plasmid pGEM-11Zf(+) (Promega, Madison, WI, USA) or pJH621 (pGEM-11Zf(+) containing a cloned XhoI-digested C. fasciculata minicircle). Hybridization was performed as described (33).
kDNA isolation
kDNA was isolated from 40 ml of synchronized cells at each 30 min interval as described (34).
Minicircle markers
A mixture of linear, covalently closed circular and nicked circular minicircles was prepared by partial ligation of XhoI linear minicircles released from pJH621 DNA. Each form was gel purified on 0.7% agarose gels for use as markers on neutral and alkaline gels.
Agarose gels and Southern blotting
Neutral agarose gels contained 0.7% agarose in 0.5 × Tris-borate-EDTA (TBE) buffer and 0.25 μg/ml ethidium bromide and were electrophoresed with a running buffer containing 0.5 × TBE buffer and 0.25 μg/ml ethidium bromide. Gels were transferred to Hybond N membranes (Amersham) and probed with a minicircle DNA probe that had been 32P-labeled by random priming of an XhoI-cut minicircle DNA. Alkaline agarose gels containing 1% agarose were soaked in 30 mM NaOH, 1 mM EDTA for 1 h before running in 30 mM NaOH, 1 mM EDTA. Alkaline agarose gels were run and transferred to Hybond N membranes essentially as described (35). Duplicate samples were run on the same gel and after transfer the membrane was cut apart to allow separate hybridization to the L- and H-strand-specific probes and to a molecular weight marker probe (32P-end labeled DNA Plus ladder, Invitrogen).
Strand-specific DNA probes
pJH621 was digested with Sal I and transcribed with SP6 RNA polymerase and 32P-labeled ribonucleoside triphosphates to produce a probe for minicircle L strands. Similarly, a probe for H strands was produced by transcription of BamHI-digested pJH621 with T7 RNA polymerase. The labeled RNA transcripts were digested with pancreatic DNase and purified over G25 MicroSpin columns before use in hybridizations.
RESULTS
kDNA replication in synchronized cultures of C. fasciculata
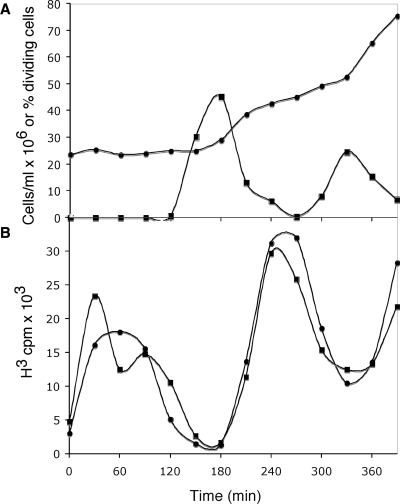
The present experiments investigate the possible basis for the delay in covalent repair of discontinuities at the replication origins of kDNA-associated minicircles until every minicircle in the network has been replicated. In order to examine the structure of kDNA network-associated minicircles during kinetoplast S phase we have synchronized C. fasciculata by release from a hydroxyurea block. Synchrony was shown by counting dividing cells (cells with two nuclei) (Figure 1A) and by monitoring both total DNA synthesis and kinetoplast minicircle DNA synthesis (Figure 1B) at 30 min intervals. The cells go through the first S phase during the first 90 min following release from the hydroxyurea block. Dividing cells are frequent from 150 to 210 min and the second S phase occurs from 210 to 300 min. Tritiated thymidine pulse labeling shows that total DNA synthesis, which is largely nuclear, and kDNA minicircle DNA synthesis occur in synchrony in the culture.
Figure 1.
DNA synthesis in a synchronous culture of C. fasciculata. Crithidia fasciculata was synchronized by release from a hydroxyurea block and sampled at 30 min intervals to determine (A) the total cell number (filled circles) and the percentage of dividing cells (filled squares) and (B) the rate of total DNA synthesis (filled circles) and the rate of kDNA synthesis (filled squares).
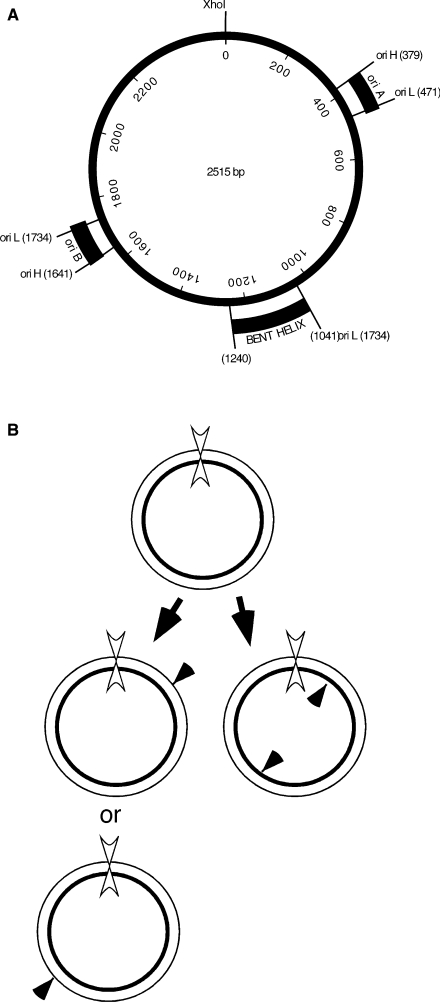
Fortunately, C. fasciculata minicircles have a nearly homogenous sequence (36), unlike minicircles in most other kinetoplastid species that can have hundreds of sequence classes. Thus with a unique restriction map available it is possible to map discontinuities in the kinetoplast-associated minicircles (23,37). Figure 2A shows a physical map of the C. fasciculata minicircle and sites of specific nicks observed previously at the replication origins of free minicircles replicated in an isolated kinetoplast system (22). The location of a static bend in the DNA, termed a bent helix, located approximately between nucleotides 1041 and 1240 is also shown. We have mapped the sites of nicks in each of the strands of the kinetoplast-associated minicircles by strand-specific probing of Southern blots of XhoI-digested kDNA networks run on alkaline denaturing gels. Cleavage of covalently closed minicircles at the unique XhoI site prior to denaturation releases unit-length strands (2.5 kb) of each type (H and L) but molecules with specific discontinuities at the replication origins will produce a 2.5 kb fragment of the circular strand and two or more fragments of defined size from the newly synthesized strand (Figure 2B).
Figure 2.
(A) Physical map of C. fasciculata minicircle DNA (36). The two-replication origins, oriA and oriB, are located 180° apart on the circular DNA. Specific nicks in nicked circular (form II) minicircles identified in free minicircles replicated in an isolated kinetoplast system define the initiation sites of the leading L strand and the initiation sites of the first H-strand Okazaki fragment at each origin (23). The nucleotide coordinates are indicated for the specific nicks in the H and L strands. The region from 1041 to 1240 nt has been shown to confer a static bend in the DNA (42) and is referred to as a bent helix. (B) Crithidia fasciculata minicircle replication schematic. Thin line, L strand; thick line, H strand; filled arrowheads, specific nicks; double arrowhead, XhoI site.
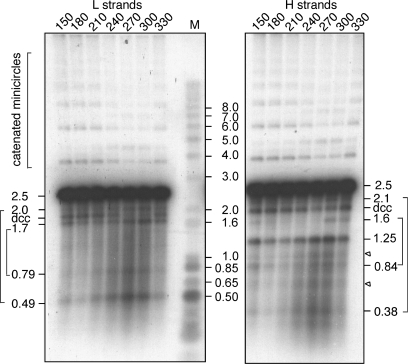
We have analyzed the strands released upon denaturation of kDNA network-associated minicircles isolated throughout kinetoplast S phase. Figure 3 shows a Southern blot of a denaturing gel analysis of duplicate samples of XhoI-digested kDNA taken at 30 min intervals spanning the second S phase. One set was probed to detect L strands and one to detect H strands. The 2.5 kb L-strand fragments correspond to linear strands derived by XhoI cleavage of either covalently closed (unreplicated) minicircles or from replicated minicircles in which the L strand was the parental covalently closed circle. Two pairs of fragments (2.0 + 0.49 and 1.7 + 0.79; Table 1 and Figure 3) are consistent with the strands having been derived from nicked or gapped minicircles with a single discontinuity at oriA or oriB. This result indicates that L-strand replication initiates at either oriA or oriB but not from both in the same molecule. In addition, we observed a ladder of catenated minicircles and a band labeled denatured covalently closed (dcc) that represents denatured covalently closed minicircles. Both of these species represent a minor class of minicircles that are resistant to cleavage by XhoI and will be addressed later.
Figure 3.
Alkaline gel analysis of XhoI-digested kDNA from samples taken from a synchronous culture at 30 min intervals during S phase. Duplicate samples were run on the same gel and transferred to a membrane that was cut into three parts for hybridization separately to probes for minicircle L and H strands and for molecular weight markers. Fragment pairs reflecting discontinuities at oriA or oriB are indicated in brackets. Minor H-strand fragments corresponding to molecular sizes of ∼1.0 and 0.65 kb are indicated by triangles. A ladder of catenated minicircles is indicated by a bracket in the upperpart of the gel. dcc, denatured covalently closed minicircles.
Table 1.
Predicted and observed XhoI fragments
| XhoI fragments | Observed | |
|---|---|---|
| L strand nicks | ||
| oriA | 471 + 2044 | 490 + 2000 |
| oriB | 781 + 1734 | 790 + 1700 |
| oriA and oriB | 471 + 781 + 1263 | n.o. |
| H strand nicks | ||
| oriA | 379 + 2136 | 380 + 2100 |
| oriB | 874 + 1641 | 840 + 1600 |
| oriA and oriB | 379 + 874 + 1250 | 380 + 840 + 1250 |
Minicircle H and L single-strand fragments produced upon XhoI digestion and alkaline denaturation of network-associated minicircles containing nicks at the replication origins. Fragment sizes are based on mapping at the nucleotide level of nicks in free form II minicircles replicated in an isolated kinetoplast system (23). The observed values are estimates based on gel migration relative to DNA markers in Figure 3 and in earlier experiments (22).
In the case of fragments derived from the H strand, two pairs of fragments (2.1 + 0.38 and 1.6 + 0.84) are consistent with the strands having resulted from cleavage of minicircles in which there is a single discontinuity at either oriA or oriB. In addition, a more abundant species corresponding to 1.25 kb half-length molecules must have resulted from replicated minicircles with a new H strand in which discontinuities are present at both oriA and oriB. Thus, in these molecules all Okazaki fragments have been covalently joined except for the fragments initiated at each of the origins. Two additional minor H-strand fragments of ∼1.0 and 0.65 kb are also observed. These minor species could possibly be derived from minicircles containing discontinuities at oriB and at a previously unknown site at either 1.0 or 0.65 kb clockwise from the XhoI site. It is unclear what significance a discontinuity at either site might have. A smear of fragments from ∼1.2 to 0.2 kb possibly reflects the presence of H-strand Okazaki fragments that have not yet been ligated.
Repair of discontinuities in replicated minicircles
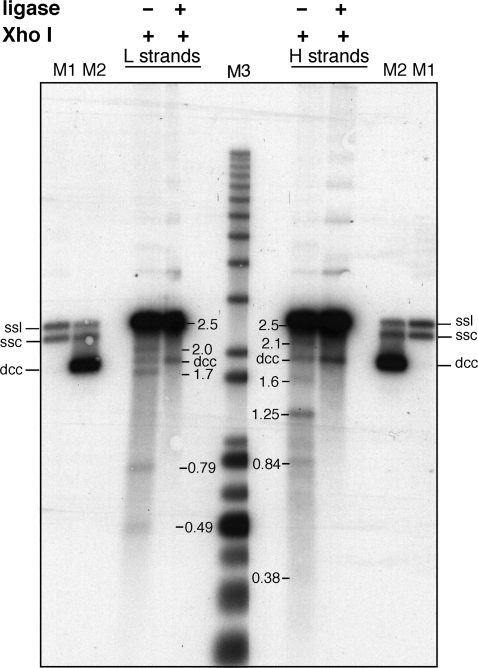
The lack of covalent closure of discontinuities at the origin/terminus of strands in replicated minicircles until just prior to network division could be the consequence of inappropriate termini for covalent closure, among other things. To address this possibility we have treated kDNA samples with Escherichia coli DNA ligase prior to XhoI digestion, alkaline gel analysis and Southern blotting. Unlike T4 DNA ligase, the E. coli enzyme cannot join a DNA 3′OH terminus to a strand containing 5′ ribonucleotides (38,39). Termini of the latter type require the removal of the ribonucleotides(s) and gap filling before they can be ligated by the E. coli ligase. The E. coli ligase can only join a simple nick in DNA with a 3′OH and 5′ deoxynucleoside monophosphate. The disappearance of the specific fragments and the increase of the denatured covalently closed circles upon ligase treatment (Figure 4) indicate that the specific discontinuities in both the L and H strands were repaired by E. coli DNA ligase. Any RNA primers must have been removed and gaps filled soon after their reattachment to the kDNA network. We conclude that the lack of covalent closure of the origin-specific nicks in vivo in the network-associated minicircles is not due to inappropriate termini for ligation.
Figure 4.
Ligation of discontinuities in kinetoplast-associated minicircles. Purified kDNA from 300 min after release from the hydroxyurea block were either treated with E. coli DNA ligase (+) or not (−) prior to digestion with XhoI and alkaline denaturing gel analysis. The gel was transferred and hybridized as in Figure 3 to identify released H and L single strands. Marker lanes: M1, minicircle single strand linears (ssl) and single strand circles (ssc); M2, denatured covalently closed minicircles (dcc) in addition to minicircle single strand linears (ssl) and single strand circles (ssc); M3, molecular weight markers.
Nature of the catenated XhoI-resistant minicircles
To address the possibility that the ladder of slower migrating minicircles seen on alkaline gels might result from head-to-tail catenated molecules rather than from chains of topologically interlocked unit-size minicircles we have analyzed a kDNA sample on a neutral gel after treatment with XhoI or XhoI followed by a type II DNA topoisomerase. The disappearance of the catenated molecules upon XhoI and topoisomerase treatment as shown in Figure 5 indicates that these molecules represent catenated, XhoI-resistant minicircles each of unit size. The basis for the resistance of this class of minicircles to cleavage by XhoI was investigated by PCR amplification and cloning of a DNA sequence spanning the region of the XhoI site in gel-isolated catenated molecules. DNA sequence analysis of four independent clones shows that these minicircles contained a T to C substitution in the XhoI site as well as two other single nucleotide substitutions, a single nucleotide deletion adjacent to the mutant XhoI site and a 10 nt insertion elsewhere (Figure 6).
Figure 5.
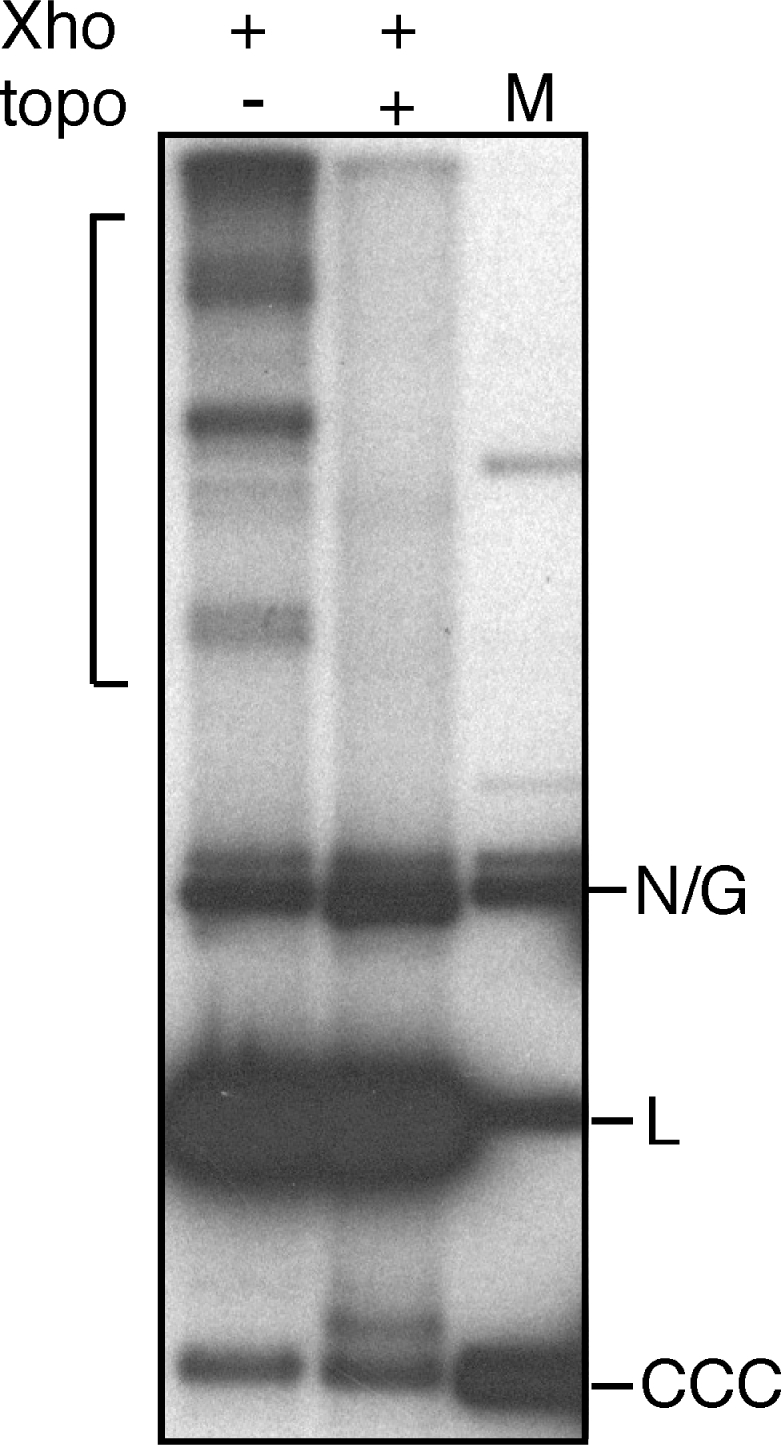
Topoisomerase decatenation of XhoI-resistant catenated minicircle molecules. Purified kDNA prepared at 300 min was digested with XhoI and one-half was additionally treated with a type II DNA topoisomerase prior to neutral gel electrophoresis and Southern blotting using a minicircle probe. Catenated minicircles are indicated by a bracket. N/G, nicked or gapped minicircle DNA; L, unit-length linear minicircle DNA; CCC, covalently closed circular minicircle DNA.
Figure 6.
Sequence alignment of PCR amplified sequences from the region surrounding the XhoI site in catenated XhoI-resistant minicircles. Cf-mo, major minicircle sequence class; 1–5, 1–11, 2–13 and 2–16 sequences come from independent clones of PCR amplification of isolated dimers and trimers of XhoI-resistant minicircles. The XhoI site in the major sequence class is indicated by an overline.
DISCUSSION
Many of the details of the replication mechanism of kinetoplast minicircles are now known, yet much less is known about the regulation of network growth and division. A key feature of the regulation of network division is the lack of closure of discontinuities at the minicircle replication origins until the time of network division (18,19). The studies presented here imply that gaps between Okazaki fragments, except within the replication origins, are largely fully repaired soon after attachment of nascent minicircles to the kDNA network. The remaining discontinuities at the replication origins in the replicated minicircles were found to be simple nicks, based on their susceptibility to closure by E. coli DNA ligase, that remain unrepaired throughout S phase.
The fragment pattern generated by cleavage of network-associated minicircles isolated during kinetoplast S phase indicate that the L strand initiates from either oriA or oriB resulting in a newly synthesized L strand of unit length on a circular H-strand template. These results confirm our earlier conclusion that both origins are utilized in replication of the C. fasciculata minicircles (23).
Replication of the H strand of the minicircle is discontinuous and initiates within the CSB-1 sequence of oriA and oriB. Although discontinuities between H-strand Okazaki fragments must be largely repaired shortly after reattachment of the nascent minicircles to the network, discontinuities remain in the newly synthesized H strands at the two origins. This result suggests that the 5′ terminus of the first Okazaki fragment is somehow different from that of subsequent Okazaki fragments since the Okazaki fragments become covalently closed whereas the 5′ terminus of the first H-strand fragment remains unligated. The abundant existence of minicircles with H-strand discontinuities at both oriA and oriB indicates that a second site-specific initiation of the discontinuously synthesized H strand must also occur at the second origin halfway around the circular DNA and likewise the 5′ terminus is prevented from being ligated. The reason for the lack of covalent closure of the nicks in the H-strand discontinuities at the origins also cannot be due to inappropriate termini for ligation since these nicks, like those in the L strands, can be repaired by treatment of isolated kDNA networks with E. coli DNA ligase. Additionally, the remaining Okazaki fragments are ligated by E. coli DNA ligase as well, indicating that they lack RNA primers and gaps between fragments.
We suggest that it is more likely that a bound protein, possibly the UMS-binding protein UMSBP, may prevent closure of the discontinuities at the replication origins. UMSBP is a good candidate for such a role since it binds specifically to CSB-3 and to the complement of CSB-1 minicircle sequences in their single-stranded forms and also binds to native minicircle origin fragments in vitro (40). Binding at the sites of the specific nicks could prevent access to the nicks by DNA ligases. In addition, binding of UMSBP to an oriA fragment was found to require both CSB-1 and CSB-3 sequences suggesting a cooperative binding of UMSBP at the two sequences within each replication origin. It is particularly of interest to note that UMSBP binding to origin sequences involves the binding of monomers through the C-terminal region of the protein and that the binding is sensitive to the redox potential (41). Oxidation of UMSBP leads to dimerization and a concomitant inhibition of binding to DNA. Thus, if UMSBP is bound to the minicircle replication origins and thereby preventing closure of the nicks in the replication origins, oxidation of UMSBP might result in the release of UMSBP allowing the covalent closure of the nicks and permitting progression to network division. Networks with accessible nicks could serve to recruit a DNA ligase to the nicks for their repair. Such a model is consistent with the observation in C. fasciculata that DNA LIG kα is observed to be associated with the kDNA only at the end of S phase when cells have begun to undergo cell division.
Finally, the observation of XhoI-resistant catenated minicircles has implications for the organization of kDNA networks. Since the XhoI-resistant minicircles are quite rare, it is remarkable that many of these molecules are catenated to one another. This observation suggests that minicircles of this minor sequence class are not distributed randomly throughout the kDNA network but instead are clustered together within the network. Asymmetric distribution of newly replicated daughter minicircles to the antipodal attachment sites might possibly contribute to such a distribution.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health grant GM53254. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health grant GM53254.
Conflict of interest statement. None declared.
REFERENCES
- 1.Simpson AG, Lukes J, Roger AJ. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 2002;19:2071–2083. doi: 10.1093/oxfordjournals.molbev.a004032. [DOI] [PubMed] [Google Scholar]
- 2.Lukes J, Guilbride DL, Votypka J, Zikova A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shlomai J. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 6.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluenz E, Shaw MK, Gull K. Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol. Microbiol. 2007;64:1529–1539. doi: 10.1111/j.1365-2958.2007.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuart K, Feagin JE. Mitochondrial DNA of kinetoplastids. Int. Rev. Cytol. 1992;141:65–88. doi: 10.1016/s0074-7696(08)62063-x. [DOI] [PubMed] [Google Scholar]
- 9.Simpson L, Neckelmann N, de la Cruz VF, Simpson AM, Feagin JE, Jasmer DP, Stuart JE. Comparison of the maxicircle (mitochondrial) genomes of Leishmania tarentolae and Trypanosoma brucei at the level of nucleotide sequence. J. Biol. Chem. 1987;262:6182–6196. [PubMed] [Google Scholar]
- 10.Stuart K, Feagin JE, Abraham JM. RNA editing: the creation of nucleotide sequences in mRNA—a minireview. Gene. 1989;82:155–160. doi: 10.1016/0378-1119(89)90040-1. [DOI] [PubMed] [Google Scholar]
- 11.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madison-Antenucci S, Grams J, Hajduk SL. Editing machines: the complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter LR, Englund PT. Kinetoplast maxicircle DNA replication in Crithidia fasciculata and Trypanosoma brucei. Mol. Cell. Biol. 1995;15:6794–6803. doi: 10.1128/mcb.15.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Englund PT. Free minicircles of kinetoplast DNA networks in Crithidia fasciculata. J. Biol. Chem. 1979;254:4895–4900. [PubMed] [Google Scholar]
- 15.Drew ME, Englund PT. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 2001;153:735–744. doi: 10.1083/jcb.153.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melendy T, Sheline C, Ray DS. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell. 1988;55:1083–1088. doi: 10.1016/0092-8674(88)90252-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Englund PT. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Morga D, Englund PT. The structure of replicating kinetoplast DNA networks. J. Cell Biol. 1993;123:1069–1079. doi: 10.1083/jcb.123.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilbride D, Englund P. The replication mechanism of kinetoplast DNA networks in several trypanosomatid species. J. Cell Sci. 1998;111:675–679. doi: 10.1242/jcs.111.6.675. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Englund PT. The rotational dynamics of kinetoplast DNA replication. Mol. Microbiol. 2007;64:676–690. doi: 10.1111/j.1365-2958.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 21.Ray DS. Conserved sequence blocks in kinetoplast minicircles from diverse species of trypanosomes. Mol. Cell. Biol. 1989;9:1365–1367. doi: 10.1128/mcb.9.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birkenmeyer L, Ray DS. Replication of kinetoplast DNA in isolated kinetoplasts from Crithidia fasciculata. J. Biol. Chem. 1986;261:2362–2368. [PubMed] [Google Scholar]
- 23.Birkenmeyer L, Sugisaki H, Ray DS. Structural characterization of site-specific discontinuities associated with replication origins of minicircle DNA from Crithidia fasciculata. J. Biol. Chem. 1987;262:2384–2392. [PubMed] [Google Scholar]
- 24.Kitchin PA, Klein VA, Fein BI, Englund PT. Gapped minicircles. A novel replication intermediate of kinetoplast DNA. J. Biol. Chem. 1984;259:15532–15539. [PubMed] [Google Scholar]
- 25.Ntambi JM, Englund PT. A gap at a unique location in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 1985;260:5574–5579. [PubMed] [Google Scholar]
- 26.Sheline C, Melendy T, Ray DS. Early intermediates in the replication of DNA minicircles in isolated kinetoplasts from Crithidia fasciculata. Mol. Cell. Biol. 1988;9:169–176. doi: 10.1128/mcb.9.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ntambi JM, Shapiro TA, Ryan KA, Englund PT. Ribonucleotides associated with a gap in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 1986;261:11890–11895. [PubMed] [Google Scholar]
- 28.Perez-Morga DL, Englund PT. The attachment of minicircles to kinetoplast DNA networks during replication. Cell. 1993;74:703–711. doi: 10.1016/0092-8674(93)90517-t. [DOI] [PubMed] [Google Scholar]
- 29.Klingbeil MM, Motyka SA, Englund PT. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell. 2002;10:175–186. doi: 10.1016/s1097-2765(02)00571-3. [DOI] [PubMed] [Google Scholar]
- 30.Sinha KM, Hines JC, Ray DS. Cell cycle-dependent localization and properties of a second mitochondrial DNA ligase in Crithidia fasciculata. Eukaryot. Cell. 2006;5:54–61. doi: 10.1128/EC.5.1.54-61.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidder GW, Dutta BN. The growth and nutrition of Crithidia fasciculata. J. Gen. Microbiol. 1958;18:621–638. doi: 10.1099/00221287-18-3-621. [DOI] [PubMed] [Google Scholar]
- 32.Pasion SG, Brown GW, Brown LM, Ray DS. Periodic expression of nuclear and mitochondrial DNA replication genes during the trypanosomatid cell cycle. J. Cell Sci. 1994;107:3515–3520. doi: 10.1242/jcs.107.12.3515. [DOI] [PubMed] [Google Scholar]
- 33.Cannon G, Heinhorst S, Weissbach A. Quantitative molecular hybridization on nylon membranes. Anal. Biochem. 1985;149:229–237. doi: 10.1016/0003-2697(85)90500-7. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro TA, Klein VA, Englund PT. Isolation of kinetoplast DNA. Methods Mol. Biol. 1999;94:61–67. doi: 10.1385/1-59259-259-7:61. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Molina H, Kalume D, Pandey A, Griffith JD, Englund PT. The role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Mol. Cell. Biol. 2006;26:5382–5393. doi: 10.1128/MCB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugisaki H, Ray DS. DNA sequence of Crithidia fasciculata kinetoplast minicircles. Mol. Biochem. Parasitol. 1987;23:253–263. doi: 10.1016/0166-6851(87)90032-6. [DOI] [PubMed] [Google Scholar]
- 37.Birkenmeyer L, Sugisaki H, Ray DS. The majority of minicircle DNA in Crithidia fasciculata strain CF-Cl is of a single class with nearly homogeneous DNA sequence. Nucleic Acids Res. 1985;13:7101–7118. doi: 10.1093/nar/13.19.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fareed GC, Wilt EM, Richardson CC. Enzymatic breakage and joining of deoxyribonucleic acid. 8. Hybrids of ribo- and deoxyribonucleotide homopolymers as substrates for polynucleotide ligase of bacteriophage T4. J. Biol. Chem. 1971;246:925–932. [PubMed] [Google Scholar]
- 39.Nath K, Hurwitz J. Covalent attachment of polyribonucleotides to polydeoxyribonucleotides catalyzed by deoxyribonucleic acid ligase. J. Biol. Chem. 1974;249:3680–3688. [PubMed] [Google Scholar]
- 40.Onn I, Kapeller I, Abu-Elneel K, Shlomai J. Binding of the universal minicircle sequence binding protein at the kinetoplast DNA replication origin. J. Biol. Chem. 2006;281:37468–37476. doi: 10.1074/jbc.M606374200. [DOI] [PubMed] [Google Scholar]
- 41.Onn I, Milman-Shtepel N, Shlomai J. Redox potential regulates binding of universal minicircle sequence binding protein at the kinetoplast DNA replication origin. Eukaryot. Cell. 2004;3:277–287. doi: 10.1128/EC.3.2.277-287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray DS, Hines JC, Sugisaki H, Sheline C. kDNA minicircles of the major sequence class of C. fasciculata contain a single region of bent helix widely separated from the two origins of replication. Nucleic Acids Res. 1986;14:7953–7965. doi: 10.1093/nar/14.20.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]