Abstract
Light-activated antisense oligodeoxynucleotides (asODNs) were developed to control the degradation of target mRNA in living cells by RNase H. A 20-mer asODN previously shown to target c-myb, a hematopoietic transcription factor, was covalently attached via a photocleavable linker (PL) to partially complementary 20-mer sense strands (sODNs). In the ‘caged’ state, the sODN blocked hybridization of the asODN to c-myb mRNA. Six asODN-PL-sODN conjugates, C1-C6, were synthesized. C5, with twelve complementary bases, gave the largest decrease in melting temperature (Tm) upon UV irradiation (ΔTm = −29°C). The most thermally stable conjugate, C6 (Tm = 84°C), gave the lowest background RNase H activity, with just 8.6% degradation of an RNA 40-mer after 1 h incubation. In biochemical assays with C6, RNA digestion increased 10-fold 10 min after UV irradiation. Finally, phosphorothioated analogs S-C5 and S-C6 were synthesized to test activity in cultured K562 (human leukemia) cells. No knockdown of c-myb mRNA or protein was observed with intact S-C5 or S-C6, whereas more than half of c-myb mRNA was degraded 24 h after photoactivation. Two-fold photomodulation of c-MYB protein levels was also observed with S-C5. However, no photomodulation of c-MYB protein levels was observed with S-C6, perhaps due to the greater stability of this duplex.
INTRODUCTION
A current challenge in chemical biology is to control the concentration or activity of gene products (e.g. mRNA, proteins) with high spatial and temporal resolution (1–14). Chemical approaches for regulating gene activity within living cells and organisms offer exciting possibilities for elucidating complex biological processes. Specifically, light-activated or ‘caged’ oligonucleotides provide unique possibilities for controlling gene expression in both space and time (15,16), but their development has been slowed by synthetic and experimental complexities. Taylor and Ordoukhanian (17) in 1995 demonstrated the triggering of DNA/DNA duplex formation with light by incorporating a bridging photoactive o-nitrobenzyl moiety within the phosphate backbone of a DNA hairpin. Near UV irradiation under ambient conditions induced a strand break that revealed a 5′-phosphate 18-mer oligonucleotide with a 9-fold greater affinity for a complementary DNA strand. While there have been many more recent examples of generating single- and double-strand breaks, and mimicking genomic lesions using photoactive oligonucleotides (18–23), there have been relatively few reports of photo-triggered hybridization.
Based on the myriad biological processes that involve DNA/DNA, DNA/RNA or RNA/RNA hybridization, we have sought to develop synthetically facile and high quantum efficiency routes for photomodulating the hybridization of a reverse complementary, or ‘antisense’ oligodeoxynucleotide (asODN) to target DNA or mRNA molecules (24). We developed a strategy for attaching the asODN to a complementary sense ODN (sODN) via a photocleavable linker (PL) (24,25). As the asODN-PL-sODN conjugate, antisense activity is transiently masked, until photoactivation reveals the asODN. Rotaru and Mokhir (26) extended this approach by employing a different, redox-active PL. Our lab showed recently that genes can be photo-regulated in zebrafish embryos by using an antisense negatively charged peptide nucleic acid (ncPNA) conjugated via a PL to a complementary 2′-OMe RNA strand (27). In the current study, we considered strategies for photoregulating gene expression in human cancer cells.
The development of light-activated oligonucleotides for controlling mRNA degradation has become an active area of study (2,20,24,28–31). For example, Matsunaga et al. (28) applied the UV-mediated trans→cis isomerization of azobenzene to control RNA digestion by RNase H. By incorporating five azobenzenes to a 20-mer sense DNA strand, the binding of a complementary asODN decreased upon UV irradiation. Based on the binding differential, ΔTm = −18.2°C, it was possible to regulate the hybridization of the asODN to a complementary RNA target. However, the requirement for five azobenzenes greatly reduced the photo-efficiency of this system.
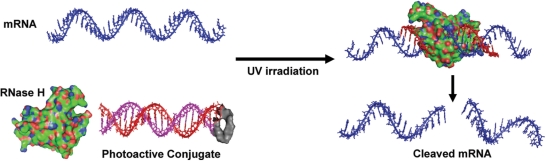
In general, efforts to prevent DNA/DNA or DNA/mRNA hybridization using sterically encumbered, photo-labile blocking groups have proven relatively ineffective (32–34), due to the large thermodynamic driving force, and specific base-pairing interactions involved in duplex formation. Use of multiple, non-specific labels requires greater UV exposure, which generates unintended side products, including cytotoxic radical species. Photoactivation inside biological samples is additionally complicated, due to issues of molecular diffusion, phototoxicity and the absorption of UV light by endogenous chromophores. Thus, we sought to employ a relatively simple strategy involving only a single photocleavable moiety. Previously, the stability of DNA hairpins relative to the corresponding DNA/RNA hybrids was shown to influence the extent of RNA degradation by RNase H (24,35). Based on this observation, we designed several light-activated asODN–PL–sODN conjugates to investigate factors important in controlling this enzymatic process (Scheme 1), both in biochemical and cellular assays.
Scheme 1.
Strategy for regulating mRNA (blue strand) digestion by RNase H (green surface rendering) using a photoactive asODN conjugate (red and magenta strands). Oligonucleotide structures were generated with Insight II (Accelrys, San Diego, USA). Representation of RNase H was generated from the PDB coordinates using PyMol (DeLano, W.L., 2002, on World Wide Web, http://www.pymol.org).
This work represents the first example in which a cancer-related gene was down-regulated using light-activated oligonucleotides. Caged oligos were targeted against the proto-oncogene c-myb (36–39) in human K562 (leukemia) cells. The c-MYB is a transcription factor that regulates cellular differentiation and proliferation and is regulated by complex mechanisms that control its repressed oncogenic activity. Control of such critical functions also suggests a potential role for MYB in leukemic transformation (40–42). MYB has been shown to represent a legitimate therapeutic target in patients with hematologic malignancies, because normal cells are more tolerant of transient MYB deprivation. Reverse complementary ODNs that target a 20-base region within c-myb mRNA, 326←345, have been shown to degrade c-myb by recruiting RNase H (36–39).
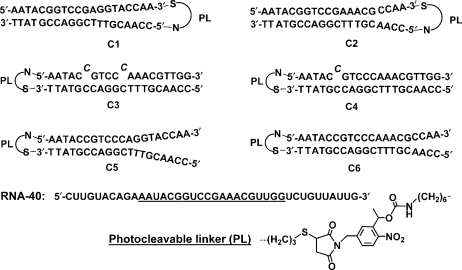
A series of six photoactive DNA conjugates (C1–C6, Figure 1) was synthesized by covalently attaching a 20-mer asODN targeting c-myb residues 326←345 to a complementary sense strand via a 1-(5-(N-maleimidomethyl)-2-nitro-phenyl)ethanol N-hydroxysuccinimide ester. This heterobifunctional PL was designed to react with thiol and amine functionalities on opposite ends (Figure 1). We varied the size and directionality of the hairpin loop, and the number and placement of complementary bases in the stem, in order to stabilize the caged asODN–PL–sODN conjugates relative to the corresponding asODN/sODN duplexes. We defined the increase in RNase H activity as photomodulation efficiency: (RNA digested after photoactivation)/(RNA digested before photoactivation). C5 and C6 showed the greatest photomodulation efficiency in biochemical RNase H assays, and were subsequently synthesized with a more nuclease-resistant phosphorothioate backbone. These conjugates, S-C5 and S-C6, were tested in K562 cells for their ability to photoregulate the digestion of c-myb mRNA by RNase H, as well as c-MYB protein knockdown. These studies point the way towards new cellular gene regulation strategies and photodynamic therapies.
Figure 1.
Sequences of the photoactive asODN-PL-sODN conjugates, C1–C6, with sense (top) strand linked via the photocleavable linker, PL, to the antisense (bottom) strand. Twenty underlined bases in the RNA 40-mer target, RNA-40, correspond to bases 326–345 of c-myb.
MATERIALS AND METHODS
General methods
All single-stranded oligonucleotides were purified with an Agilent 1100 HPLC system using a reverse-phase analytical HPLC column (ZORBAX 300Extended-C18, 4.6 × 250 mm, 5 μm beads). A specialized oligonucleotide column (ZORBAX Oligo, 9.4 × 250 mm, 5 μm beads) was used to purify all photoactive conjugates. Concentrations of oligonucleotide conjugates were measured in water at 260 nm (ε260 = 4.3 × 105 M−1 cm−1) using an Agilent 8453 UV/Vis spectrophotometer. Melting studies were carried out with a Beckman Coulter DU 800 UV/Vis spectrophotometer with the DU 800 Kinetics Package, including a 6-cell holder. Gels of RNA digestion were imaged using an Amersham Biosciences Storm 840 phosphorimager and quantified with ImageQuant software (ImageQuant™ TL v2005, GE Healthcare). All photoirradiation experiments with pure oligonucleotide samples were carried out with a xenon lamp (450 W) and monochromator (20 nm slit centered at 355 nm, 36 mW/cm2 at the sample). K562 cells were typically irradiated for 5 min with a 12” × 12” tabletop Spectroline ultraviolet transilluminator (Spectronics Corporation, TL-365R, 9 mW/cm2 at peak intensity, 365 nm). The duration of UV illumination required for complete photocleavage was determined through separate experiments involving 32P-labeled asDNA-PL-sDNA constructs (25). Standard 1xRNase H reaction buffer was defined as 20 mM Tris–HCl, 20 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM DTT, pH 8.0. The Tm and circular dichroism (CD) measurements used the same RNase H buffer, but the DTT became oxidized (forming disulfide) during several cycles of heating and cooling.
Synthesis of photoactive DNA conjugates, C1–C6
The synthesis of DNA conjugates C1–C6 and the PL was performed following established protocols (25). All DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT; Coralville, IA). The general procedure for synthesizing photoactive DNA conjugates is summarized as follows: Thiol-modified oligonucleotides (10 nmol) were reduced with 0.1 M DTT in 200 µl of 0.1 M phosphate buffer (pH 7.2) for 4 h, followed by removal of excess DTT on a NAP-10 column (pre-packed with Sephadex™ G-25) and purification by RP-HPLC (gradient of 0→30% acetonitrile in 0.05 M TEAA buffer in 30 min at 25°C). Each purified oligonucleotide with a free thiol group was lyophilized.
Desalted oligonucleotides modified with an amino group were purchased from IDT and used without further purification. The amine-modified oligonucleotide (20 nmol) was first dissolved in 200 µl of 0.1 M phosphate buffer (pH 7.2) and warmed to 60°C. The PL (1–2 mg in 100 µl DMSO) was added. The mixture was incubated at 35°C for 40 min, the oligonucleotide was pre-purified with a NAP-10 column and purified by RP-HPLC (gradient of 5→30% acetonitrile in 0.05 M TEAA buffer at 40°C) in about 50% yield based on integrating the area under each peak in the HPLC trace. All sample manipulations were carried out in reduced light or red light, where possible.
The purified sense and antisense oligonucleotides were mixed in 300 µl of 0.1 M phosphate buffer and 0.3 M sodium chloride (pH 7.2) at room temperature (rt) for 4 h. The conjugate was pre-purified with a NAP-10 column and dual-purified by HPLC using the ZORBAX Oligo column: buffer A, 0.02 M phosphate buffer, 5 M urea, 15% acetonitrile, pH 7; buffer B, A + 2 M NaCl; linear gradient of 5→45% B in 40 min at 40°C, 2 ml/min. HPLC traces showing purification of C1–C6 and gels of purified, 32P-labeled conjugates are provided in the Supplementary Material. The HPLC-purified product was collected and dialyzed with molecular porous membrane tubing (Spectrum, 6000–8000 MW) for 3 days at rt, then lyophilized. The concentration of photoactive conjugate was determined by heating each oligonucleotide solution in water (molecular biology grade, Eppendorf) to 80°C and measuring the UV absorbance at 260 nm. MALDI mass spectrometric data for C1–C6 are provided in Table 1.
Table 1.
Mass characterization, melting temperatures, and photomodulation efficiencies for conjugates C1–C6, showing decrease in thermal stability and increase in RNase H activity upon photoirradiation
| Photoactive conjugate | Mass (calc) | Mass (expt) | Tma (°C) | Tma (°C) (cleaved) | ΔTm (°C) | Photomodulation efficiencyb |
|---|---|---|---|---|---|---|
| C1 | 12838.2 | 12862.5 | 74 | 51 | −23 | 1.5 |
| C2 | 12807.1 | 12803.4 | 81 | 63 | −18 | 3.5 |
| C3 | 12829.4 | 12858.6 | 68 | 47 | −21 | 1.2 |
| C4 | 12789.4 | 12770.0 | 79 | 62 | −17 | 3.4 |
| C5 | 12838.4 | 12838.1 | 80 | 51 | −29 | 4.4 |
| C6 | 12807.4 | 12819.2 | 84 | 63 | –21 | 6.0 |
aMelting temperatures were determined from the peak of the first derivative plot of Abs260 versus temperature in standard RNase H buffer.
bPhotomodulation efficiency is the n-fold increase in RNA-40 cleavage elicited by photoactivation, observed in 1 h RNase H assay with 1 U enzyme.
Synthesis of S-C5 and S-C6
Synthesis of phosphorothioated DNA conjugates S-C5 and S-C6 was performed as described for C1–C6, with minor modifications (25). The amine-terminated sense strands and thiol-terminated antisense strands, comprised of DNA phosphorothioate oligonucleotides, were synthesized at the UPenn Nucleic Acid Facility and purified by RP–HPLC with a linear gradient of 5→30% acetonitrile in 0.05 M TEAA in 30 min at 40°C. HPLC traces showing purification of conjugates S-C5 and S-C6 are provided in the Supplementary Material.
Thermal denaturation and CD studies
Thermal denaturation studies were performed on conjugates (1 µM) and duplexes (1 µM) of sense and antisense strands in standard RNase H buffer. The solution was heated to 90°C for 5 min, and allowed to cool gradually to 25°C. Samples were monitored at 260 nm, while heating or cooling at a rate of 0.5°C/min. Tms were determined from the peak of the first derivative plot of Abs260 versus temperature.
CD spectra were collected on an Aviv 62DS spectropolarimeter in the wavelength range of 200–320 nm at 25°C using a 1 mm quartz cell. Oligonucleotide solutions (conjugate or corresponding 1:1 asODN/sODN duplex) were 30 μM in 1×RNase H buffer.
RNase H assays
The RNA 40-mer target sequence (RNA-40, Figure 1) in HPLC-pure form, was purchased from IDT. RNase H and reaction buffer were purchased from EPICENTRE Biotechnologies (Madison, WI, USA). The standard procedure for the RNase H assay was as follows: the conjugate was annealed in 1× RNase H buffer by heating to 95°C, slowly cooling to 70°C, remaining at 70°C for 10 min to melt non-specific DNA structures, and continuing with slow cooling to 37°C. [γ-32P]-labeled RNA oligonucleotide (10- to 200-fold excess) was added and incubated at 37°C for 20 min to allow RNA/DNA duplex formation. RNase H (0.2–3 U) was added to the mixture and incubated at 37°C. Total reaction volume was 10 µl and final concentrations of DNA (conjugate or asODN), and RNA-40 were 0.01 µM and 2 µM, respectively.
To measure RNA degradation after photoactivation, the DNA conjugates were UV-illuminated by Xe lamp, then [γ-32P]-labeled RNA oligonucleotide was added and incubated at 37°C for 20 min to allow RNA/DNA duplex formation. RNase H assays were performed as described earlier.
For studies examining the effect of the RNA/conjugate ratio on enzyme efficiency, the concentrations were 0.01 µM for DNA conjugate and 0.1 µM (10×), 0.5 µM (50×) and 2.0 µM (200×) for RNA-40. Time points were taken at 10 and 60 min by sampling 4 µl of the reaction mixture or at 10, 30 and 60 min by sampling 3 µl of the reaction mixture, adding 6 or 7 µl loading buffer (50 mM EDTA, 90% formamide, total volume = 10 μl), and heating to 95°C for 3 min to terminate the reaction.
All of the resulting solutions were subject to electrophoresis on a 20% polyacrylamide gel containing 7 M urea. Intensity values were integrated in ImageQuant for each band with automated lane and band finding using a local method background correction in the gel lane. The relative amount of RNA digestion was determined by dividing the intensity of the band corresponding to cleaved RNA by the total intensity of the cleaved and uncleaved RNA bands.
Cell culture and transfection of oligonucleotides
K562 cells (American Type Culture Collection, Manassas, VA, USA, catalog no. CCL-243™) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 0.5% penicillin/streptomycin. The cells were maintained at 37°C in a humidified incubator (95% humidity) and 5% CO2. Culture media was changed every 2 days or according to the rapidity of cell growth.
Cells were transfected with ODNs using an Amaxa Nucleofector Kit (Amaxa, Inc., Gaithersburg, MD, USA) according to manufacturer's instructions. Typically, 3 × 106 cells were electroporated with 5 µg of ODN for each condition. The cells were first washed with phosphate-buffered saline (PBS) and then resuspended in 100 µl of nucleofection solution. The ODN was added to the cell suspension which was then nucleofected using Program Q-29. Immediately after, the cells were transferred to 5 ml of culture medium and placed in the incubator. Three hours after nucleofection, one batch of cells was exposed to UV irradiation for 5 min by placing the 6-well plates (TPP 92406, Sigma) containing the cells on top of the illuminator. The plastic plates transmitted 70% of UV light at 360 nm, as determined by UV/Vis spectrometer. The cells in each well were then washed in PBS, and resuspended in 3 ml of culture media for 24 h.
Quantitative real-time PCR (QRT–PCR)
QRT–PCR was carried out as described by Livak and Schmittgen (43). In summary, total RNA from cell pellets was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Reverse transcription was carried out with 1 μg RNA aliquots using iScript reverse transcription kit (Bio-Rad Laboratories, Hercules, CA). One microliter of cDNA was used as template for the QRT–PCR reaction. Taqman Universal PCR master mix (Applied Biosystems, Foster City, CA, USA) was used for the PCR reaction and detection using iCycler iQ Real-Time PCR Detection System (BioRad). The assay was performed as in Kalota et al. (44) For amplifying c-myb product, the following primers were used: forward, dGAAGGTCGAACAGGAAGGTTATCT; and reverse, dGTAACGCTACAGGGTATGGAACA. The c-myb probe was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and Black Hole quencher (BHQ) at the 3′ end. The probe sequence was 5′-TCAAAAGCCAGCCAGCCAGCAGTG.
GAPDH, a common housekeeping gene, was employed as a reference for QRT–PCR, and amplified with the following primers: forward, 5′-GACAGTCAGCCGCATCTTCTT; and reverse, 5′-CCAATACGACCAAATCCGTTGAC. The GAPDH probe was labeled with FAM at 5′ end and BHQ at 3′ end. The probe sequence was 5′-CGTCGCCAGCCGAGCCACATCG.
All reactions were performed in triplicate with 1 µl cDNA. The volume of reaction mixture was 15 µl. The reaction mixture was pre-incubated at 50°C for 2 min. PCR cycling conditions were as follows: denaturation at 95°C for 10 min, followed by 39 cycles of 92°C for 15 s and 60°C for 45 s. Analysis of QRT–PCR data was based on comparison of the target transcript PCR signal in a treatment group to signal measured in an untreated control. Analysis was done using the 2−ΔΔCT method as described by Livak and Schmittgen (43).
Protein isolation and western blotting
Twenty-four hours after nucleofection, cells were harvested, washed in PBS and pelleted. The cell pellets were lysed using 50 µl Triple-Lysis Buffer (50 mM Tris, 150 mM NaCl, 0.02% sodium azide, 0.1% SDS and 1% Igepal; Sigma, St Louis, MO, USA) by incubating on ice for 30 min with vortexing every 10 min. The cell lysates were spun at maximum speed in a microfuge at 4°C for 20 min. The supernatant thus extracted was used for western blotting.
Protein concentration in each sample was measured using a Bradford protein assay (Bio-Rad). Samples (100 µg) were loaded onto a 10% polyacrylamide gel (100 V, 90 min), then transferred to a PVDF membrane (20 V, 60 min) using a semi-dry blotting system (Bio-Rad). A 5% solution of non-fat dry milk was used as a blocking reagent. The membrane was incubated overnight at 4°C with primary antibody against c-MYB (clone 1-1 from Upstate, Lake Placid, NY) at 1:1000 dilution. The membrane was washed with TBS-T three times and probed with HRP-conjugated anti-mouse secondary antibody (GE Healthcare, UK) at 1:1000 dilution at rt for 1 h. Blots were developed using enhanced chemiluminescence ECL+ western blotting detection kit (GE Healthcare, UK). Membranes were then stripped using Re-Blot stripping solution (Chemicon, Temecula, CA, USA) for 10 min at rt. After blocking, the membrane was incubated with anti-GAPDH antibody (Cell Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C at 1:1000 dilution, washed in PBS and then incubated with HRP-conjugated anti-rabbit secondary antibody (GE Healthcare, UK). Chemiluminescence was detected using the ECL kit (GE Healthcare, UK). Quantification of the bands was carried out using ImageQuant.
RESULTS AND DISCUSSION
Design and optimization of photoactive hairpins
Our previous work with C5 (24) identified two routes for improving the photomodulation efficiency: (i) make the conjugate more kinetically or thermodynamically resistant to binding to the target mRNA, thereby lowering the RNase H background activity; (ii) destabilize the photoactivated asODN/sODN duplex, thereby promoting asODN/mRNA hybridization, and increasing the percentage of RNA-40 degradation by RNase H.
In order to lower background RNase H activity, we sought to modify the eight unpaired bases in the stem of C5. The accessibility of these bases could have the undesired effect of allowing hybridization of asODN–PL–sODN to RNA-40. C1 and C2 were designed to reorient the loop and hybridize the ends of the hairpin stem, which we reasoned would lower background RNase H activity. C1 had the same 12 complementary bases as C5, but moving the PL increased the size of the loop by 16 nucleotides. C2 had four additional complementary bases, which created a loop with eight nucleotides. Photoactive oligos C3 and C4 were designed with two and one mismatches in the sense strand, respectively. The mismatches were chosen based on calculations on the HyTher™ server (45), which helped to identify sites that would particularly destabilize the asODN/sODN duplex. In this way, we hoped to increase the thermal stability of C3 and C4 relative to their photoactivated duplexes, thereby increasing ΔTm(conjugate-duplex). In addition, C4 was expected to be particularly stable and show low RNase H background activity, based on its 19 bp. The significant complementarity in C3 and C4 between the antisense and sense strands eliminated overhangs that might hybridize with RNA-40. Finally, C6 was designed to be more stable than C5, with the addition of four complementary base pairs in the stem. We note that C6 is a cognate of C2, with the same 16 complementary bases, but in the reverse orientation relative to the loop. Compared to C5, the conjugate C6 had only four non-complementary bases and was expected to associate less strongly with RNA-40, and therefore decrease the background RNase H activity.
To test the thermal stability of these conjugates relative to the corresponding asODN/sODN duplexes, UV absorbance melting transitions were measured in standard RNase H buffer. As shown in Table 1, with an increase in the loop from 8 bases (C2) to 16 bases (C1), the Tm of the conjugate decreased 7°C. Two base mismatches destabilized C3 by 10°C relative to C4, with only one mismatch. Interestingly, C6 had the highest Tm (Tm = 84°C), which was 3°C higher than C2 with identical matching base pairs. This provides additional evidence that a small loop formed by the PL in joining asODN and sODN strands is most effective at stabilizing the conjugate. Upon irradiation, all duplexes became less stable, with Tms lowered by 17–29°C.
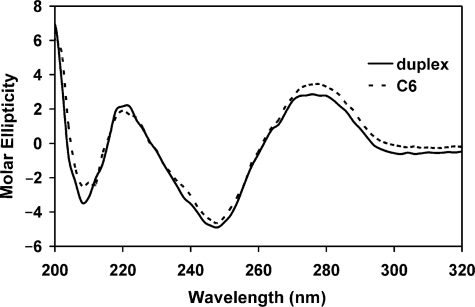
CD spectroscopy provided a convenient method for assessing the conformational structure of the oligonucleotides. Figure 2 shows the CD signal for C6, which was representative of the conjugates. The conjugates’ optical signature agreed very well with the oligonucleotide duplexes of the same sequence. This indicated that the PL had little effect on the fully base-paired helical structure.
Figure 2.
Molar ellipticity ([θ]× 10−5deg·cm2·dmol−1) of C6 and its corresponding duplex (30 μM in 1×RNase H buffer) measured by CD spectroscopy.
Photomodulation of RNA digestion by RNase H with photoactive conjugates
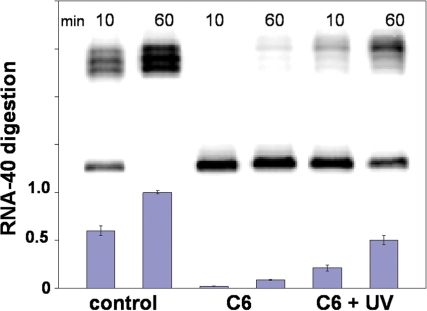
RNase H activity was studied with RNA oligonucleotide, RNA-40 (40-mer), as shown in Figure 1. Digestion of the RNA target was compared between solutions containing the control single-stranded asODN (CCAACGTTTCGGACCGTATT) or a photoactive asODN–PL–sODN conjugate. Figure 3 shows the typical gel from an RNase H assay experiment with 2 µM [γ-32P]-labeled RNA-40, 0.01 µM irradiated C6, and 1 U RNase H in standard RNase H buffer at 37°C. The first two lanes are the cleavage pattern with control single-stranded asODN at 10 and 60 min, followed by C6 at 10 and 60 min. The last two lanes show the cleavage pattern with C6 at 10 and 60 min after irradiation. The bar graph below the gel in Figure 3 shows the relative amount of RNA digestion observed under each condition, where a value of 1 equals complete digestion. Using C6 in an 1-h experiment, 50% of the total RNA-40 target was digested after irradiation, as compared to a background level of 8.6% without irradiation and ∼100% RNA degradation for the control single-stranded asODN. This corresponded to a roughly 6-fold increase using 1 U of enzyme, 1 h after photoactivation. Photomodulation efficiency was even higher early in the reaction, with 10-fold higher RNase H activity (2→21%) observed at 10 min. The UV light used in these experiments had no effect on the enzyme or RNA stability. Control experiments confirmed that RNase H hydrolyzed RNA-40 only when hybridized with asODN.
Figure 3.
Denaturing PAGE (20%) analysis of RNA-40 digestion with 2 µM [γ-32P]-labeled RNA-40, 0.01 µM photo-activated C6 and 1 U RNase H in RNase H buffer at 37°C. Lanes 1 and 2, control asODN at 10 and 60 min; Lanes 3 and 4, C6 at 10 and 60 min; Lanes 5 and 6, C6 at 10 and 60 min that had been activated with UV light (355 nm, 36 mW/cm2) for 10 min. Each blue bar is the average of three separate trials.
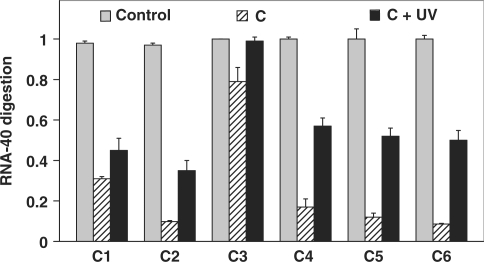
The ability of each conjugate to photomodulate RNA-40 digestion was tested under the conditions of 2 µM [γ-32P]-labeled RNA-40, 0.01 µM conjugate and 1 U RNase H at 37°C. As shown in Figure 4, loop size had a large effect on RNA-40 digestion by RNase H before irradiation: 31% for C1 compared to 9.8% for C2, indicating roughly 3-fold less digestion with the smaller loop. And upon irradiation, the photomodulation efficiency in an 1-h experiment was 1.5 for C1 and 3.5 for C2 with high background level for C1 before irradiation. This difference is presumed to be due to the difference in thermal stability between these two conjugates: ΔTm C2–C1 = 7°C. For C3, just 1.2-fold photomodulation was observed, with much higher background of 80% for C3 before irradiation. This result is explained by the C3's Tm, Tm = 68°C, while the Tm for the target duplex (asODN/RNA-40) was 71°C. Thus, the equilibrium disfavored the conjugate relative to the asODN/RNA-40 duplex. With only one mismatch in C4, the thermal stability (Tm = 79°C) was greatly improved. When compared to the even more thermally stable conjugates C2, C5 and C6, greater enzyme background activity was observed for C4, which led to a lower photomodulation efficiency of 3.4. In these biochemical assays, background activity was strongly correlated with conjugate Tm. Among the six photoactive conjugates, the most thermally stable was C6, Tm = 84°C (Table 1). Largely based on this stability, C6 gave the best photomodulation efficiency, with a 6-fold increase in RNA digestion in the 1-h enzymatic assay. Results with C6 clearly demonstrated that stabilizing the conjugate gave lower levels of background RNA digestion by RNase H.
Figure 4.
RNase H assay showing the digestion of RNA-40 (2 µM) in 60 min at 37°C with C1–C6 (0.01 µM) and 1 U RNase H. Each control column shows the hydrolysis of RNA-40 with control asODN. UV light (355 nm, 36 mW/cm2) was used to irradiate the samples. Error bars signify standard deviation from three separate trials.
RNase H concentration dependence on RNA digestion photomodulation efficiency
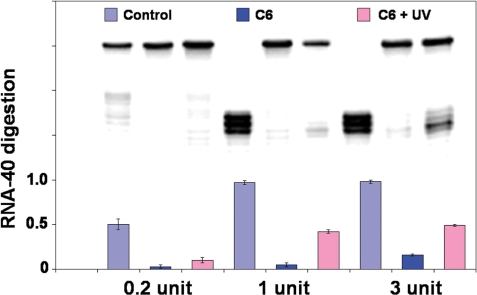
The effect of the amount of RNase H in solution on RNA degradation was also tested using C6 (2 µM [γ-32P]-labeled RNA-40, 0.01 µM C6 at 37°C). The gel trace in Figure 5 showed the pattern of RNA-40 digestion using photoactive conjugate C6 with different amounts of RNase H in a 30 min assay at 37°C. The amount of enzyme had little effect on the control reaction, but had a great influence on the reactions with C6. The gels were quantified using ImageQuant, with the results of RNA digestion shown in the bar graph for each lane (Figure 5). In 30 min, virtually all RNA-40 in the control experiment with the single-stranded asODN was digested using 1 U and 3 U RNase H, while for the conjugate C6, there was roughly 3-fold more RNA-40 digestion using 3 U of enzyme (16%) than 1 U of enzyme (5%). After irradiation of C6, relative amounts of RNA-40 digestion were similar, 42% with 1 U RNase H and 49% for 3 U RNase H. The thermal stability data cannot predict exactly the differences in RNA digestion, where only the amount of enzyme is varied. However, these studies are relevant to biological experiments, as the amount of RNase H can vary considerably between different cell types, tissues and species.
Figure 5.
Denaturing PAGE (20%) analysis of RNA-40 digestion in 30 min by 0.2, 1 or 3 U of RNase H with 2 µM [γ-32P]-labeled RNA-40, 0.01 µM C6 in RNase H buffer at 37°C. Lanes 1–3, control asODN, C6 and cleaved C6 with 0.2 U RNase H; Lanes 4–6, control asODN, C6 and cleaved C6 with 1 U RNase H; Lanes 7–9, control asODN, C6 and cleaved C6 with 3 U RNase H. C6 was photocleaved with UV light (355 nm, 36 mW/cm2) for 10 min. The columns show the ratio of degraded RNA to total RNA, data from two or three independent trials.
RNA cleavage by RNase H is a three-component system requiring the presence of target RNA, the asODN, and RNase H. However, experiments have shown that the process of RNA degradation is more complicated than simple stepwise formation of asODN/RNA duplex, followed by RNase H cleavage (46). One important difference is that RNase H has been shown to be an active participant in DNA/RNA duplex formation (47). Detailed thermodynamic measurements and gel mobility shift assays showed no hybrid duplex formation with some structured asODNs, especially with hairpin structures, while target RNA could still be cleaved by RNase H. This suggested that RNase H can promote the formation and cleavage of DNA/RNA duplexes, even under conditions where RNA/DNA hybridization is thermodynamically disfavored (46). These previous studies help to explain why RNase H activity persisted in the biochemical assays even for conjugates with Tms over 80°C.
Effect of initial RNA/DNA ratio on photomodulation efficiency
Different initial ratios of RNA/DNA were studied for photomodulation of RNA-40 digestion with a fixed concentration of C6 (0.01 µM) and 1 U RNase H in RNAse H buffer at 37°C. When the ratio of RNA-40/C6 was 200, 50 and 10, RNA-40 cleavage was 8.6%, 12% and 13% in 1-h experiments. With irradiated samples, the cleavage was 50%, 36% and 39% with an increase of about 6-fold, 3-fold and 3-fold, respectively. DNA conjugate C2 gave similar results, with a 3.5-fold and 2.9-fold increase in RNA-40 digestion after irradiation, with RNA-40/C2 ratios of 200 and 10, respectively. Under the same conditions, in this range of enzyme concentrations, RNA-40 was completely degraded by RNase H in the presence of the control asODN. These data indicated higher photomodulation efficiencies at the largest RNA/DNA ratios. Thus, light-activated oligodeoxynucleotides are likely to be most effective in cells where both the concentration of RNase H and the effective (accessible) concentration of target RNA are high.
Cell studies
To determine the effects of the photoactivatable conjuages in living cells, conjugates S-C5 and S-C6 were both targeted to the c-myb proto-oncogene mRNA (GenBank accession number: M15024) by nucleofection into K562 cells (see Materials and Methods section). K562 cell viability was determined post-nucleofection and after 5 or 10 min UV light exposure. UV irradiation itself did not appear to be toxic. However, nucleofection of conjugates followed by a 10 min UV exposure was not tolerated by the cells (>75% cell death calculated using Trypan Blue exclusion). Therefore, a less harsh treatment regimen was developed. Cells were allowed to recover in the incubator for 3 h after nucleofection, after which the cells were UV irradiated for 5 min. This strategy appeared well tolerated (<50% cell death) and was subsequently adopted for the experiments reported below.
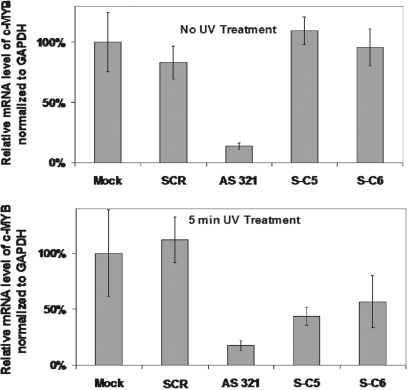
Figure 6 shows that among the two controls and three experiments, only AS 321, the 20-mer fully phosphorothioated asODN, 5′- CCAACGTTTCGGACCGTATT, targeting the same c-myb 326←345 sequence, was effective in decreasing mRNA levels. Relative to the Mock (buffer-nucleofected) control, 86% of c-myb mRNA target was eliminated 24 h after nucleofection with AS 321. Fully phosphorothioated, scrambled ODN (SCR, GAATGTGACATTTCGACACG) showed no biological activity, which confirmed sequence specificity in the interaction with target c-myb mRNA. Pre-irradiation, S-C5 and S-C6 showed the same level of c-myb mRNA as the Mock control, which indicated no discernible background activity for these caged constructs. As a separate control, in the Mock nucleofected cells that were UV-irradiated for 5 min, no statistically significant differences in c-myb mRNA levels were observed. This indicated that the UV light alone was not responsible for down-regulating c-myb. And, cells nucleofected with AS 321 showed identical mRNA ‘knockdown’, pre- and post-UV irradiation, which indicated that there were no particular synergistic effects between the antisense molecule and UV light. However, c-myb mRNA levels were substantially reduced in cells that had been nucleofected with S-C5 (56% knockdown) or S-C6 (43% knockdown), and subsequently UV-irradiated for 5 min. It was expected that the level of knockdown would be less for the photoactivated conjugates than AS 321, as the sODN remained within the cell and presumably competed with target mRNA for hybridization to the asODN. In zebrafish embryos, we found that 5-fold more caged ncPNA than native ncPNA was required to achieve similar gene knockdown after photoactivation (27).
Figure 6.
Quantitative real-time PCR (QRT–PCR) assay performed on RNA isolated from nucleofected K562 cells. Data presented are a ratio of the number of copies of c-myb mRNA relative to GAPDH mRNA, which corrects for variations in cell number and viability, and indicates c-myb-specific targeting. Mock cells were nucleofected with buffer in the absence of DNA, and other data were normalized to the Mock data, for which the ratio of gel intensities (c-myb/GAPDH) was set to 100%. mRNA analysis was done 24 h post-nucleofection. Error bars show standard deviation from the average of data from at least three independent trials.
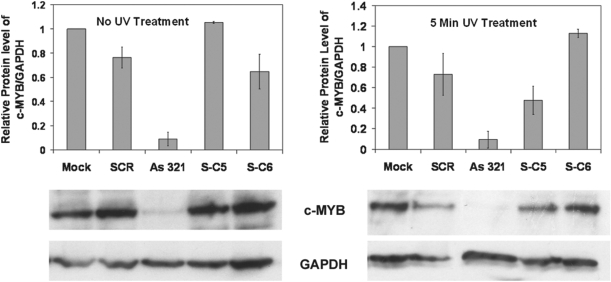
Figure 7 shows data on cellular c-MYB protein levels. Western blots provided more variable data than RT–PCR, but confirmed many of the trends seen for mRNA levels. As was done similarly for Figure 6, all c-MYB levels were corrected for variations in cell number and viability by taking a ratio of the c-MYB and GAPDH intensities. A decrease in c-MYB relative to GAPDH indicated gene-specific targeting effects. Most c-MYB protein knockdown was observed for AS 321, but S-C5 also showed substantial ability to photoregulate c-MYB levels within nucleofected K562 cells. The c-MYB protein was reduced by 52% (compare lanes for S-C5, pre- and post-UV treatment). In contrast, western blots for cells nucleofected with S-C6 indicated no obvious c-MYB protein knockdown when UV light was applied. This somewhat surprising result is discussed in greater detail below.
Figure 7.
c-MYB protein analysis by western blot performed on nucleofected K562 cells, either in the dark or treated with UV light for 5 min. Graph shows relative protein levels of c-MYB to GAPDH as quantified from western blots using ImageQuant. Mock cells were nucleofected with buffer in the absence of DNA, and other data were normalized to the Mock data, for which the ratio of gel intensities (c-MYB/GAPDH) was set to 1. Protein analysis was done 24 h post-nucleofection, with data from two independent trials. Error bars show deviation from the average.
The thermal stability studies improved our understanding of RNA/DNA duplex formation and RNA hydrolysis in biochemical assays and could also be applied to understanding RNA cleavage within the cell. It seems reasonable that less stable asODN/sODN duplexes (i.e. uncaged S-C5), which produced higher background levels of RNase H activity in biochemical assays, would perform better in complex biological systems. Although it is difficult to quantify precisely the concentration of accessible c-myb mRNA within the cell, the effective mRNA concentration was likely to be much lower for most constructs and target sequences. We hypothesize that having a shorter 12-mer sODN (as in S-C5) was advantageous within the cell, where the asODN in a less stable asODN/sODN duplex could compete more effectively for a highly structured and relatively inaccessible mRNA target. Supporting this hypothesis were results with caged ncPNAs in zebrafish embryos, which indicated first that a very short (8-mer) sODN strand was very effective at blocking antisense activity (27). Importantly, the low Tm of the 8-mer sODN/ncPNA photo-generated duplex favored binding of the antisense ncPNA to the target mRNA and showed enhanced biological activity, relative to a more stable 12-mer sODN/ncPNA duplex.
In K562 cells, fine-tuning the thermal stability of the conjugate and photo-generated asODN/sODN duplex will also be required to achieve maximal photomodulation efficiency. From the data reported herein, it is apparent that the conjugates S-C5 and S-C6 were sufficiently stable to minimize background RNase H activity. Thus, ongoing efforts will focus on engineering photoproducts with less thermally stable asODN/sODN duplexes, in order to maximize the activity of the asODN. In this way, it should be possible to reduce c-MYB protein and c-myb mRNA to levels similar to those achieved with AS 321.
Many recent efforts to increase the specific biological activity of asODNs have focused on improving cell delivery and nuclease resistance (48). Maximizing activity and resistance to nucleases within the cell can be enhanced considerably by modifying the backbone of the oligonucleotide, as exemplified by phosphorothioate (49), peptide (50), morpholine (51) and ‘locked’ (3) nucleic acids. Thus, strategies described herein using phosphorothioated DNA conjugates for cellular studies can be readily applied to the development of related antisense molecules for biotechnological and cellular applications.
CONCLUSIONS
This study represents the first example of targeting a cancer-related gene using light-activated oligonucleotides. DNA conjugates were designed to regulate DNA/DNA and DNA/RNA duplex formation using a single photoactive moiety. More than 2-fold down-regulation of c-myb mRNA and c-MYB protein in human K562 (leukemia) cells was achieved using the conjugate S-C5, which validated the utility of this approach.
We explored the effects of varying the size of the hairpin loop, and the number and position of complementary base pairs. The stability of the asODN–PL–sODN conjugates was modified and compared to the corresponding asODN/sODN duplexes. The photomodulation efficiency of RNA digestion by RNase H was investigated with six different photoactive conjugates, and the amount of RNase H (0.2, 1 or 3 U), the ratio of RNA/DNA, and the time (10 or 60 min) were varied for this enzymatic reaction. Results from this work showed that the conjugates were stabilized by smaller hairpin loops; best results were obtained when the loop was formed by only the PL, with no intervening oligonucleotides. Base pair mismatches had a surprisingly large effect on destabilizing photoactive conjugates and related asODN/sODN duplexes. Biochemical assays with RNase H clearly showed that stabilizing the conjugate helped to minimize background RNA cleavage. Among the six photoactive conjugates, C6 was the most thermally stable and gave the lowest background levels of RNA digestion and greatest photomodulation of RNA digestion. In these assays, the thermal stability of the DNA conjugate proved to be an important factor affecting RNA hydrolysis by RNase H. C6 showed 10-fold and 6-fold photomodulation of RNase H activity at 10 and 60 min, respectively. This range of photomodulation efficiencies should be useful for a variety of cellular and biotechnological applications.
Phosphorothioated versions of the conjugates with the highest photomodulation efficiencies, C5 and C6, were synthesized and introduced by nucleofection into K562 cells. Significant knockdown of c-myb mRNA (56%) and c-MYB protein (52%) was observed 24 h after photoactivation of S-C5. Interestingly, no effect on c-MYB protein level was observed with the more stable conjugate S-C6, although c-myb mRNA levels in cells transfected with S-C5 or S-C6 were similar under pre- and post-UV illumination conditions. Most likely, this was due to the greater duplex stability of S-C6, which inhibited binding to c-myb mRNA. The antisense strand of the less stable S-C5 duplex may have had a greater propensity to inhibit ribosomal protein synthesis, in addition to recruiting RNase H.
The exact reasons for the difference in mRNA and protein knockdown that was observed for S-C6 are still unresolved and will be investigated using other photoactive oligonucleotides. However, we postulate that very stable photoactive conjugates, such as S-C6, are more likely to give small and inconsistent photomodulation effects in biological systems. As stated previously, more convincing biological results were obtained in living zebrafish embryos when a negatively charged peptide nucleic acid antisense strand was conjugated to very short (8-mer) sODN (27). Ongoing efforts to target genes in K562 cells by this approach will focus on using shorter blocking sODNs, in order to maximize the activity of the asODN after photoactivation.
We conclude that this class of light-activated antisense molecules provides a versatile method for modulating the activity of RNase H. Light-activated oligonucleotides can be readily adapted and applied to other biological processes that involve DNA/DNA, DNA/RNA and RNA/RNA hybridization.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Jim Eberwine, Feng Gai and Eric Meggers for access to instrumentation. Adam Peritz synthesized phosphorothioated oligodeoxynucleotides for S-C5 and S-C6 in the UPenn Nucleic Acid Facility. I.J.D. and X.T. were supported by a Camille and Henry Dreyfus New Faculty Award, the Penn Genomics Institute, and Institute for Medicine and Engineering. National Institute of Health grants (R01-CA101859, P01-CA72765) and a translational research grant (6050-07) from the Leukemia and Lymphoma Society went to A.M.G. A training grant from National Institutes of Health (T32-DK07780) went to J.S. Funding to pay the Open Access publication charges for this article was provided by the Dreyfus Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shi Y, Koh JT. Light-activated transcription and repression by using photocaged SERMs. Chem. Biochem. 2004;5:788–796. doi: 10.1002/cbic.200300823. [DOI] [PubMed] [Google Scholar]
- 2.Shah S, Rangarajan S, Friedman SH. Light-activated RNA interference. Angew. Chem., Int. Ed. Engl. 2005;44:1328–1332. doi: 10.1002/anie.200461458. [DOI] [PubMed] [Google Scholar]
- 3.Shoham S, O’Connor DH, Sarkisov DV, Wang S.S.-H. Rapid neurotransmitter uncaging in spatially defined patterns. Nat. Methods. 2005;2:837–843. doi: 10.1038/nmeth793. [DOI] [PubMed] [Google Scholar]
- 4.Rothman DM, Petersson EJ, Vazquez ME, Brandt GS, Dougherty DA, Imperiali B. Caged phosphoproteins. J. Am. Chem. Soc. 2005;127:846–847. doi: 10.1021/ja043875c. [DOI] [PubMed] [Google Scholar]
- 5.Pollitt SK, Schultz PG. A photochemical switch for controlling protein-protein interactions. Angew. Chem., Int. Ed. Engl. 1998;37:2104–2107. doi: 10.1002/(SICI)1521-3773(19980817)37:15<2104::AID-ANIE2104>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Wu N, Deiters A, Cropp TA, King D, Schultz PG. A genetically encoded photocaged amino acid. J. Am. Chem. Soc. 2004;126:14306–14307. doi: 10.1021/ja040175z. [DOI] [PubMed] [Google Scholar]
- 7.Lima SQ, Miesenbock G. Remote control of behavior through genetically targed photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Adesnik H, Nicoll RA, England PM. Photoinactivation of native AMPA receptors reveals their real-time trafficking. Neuron. 2005;48:977–985. doi: 10.1016/j.neuron.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 9.Ando H, Furuta T, Tsien RY, Okamoto H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001;28:317–325. doi: 10.1038/ng583. [DOI] [PubMed] [Google Scholar]
- 10.Ando H, Furuta T, Okamoto H. Methods in Cell Biology. Elsevier Inc.; 2004. pp. 159–171. [DOI] [PubMed] [Google Scholar]
- 11.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers JJ, Gouda H, Young DM, Kuntz ID, England PM. Photochemically knocking out glutamate receptors in vivo. J. Am. Chem. Soc. 2004;126:13886–13887. doi: 10.1021/ja048331p. [DOI] [PubMed] [Google Scholar]
- 13.Monroe WT, McQuain MM, Chang MS, Alexander JS, Haselton FR. Targeting expression with light using caged DNA. J. Biol. Chem. 1999;274:20895–20900. doi: 10.1074/jbc.274.30.20895. [DOI] [PubMed] [Google Scholar]
- 14.Minden J, Namba R, Mergliano J, Cambridge S. Photoactivated gene expression for cell fate mapping and cell manipulation. Sci. STKE. 2000;2000:PL1. doi: 10.1126/stke.2000.62.pl1. [DOI] [PubMed] [Google Scholar]
- 15.Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew. Chem., Int. Ed. Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 16.Tang X, Dmochowski IJ. Regulating gene expression with light-activated oligonucleotides. Mol. BioSyst. 2007;3:100–110. doi: 10.1039/b614349k. [DOI] [PubMed] [Google Scholar]
- 17.Ordoukhanian P, Taylor J.-S. Design and synthesis of a versatile photocleavable DNA building block. Application to phototriggered hybridization. J. Am. Chem. Soc. 1995;117:9570–9571. [Google Scholar]
- 18.Ordoukhanian P, Taylor J.-S. Caged single and double strand breaks. Bioconjug. Chem. 2000;11:94–103. doi: 10.1021/bc9900993. [DOI] [PubMed] [Google Scholar]
- 19.Dussy A, Meyer C, Quennet E, Bickle TA, Giese B, Marx A. New light-sensitive nucleosides for caged DNA strand breaks. Chem. Biochem. 2002;3:54–60. doi: 10.1002/1439-7633(20020104)3:1<54::AID-CBIC54>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Chaulk SG, MacMillan AM. Caged RNA: photo-control of a ribozyme reaction. Nucleic Acids Res. 1998;26:3173–3178. doi: 10.1093/nar/26.13.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Taylor J.-S. A caged ligatable DNA strand break. J. Am. Chem. Soc. 1999;121:11579–11580. [Google Scholar]
- 22.Pirrung MC, Zhao X, Harris SV. A universal, photocleavable DNA base: nitropiperonyl 2′-deoxyriboside. J. Org. Chem. 2001;66:2067–2071. doi: 10.1021/jo001594r. [DOI] [PubMed] [Google Scholar]
- 23.Lenox HJ, McCoy CP, Sheppard TL. Site-specific generation of deoxyribonolactone lesions in DNA oligonucleotides. Org. Lett. 2001;3:2415–2418. doi: 10.1021/ol016255e. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, Dmochowski IJ. Controlling RNA digestion by RNase H with a light-activated DNA hairpin. Angew. Chem., Int. Ed. Engl. 2006;45:3523–3526. doi: 10.1002/anie.200600954. [DOI] [PubMed] [Google Scholar]
- 25.Tang X, Dmochowski IJ. Synthesis of light-activated antisense oligodeoxynucleotide. Nat. Protoc. 2007;1:3041–3048. doi: 10.1038/nprot.2006.462. [DOI] [PubMed] [Google Scholar]
- 26.Rotaru A, Mokhir A. Nucleic acid binders activated by light of selectable wavelength. Angew. Chem. Int. Ed. 2007;46:6180–6183. doi: 10.1002/anie.200700641. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, Dmochowski IJ. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 2007;129:11000–11001. doi: 10.1021/ja073723s. [DOI] [PubMed] [Google Scholar]
- 28.Matsunaga D, Asanuma H, Komiyama M. Photoregulation of RNA digestion by RNase H with azobenzene-tethered DNA. J. Am. Chem. Soc. 2004;126:11452–11453. doi: 10.1021/ja0471976. [DOI] [PubMed] [Google Scholar]
- 29.Keiper S, Vyle JS. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew. Chem., Int. Ed. Engl. 2006;45:3306–3309. doi: 10.1002/anie.200600164. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 31.Ting R, Lermer L, Perrin DM. Triggering DNAzymes with light: A photoactive C8 thioether-linked adenosine. J. Am. Chem. Soc. 2004;126:12720–12721. doi: 10.1021/ja046964y. [DOI] [PubMed] [Google Scholar]
- 32.Ghosn B, Haselton FR, Gee KR, Monroe WT. Control of DNA hybridization with photocleavable adducts. Photochem. Photobiol. 2005;81:953–959. doi: 10.1562/2004-11-15-RA-373. [DOI] [PubMed] [Google Scholar]
- 33.Tang X, Dmochowski IJ. Phototriggering of caged fluorescent oligodeoxynucleotides. Org. Lett. 2005;7:279–282. doi: 10.1021/ol047729n. [DOI] [PubMed] [Google Scholar]
- 34.Iwase R, Kitani A, Yamaoka T, Murakami A. Synthesis of antisense oligonucleotides containing photocleavable protecting groups on the thymine bases and their photoinduced duplex formation. Nucleic Acids Res. Supp. 2003;3:61–62. doi: 10.1093/nass/3.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Bourdelat-Parks B, Boatright JH, Wartell RM. Targeting degradation of RNA by RNase H using DNA hairpins. Biochemistry. 2003;42:10945–10954. doi: 10.1021/bi034897z. [DOI] [PubMed] [Google Scholar]
- 36.Kalota A, Shetzline SE, Gewirtz AM. Progress in the development of nucleic acid therapeutics for cancer. Cancer Biol. Ther. 2004;3:4–12. doi: 10.4161/cbt.3.1.517. [DOI] [PubMed] [Google Scholar]
- 37.Opalinska JB, Kalota A, Gifford LK, Lu P, Jen K.-Y, Pradeepkumar PI, Barman J, Kim TK, Swider CR, et al. Oxetane modified, conformationally constrained, antisense oligodeoxyribonucleotides function efficiently as gene silencing molecules. Nucleic Acids Res. 2004;32:5791–5799. doi: 10.1093/nar/gkh893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gifford LK, Opalinska JB, Jordan D, Pattanayak V, Greenham P, Kalota A, Robbins M, Vernovsky K, Rodriguez LC, et al. Identification of antisense nucleic acid hybridization sites in mRNA molecules with self-quenching fluorescent reporter molecules. Nucleic Acids Res. 2005;33:e28/21–e28/29. doi: 10.1093/nar/gni024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Opalinska JB, Machalinski B, Ratajczak J, Ratajczak MZ, Gewirtz AM. Multigene targeting with antisense oligodeoxynucleotides: An exploratory study using primary human leukemia cells. Clin. Cancer Res. 2005;11:4948–4954. doi: 10.1158/1078-0432.CCR-05-0106. [DOI] [PubMed] [Google Scholar]
- 40.Ness SA. Myb binding proteins: regulators and cohorts in transformation. Oncogene. 1999;18:3039–3046. doi: 10.1038/sj.onc.1202726. [DOI] [PubMed] [Google Scholar]
- 41.Kowenz-Leutz E, Herr P, Niss K, Leutz A. The homeobox gene GBX2, a target of the myb oncogene, mediates autocrine growth and monocyte differentiation. Cell. 1997;91:185–195. doi: 10.1016/s0092-8674(00)80401-8. [DOI] [PubMed] [Google Scholar]
- 42.Weston K. Reassessing the role of C-MYB in tumorigenesis. Oncogene. 1999;18:3034–3038. doi: 10.1038/sj.onc.1202728. [DOI] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Kalota A, Karabon L, Swider CR, Viazovkina E, Elzagheid M, Damha MJ, Gewirtz AM. 2′-Deoxy-2′-fluoro-β-D-arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persisten, gene silencing. Nucleic Acids Res. 2006;34:451–461. doi: 10.1093/nar/gkj455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peyret J, SantaLucia J. HyTher 1.0. Detroit, MI: Wayne State University; 1998. [Google Scholar]
- 46.Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. A critical survey o the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J. Biochem. Biophys. Methods. 2001;48:189. doi: 10.1016/s0165-022x(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Wartell RM. RNase H1 can catalyse RNA/DNA formation and cleavage with stable hairpin or duplex DNA oligomers. Biochemistry. 1998;37:5154–5161. doi: 10.1021/bi9730801. [DOI] [PubMed] [Google Scholar]
- 48.Rubenstein M, Tsui P, Guinan P. A review of antisense oligonucleotides in the treatment of human disease. Drugs Future. 2004;29:893–909. [Google Scholar]
- 49.Stec WJ, Zon G, Egan W, Stec B. Automated solid-phase synthesis, separation, and stereochemistry of phosphorothioate analogues of oligodeoxyribonucleotides. J. Am. Chem. Soc. 1984;106:6077–6079. [Google Scholar]
- 50.Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA, Ricca DJ, Hassman CF, Bonham MA, Au KG, et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 51.Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]