Abstract
The active center clefts of RNA polymerase (RNAP) from the archaeon Pyrococcus furiosus (Pfu) and of yeast RNAP II are nearly identical, including four protruding loops, the lid, rudder, fork 1 and fork 2. Here we present a structure–function analysis of recombinant Pfu RNAP variants lacking these cleft loops, and analyze the function of each loop at different stages of the transcription cycle. All cleft loops except fork 1 were required for promoter-directed transcription and efficient elongation. Unprimed de novo transcription required fork 2, the lid was necessary for primed initial transcription. Analysis of templates containing a pre-melted bubble showed that rewinding of upstream DNA drives RNA separation from the template. During elongation, downstream DNA strand separation required template strand binding to an invariant arginine in switch 2, and apparently interaction of an invariant arginine in fork 2 with the non-template strand.
INTRODUCTION
Multi-subunit RNA polymerases (RNAPs) catalyze RNA synthesis from a DNA template during gene transcription. The eukaryotic nucleus contains three RNA polymerases, called RNAP I, II and III, whereas bacterial and archaeal cells contain only one RNAP. Analysis of gene regulation requires a detailed structure-based understanding of the transcription mechanism. During the transcription cycle, the polymerases first assemble with initiation factors on promoter DNA (closed complex formation). The complex then unwinds the DNA double helix (open complex formation). The polymerase begins to synthesize short RNA oligonucleotides that are often released (abortive transcription). When the RNA product reaches a certain length, the enzyme enters the elongation phase, characterized by a stable, processive elongation complex. The polymerase then elongates the RNA chain, unwinds downstream DNA and rewinds upstream DNA. Finally, the RNA transcript and the DNA are released during termination.
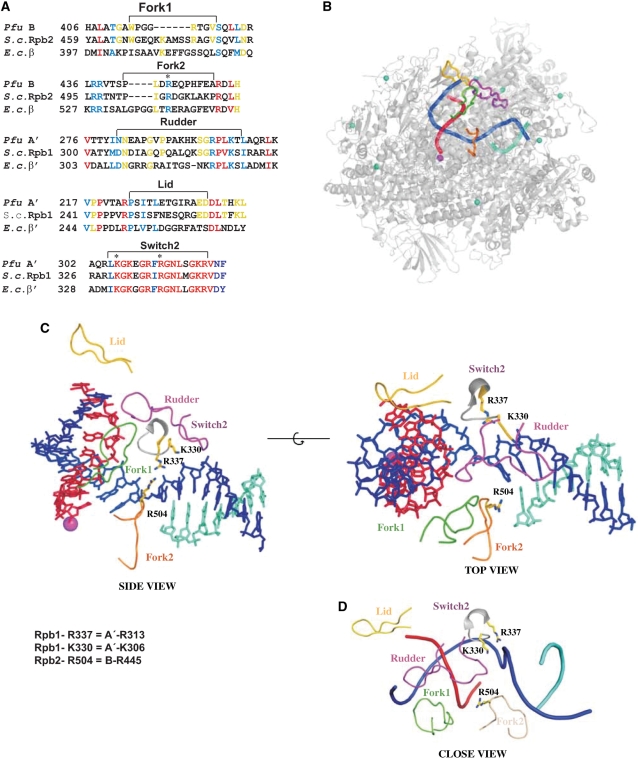
Detailed crystallographic structures are available for yeast RNAP II and bacterial RNAPs (1–5) and enable mechanistic studies of the transcription cycle by designing mutations. Four prominent loops were revealed above the active site in the polymerase cleft, named the rudder, lid, fork loop 1 and fork loop 2 (2,6). Whereas the rudder and lid protrude from the mobile clamp of the polymerase, the two fork loops are located on the opposite side of the cleft (Figure 1).
Figure 1.
Design of the loop deletions and single-point mutations. (A) Internal deletion of B and A′ subunits. Primary sequence alignment between the four loops fork1, fork 2, rudder and lid from P. furiosus, S. cerevisiae and E. coli (CULSTAL W). Invariant, conserved and weakly conserved residues are colored in red, blue and yellow, respectively. The extent of deletions and the single-point mutations introduced in fork loop 2 and switch 2 are indicated and highlighted by asterisks. (B) Surface representation of the S. cerevisiae RNAP II elongation complex (8). Template DNA, non-template DNA, RNA, Mg2+ and Zn2+ ions are shown in blue, cyan, red, magenta and light blue, respectively. Lid, rudder, fork 1 and fork 2 are represented in yellow, magenta, green and ochre, respectively. (C) Close up view of the RNAP II active site. Single-point mutations are shown. The same color code as in (B) was used. The residues corresponding in the archaeal enzyme to the mutated putative Rpb1 and Rpb2 key residues are indicated. (D) Close-up view of nucleic acids in the yeast RNAP II active center highlighting the position of single-point mutations analyzed in this study, the color code is like in (B).
The rudder and lid were suggested to maintain the upstream end of the hybrid and the bubble (2,6–8). Functional roles of the rudder and lid were analyzed in the bacterial enzyme (9–11). Mutagenesis of the rudder showed that this element stabilizes the elongation complex but that it is not involved in maintaining the hybrid length (9). The lid was suggested to help separate RNA from DNA at the upstream end of the hybrid (2,3,6,7) but a mutant bacterial RNA polymerase lacking the lid could displace RNA normally (10). It was suggested that fork loop 2 blocks the path of the non-template strand before the active site, and thereby helps to separate the DNA strands at the downstream edge of the bubble (6,8). In yeast RNAP II, mutations in the proximity to fork loop 2 have been shown to lower the polymerization rate but no mutational in vitro studies on the rudder, lid or fork loop 1 (12) have been reported.
Recently, recombinant forms of archaeal RNAPs became available, which enable rapid site-directed mutagenesis (13,14). The archaeal enzymes are closely related in sequence to eukaryotic RNAP II (15). All yeast RNAP II subunits have counterparts in the archaeal enzyme, except the small peripheral subunit Rpb8. In the Pyrococcus furiosus (Pfu) enzyme, 38% of the amino acid residues are identical with yeast RNAP II (16,17). The similarity of the archaeal and eukaryotic transcription machineries extends to protein–protein interactions of the polymerase subunits (13,16) and initiation factors that are required for promoter binding (18). The archaeal initiation factors TBP, TFB and TFE have homologs in the eukaryotic RNAP II apparatus, named TBP, TFIIB and TFIIE, respectively. Whereas the factors TBP and TFB are sufficient to bind and open promoter DNA in the Pyrococcus system at 70°C (19,14) the eukaryotic machinery requires in addition TFIIF to bind the promoter, and TFIIE/TFIIH to open DNA. Despite these differences, the recent success in obtaining highly active recombinant Pfu RNAP opens up the possibility to rapidly prepare and functionally analyze mutant RNAP II-like enzymes (14).
Here, we analyzed the function of four recombinant archaeal RNA polymerase deletion mutant enzymes, each lacking one of the four cleft loops. Together with an analysis of three additional mutant enzymes carrying selected point mutations in fork loop 2 and switch region 2, another active center element that was thus far not studied by mutagenesis, our results unravel the functional significance of these elements at various stages of the transcription cycle. In addition, we have used different nucleic acid scaffolds to elucidate the initiation–elongation transition, one of the most dynamic and least understood aspects of the transcription cycle.
MATERIALS AND METHODS
Primer sequences
The sequence of primers used for mutagenesis and PCR are provided in the Supplementary Data.
Construction of subunit B (rpb2) and subunit A′ mutants by site-directed mutagenesis
The rudder and lid domains of subunit A′ and fork loop 1 and fork loop 2 of subunit B were deleted using a two-round, four-primer technique. In round 1, two PCR products were generated containing the DNA region upstream and downstream from the deletion in separate reactions. Each PCR was performed using genomic DNA as template, end primers (FwdA and RevD) and a pair of primers flanking the internal sequence to be deleted (RevB and FwdC). The sequences of all primers are provided in the SupplementaryData. The resulting products were analyzed by agarose gel electrophoresis and purified using a QIAquick spin Gel Extraction Kit. Purified DNA fragments were added to a second round of PCR. Fusion of the two intermediates was achieved as a result of overlapping complementary regions in the products left and right to the deleted sequence formed in round 1. The products paired during the annealing phase of PCR round 2 and were amplified by the addition of primers complementary to the end of each single-stranded DNA fragment (primers FwdA and RevD). For the generation of single-point mutants, two complementary primers (B-R445A-RevB and B-R445A-FwdC for B-R445A; A′-R313A-RevB and A′-R313A-FwdC for A′-R313A; A′-K306A-RevB and A′-K306A-FwdC for A′-K306A) were used along with end primers (FwdA and RevD) to substitute arginine or lysine with alanine. After analysis of the PCR products by agarose gel electrophoresis, the resulting mutant DNA was purified and ligated into pET151/D-TOPO and transformed into Escherichia coli.
Overexpression of recombinant subunits
The subunits were expressed in BL21(DE3) Codon Plus TM by inducing exponentially grown cultures over night at 20°C with IPTG. The his6-tagged mutated subunits B and A′ were (like their wild-type counterparts) highly insoluble and expressed as inclusion bodies in E. coli cells. They were solubilized in 6 M guanidine HCl and immobilized on a NiNTA column. The proteins were refolded on column by washing with a decreasing gradient of urea. The renatured subunits were eluted with imidazole and used for reconstitution of the RNAP as described previously (14).
Reconstitution of mutant and wild-type Pyrococcus RNAP
The RNAPs containing mutated and wild-type components were reconstituted from 11 bacterially produced subunits after denaturation in TB buffer containing 6 M urea and stepwise dialysis against TB buffer containing 3 M urea and no urea. The renatured RNAP assemblies were purified by Superdex 200 chromatography as described previously (14). The protein eluting as a homogenous peak from the Superdex 200 column was analyzed by SDS PAGE and in specific run-off transcription assays using the Pyrococcus gdh promoter as template. Fractions containing active RNAP were combined and used for transcriptional analyses.
Cell-free transcription reactions
Promoter-independent assay. Reactions were performed in a total volume of 100 µl containing 9 nM RNAP (or mutant derivative) 900 µM ATP, 90 µM UTP, 0.15 MBq(α-32P)-UTP (110 TBq/mmol) and 3 µg of poly[(dA-dT)] as template. Reactions were incubated for 30 min at 70°C and counts insoluble in 5% TCA (w/v) were determined.
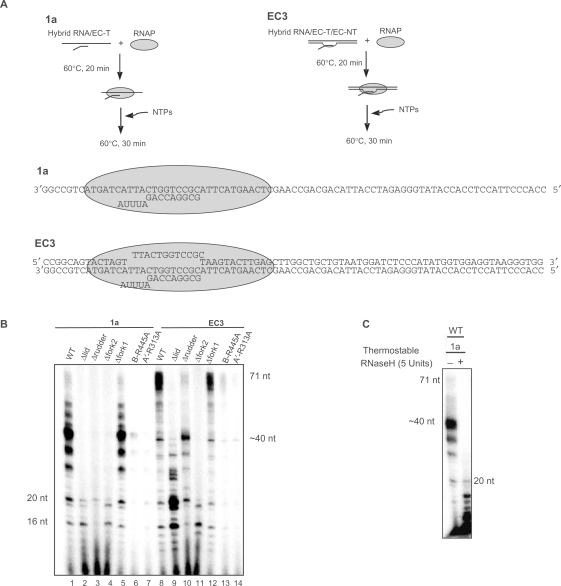
Promoter-directed assays. Specific in vitro transcription reactions were essentially conducted as described by (20). XbaI-digested plasmid pUC19 containing the gdh promoter region from −95 to +163 was incubated with 35 nM TBP, 30 nM TFB and 9 nM endogenous RNAP, recombinant RNAP or mutant derivative in 25 µl TB (40 mM Na-HEPES, pH 7.3, 250 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 5 mM -mercaptoethanol, 0.1 mg/ml BSA). NTPs were added to 500 µM ATP, GTP, CTP and 10 µM UTP and 0.15 MBq(α-32P)UTP (110 TBq/mmol). The reactions were assembled at 4°C and started by transfer to 70°C unless otherwise indicated. The 172 nt run-off transcripts and various abortive transcripts were analyzed in 6, 28 or 20–28% denaturing polyacrylamide gels as indicated and visualized by phosphoimaging. To construct the pre-opened bubble (Figure 5) the template and non-template strand (10 µM each) were incubated for 2 min at 92°C and cooled down slowly over night to room temperature. This hybrid (10 µM) was used as template in cell-free transcription reactions. The RNA–DNA template strand hybrid (1a; Figure 6A) mimicking an elongation complex was assembled by heating 10 µM RNA and 5 µM template strand in a volume of 100 µl for 2 min at 92°C and cooled down slowly overnight to room temperature. A 0.5 µl of this mixture was used as template in the transcription assays shown in Figure 6C. Template ECR3 (Figure 6A) was assembled by the same procedure and contained in addition to the hybrid RNA–DNA template strand 5 µM of the corresponding non-template strand.
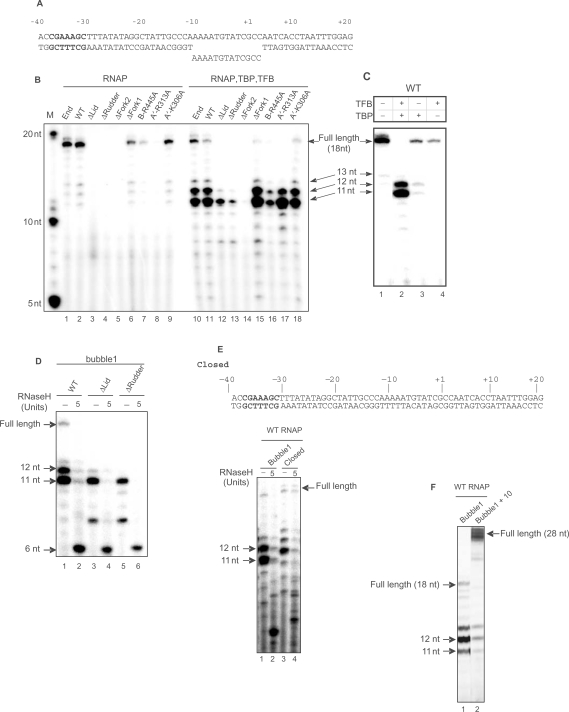
Figure 5.
Initiation from a pre-melted mismatch bubble. (A) Template of the gdh promoter containing a mismatch in the region from −10 to +3 (bubble 1). The TATA box sequence is underlined and the the BRE sequence is shown in bold letters. (B) Three loops lid, rudder and fork 2 and the conserved arginine residue 313 in switch 2 are required for the synthesis of full-length transcripts. The various RNAP fractions and mutants were pre-incubated in transcription reactions with the template shown in (A) for 30 min at 70°C shown in the absence (left panel) and presence of TBP (35 nM) and TFB (30 nM) (right panel). Transcription was started by the addition of NTPs and the reactions were incubated for further 30 min at 70°C. RNA products were analyzed on a 28% polyacrylamide gel. The lane labeled M indicates RNA markers. (C) The +11 to +12 transcripts are induced by the TBP–TFB complex. Transcripts from bubble 1 formed in the presence of the individual components indicated on top of the lanes were analyzed as in (B) Note that a weak barrier is also imposed in the presence of TBP alone (lane 3) and that TFB has an inhibitory effect on the synthesis of the full-length transcript (compare lanes 1 and 4) most likely by binding to free RNAP. (D) The 11- to 12-mer and full length transcripts are RNase H sensitive. Transcripts formed on bubble 1 were digested for 15 min at 37°C with RNase H and analyzed as in (B). (E) RNA displacement requires upstream rewinding of the template strand. The double-stranded closed template containing no mismatch (upper panel) and bubble 1 were transcribed in the presence of transcription factors and treated for 15 min at 37°C with 5 units of RNase H as indicated. (F) The length of downstream duplex DNA affects promoter escape. Transcripts from bubble 1 and from a template with the same mismatch region but extended at the downstream end by 10 nt of duplex DNA were analyzed as in (B).
Figure 6.
Lid, rudder, fork 2 and the conserved arginine residue 313 in switch 2 are required for normal elongation. (A) Experimental design and templates, template 1a contains the template strand hybridized with 9 nt of a 14 nt RNA primer, template ECR3 contains a 12 bp heteroduplex hybridized with the RNA primer. (B) Analysis of the effect of mutations on elongation. Mutants and WT enzymes were incubated with the templates indicated in the absence of TBP and TFB at 60°C and RNA products synthesized were analyzed on 20–28% polyacrylamide gradient gels. (C) The archaeal enzyme forms extended DNA–RNA hybrids on the single-stranded template. Transcription reactions were conducted at 60°C and digested with thermostable RNase H at 70°C as indicated in the Figure.
Abortive transcription reactions
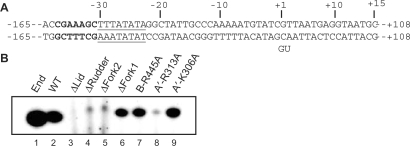
300 ng double stranded template gdh-C15 (21); sequence of the promoter region (shown in Figure 4A) was incubated with transcription factors and Pyrococcus RNAP as described under cell-free transcription reactions. In the dinucleotide primed reactions shown in Figure 4 the reaction was started with 40 µM GpU and 10 µM UTP and 0.15 MBq(α-32P)UTP (110 TBq/mmol).
Figure 4.
The loops rudder, lid and fork 2 and the conserved arginine residue 313 in switch 2 are required for synthesis of short abortive transcripts. (A) A template containing a modified sequence of the gdh promoter gdh-C15(−165 to +108; 21) was used to detect initial RNA synthesis. The TATA box is underlined, the BRE element is indicated by bold letters and the transcription start site is indicated by +1. (B) Synthesis of a 3 nt abortive transcript in dinucleotide primed reactions. Abortive RNA products were separated in a 28% polyacrylamide gel.
Electrophoretic mobility shift assays
DNA fragments spanning the gdh promoter region from −60 to +37 were used as probes in assays with mutated and wild type RNAP as described previously (16). Binding reactions with the gdh promoter contained in addition TBP and TFB. The binding reactions (25 µl) contained 9 nM endogenous or 27 nM recombinant RNAP (or mutant derivatives), 100 nM TBP, 100 nM TFB, 0.5 nM gdh promoter and 1 µg of poly[dI-dC)] as non-specific competitor DNA.
Permanganate footprinting
Thymidine residues in the open complexes formed by wild-type and mutant forms of RNAP in pre-initiation complexes of the gdh promoter containing TBP and TFB were detected by treatment with potassium permanganate (KMnO4) as described in the legend of Figure 3.
Figure 3.
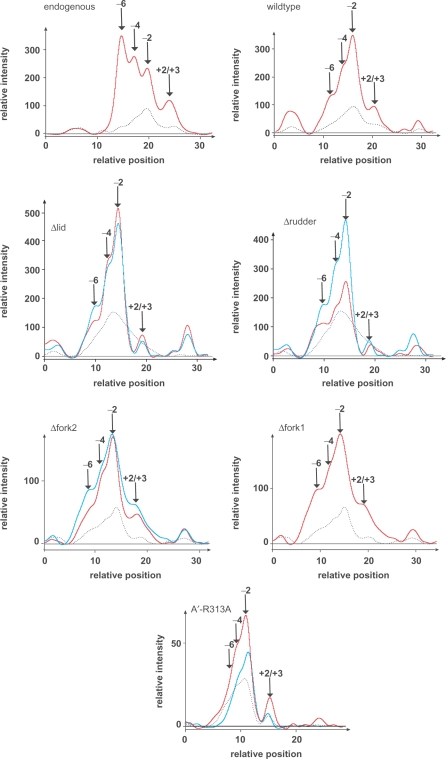
The cleft loops fork 1, fork 2 and lid and R-313 in switch 2 are not essential for the formation of open complexes. The various RNAP preparations and RNAP mutants as indicated in the figure were incubated with the gdh promoter containing a 5′ end-labeled template strand in the presence of TBP and TFB for 10 min at 70°C to allow bubble opening and were probed with KMnO4for 5 min at 70°C (21). Modified thymine residues in single-stranded DNA were resolved after piperidine cleavage by electrophoresis in 6% sequencing gels. Phosphoimager traces of the KMnO4 footprinting profiles of the endogenous recombinant and cleft loop polymerase mutants are shown. The dotted line represents the modification pattern in control reactions without RNAP, the red line the permanganate footprint. The blue line represents the signal obtained with WT enzyme in a control reaction on the same gel. The footprint of the endogenous enzyme extends from −6 to +3. This enzyme contains, in contrast to the reconstituted polymerase, nearly stoichiometric amounts of the subunits E′ and F (the archaeal Rpb7/Rpb4 homologs). The presence of the E′ subunit in the endogenous enzyme is responsible for the extension of the permanganate footprint to the upstream end (14). The footprints of the reconstituted and mutant enzymes Δlid, Δfork 2 and Δfork 1 extend from +3 to −4. Note that formation of the footprint by the Δrudder enzyme is impaired, in particular at the downstream end.
RESULTS
Design and preparation of Pfu RNA polymerase variants
Due to the high sequence conservation between the Pfu polymerase and yeast RNAP II, the Pfu lid, rudder, fork loop 1 and fork loop 2 were easily identified in a sequence alignment of the largest subunits (Figure 1). Whereas the loops themselves are not well conserved, the regions immediately preceding and following the exposed loops are highly conserved, enabling unambiguous definition of the loop borders. The complete RNAP II elongation complex structure (8) guided the design of four Pfu RNAP deletion mutants that lack the lid (residues 224–236 in subunit A′), rudder (residues 281–300 in subunit A′), fork loop 1 (residues 413–420 in subunit B) and fork loop 2 (residues 442–452 in subunit B). The deletions were designed such that small stretches of amino acids were left intact and the ends resulting from truncation were connected without perturbation of the structure in the surrounding protein region (non-disruptive mutations).
In addition, we prepared three enzyme variants carrying alanine point mutations. One mutant modifies a conserved basic residue in fork loop 2, R445 in the B subunit, corresponding to Rpb2 residue R504 in RNAP II, and two mutants target basic residues in switch 2, K306 and R313 in the A′ subunit, which correspond to Rpb1 residues K330 and R337, respectively, in RNAP II. These two residues are highly conserved in the switch 2 element across the three domains of life (Figure 1A, lower panel). Structural analyses of a yeast RNAP II elongation complex suggested that the basic amino acids R337 and K330 might be involved in pulling the template strand upwards in the active site and could be therefore critical for DNA strand separation in the active center (8). A close-up view showing the position of the mutated basic amino acids relative to the nucleic acids in the active center is provided in Figure 1D. The four recombinant deletion enzymes, here referred to as Δlid, Δrudder, Δfork1 and Δfork2, and the three single-point mutation enzymes, termed B-R445A, A′-K306A, and A′-R313A, were reconstituted as described (14). The seven purified variants showed a size-exclusion chromatography profile identical to the wild-type recombinant RNAP (data not shown).
Fork 1 is the only cleft loop not required for promoter-dependent transcription
The functional properties of the Pfu RNAP variants were analyzed and compared to the properties of purified endogenous RNAP (20) and recombinant wild-type enzyme. In a non-specific transcription assay that uses poly-d(A–T) as a template, all mutant enzymes showed low activities close to background levels and also the activity of the reconstituted enzyme was low compared to that of the endogenous enzyme (data not shown). Therefore, the activity of the mutant enzymes was not to be quantitatively determined in this assay and the mutant enzymes were rather analyzed in more sensitive assays measuring the synthesis of distinct RNA products.
To analyze if the variants have an overall defect in promoter-dependent transcription, we subjected them to a promoter-specific transcription assay that uses as a template the strong Pfu glutamate dehydrogenase (gdh) promoter (20). The variants were incubated with DNA, TFB and TBP, and reactions were initiated by addition of nucleoside triphosphates (NTPs). The run-off RNA product was efficiently synthesized by the endogenous and wild-type recombinant RNAPs, providing a positive control (Figure 2). The ▵fork1 RNAP was highly active, showing that fork loop 1 does not have an essential role in transcription. In contrast, Δlid, Δrudder and Δfork2 showed no transcription activity (Figure 2), pointing to an essential function of the lid, rudder and fork 2. Of the single-point mutants, B-R445A and A′-K306A showed reduced but still significant activity, whereas Á-R313A was totally inactive. Taken together, our results confirm the functional importance of the rudder and lid, which was previously reported for the bacterial enzyme, and additionally show that fork 2 and switch 2 are required for normal polymerase function.
Figure 2.
The cleft loops lid, rudder, fork 2 and the conserved arginine residue 313 in switch 2 are required for promoter-dependent initiation. Promoter-dependent assays. Equal amounts (100 ng) of Superdex fractions of reconstituted RNAP mutants and of endogenous and reconstituted RNAP were incubated in standard transcription reactions in the presence of TBP (35 nM) and TFB (30 nM) with the gdh promoter (6.5 nM) as template. RNA products were analyzed on 6% polyacrylamide gels.
The rudder is required for open complex formation
The mutant variants were incubated at 37°C with TBP, TFB, a DNA-fragment containing the gdh promoter, and competitor DNA, and were separated in native gels. All variants formed stable closed complexes, except the ▵lid variant, which apparently formed large aggregates of unknown composition (data not shown). However, the Δlid enzyme formed short transcripts in the presence of the TBP–TFB complex on a pre-melted template (Figure 5B, lane 12) indicating that its ability to interact with DNA and the promoter-bound TBP–TFB complex was not generally impaired. In order to analyze the role of the mutated elements in open complex formation we used KMnO4 footprinting, which identifies thymine residues in single-stranded DNA within melted regions. All cleft loop deletion mutants except ▵rudder produced a footprint around the transcription start site at 70°C, the switch 2 mutant R313A was also able to melt DNA in the promoter region (Figure 3). In particular, positions +3/+2, −2 and −4 were accessible to permanganate, indicating that the DNA strands are melted around the transcription start site (position +1). Promoter melting in the −4/+3 region is sufficient for specific initiation at 70°C (14). These results indicate that the rudder of the archaeal enzyme has an important role in DNA strand separation and/or maintenance of melted DNA as in the bacterial counterpart (9).
The lid, rudder and fork2 are essential for primed transcription
To investigate whether the initial phase of transcription is affected by our loop deletions and point mutations, we carried out an abortive transcription assay, using the gdh promoter as a template and the RNA dinucleotide 5′-GpU-3′ as a primer (Figure 4A). By addition of the radioactive nucleotide [α-32P]-UTP, we tested whether the polymerase variants were able to elongate the GpU primer to GpUpU. In the absence of TBP and TFB, none of the mutant enzymes was active (data not shown). In the presence of TBP and TFB, endogenous and recombinant Pfu RNAPs, the Δfork1 variant and variants R445A and K306A readily synthesized the trinucleotide product (Figure 4B). The Δrudder, Δfork2 and R313A variants were highly defective, Δrudder and Δfork2 synthesized a product of different mobility indicating that they were also unable to perform the primed reaction. The ▵lid enzyme, however, was totally inactive (Figure 4B, lane 3), suggesting that the lid plays an essential role in stabilizing the initially transcribing complex. To investigate whether increasing primer lengths would support abortive transcription by the ▵lid enzyme, the reactions were performed in the presence of a tri-, tetra- or a pentanucleotide RNA primer. In contrast to the bacterial Δlid enzyme that can synthesize longer transcripts in the presence of longer priming RNAs (11) the archaeal ▵lid enzyme was unable to elongate tetra- and pentanucleotide RNAs (data not shown).
Fork 2 is essential for unprimed de novo transcription
In a second approach to study initial RNA synthesis, we used a gdh template with a ‘pre-melted’ mismatch bubble (bubble1) at positions −10 to +3, mimicking DNA in the open complex (Figure 5A). In the absence of transcription factors, the endogenous polymerase, the recombinant enzyme, the Δfork1 mutant, and the single-point mutants R445A and K306A produced an 18 nt transcript as the major RNA product, compared to a full-length run-off product of 20 nt length (Figure 5B). This indicates that a pre-melted bubble can be bound by the polymerase and direct de novo RNA synthesis. It is not surprising that the lid is inactive in this unprimed transcription assay, as it also fails to elongate RNA primers. In addition, the rudder, fork 2 and the arginine in switch 2 are essential for unprimed de novo transcription.
To investigate whether defects in de novo transcription may be compensated by initiation factors, we repeated the experiment in the presence of TBP and TFB. This showed that the lid, rudder and the invariant arginine in switch 2 were not strictly required for de novo RNA synthesis in the context of an initiation complex. However, the ▵fork 2 variant remained totally inactive, showing that this loop is essential for de novo transcription even within the initiation complex. The structural basis for this observation remains unknown.
DNA rewinding drives RNA displacement
A surprising result from the de novo transcription assays was that inclusion of TFB and TBP induced the accumulation of shorter, 11- and 12-mer RNA products, and strongly reduced the amount of 18-mer product (Figure 5B). The shorter products resulted from the presence of the TBP–TFB complex (Figure 5C), which apparently formed a barrier to further progression of the early transcribing complex. To investigate the nature of this barrier, we tested whether the RNA product was properly separated from the DNA template strand by treating the reaction products with RNase H that specifically degrades RNA in a DNA–RNA hybrid. Indeed, the products were RNase H sensitive (Figure 5D), suggesting that RNA strand separation did not occur. To show that the failure of RNA displacement was due to the mismatch bubble design, we repeated the assay with a closed DNA. In this case, the longer RNA product was RNase H resistant, showing that it was properly displaced from the template (Figure 5E). However, short RNAs were formed also on the not displaced closed template (Figure 5E, lane 4) indicating that the barrier is caused by the TBP/TFB complex independent of the presence of the mismatch bubble. The 1 nt upstream shift of the block in the closed template (Figure 5E) suggests an altered TBP/TFB interaction with the pre-melted bubble (unpublished data of Christine Richter and Winfried Hausner suggest that transcription initiates at the bubble template at +2 and at the closed template at +1).
Our finding that the full-length transcript is RNase H sensitive on the mismatch bubble indicates that the mismatch bubble template prevents RNA displacement from the DNA template strand, and indicate that RNA displacement requires upstream rewinding of the DNA duplex, which cannot occur when the DNA strands are non-complementary, as in the mismatch bubble. Since the lid is located at the upstream end of the DNA–RNA hybrid, one may imagine that deletion of the lid would allow for continued growth of a persistent hybrid. However, the ▵lid enzyme produced 11- and 12-mer and shorter RNA products but no full-length transcripts (Figure 5B), showing that the lid was not the cause of the observed elongation barrier. Taken together, these data are consistent with the idea that DNA–RNA strand separation is driven by successful competition of the DNA non-template strand with the RNA for the DNA template strand. When we used the mismatch bubble containing longer segments (e.g. 10 nt in Figure 5F) of downstream duplex DNA, the processivity of the WT enzyme was greatly increased and the arrest at +11/12 less pronounced (Figure 5F). This finding indicates that a certain length of DNA downstream duplex is required for efficient displacement of TFB/TBP, at least in our system.
Invariant arginines in fork 2 and switch 2 cooperate in DNA unwinding
After completion of the initiation–elongation transition, the polymerase alone can elongate the RNA chain. To investigate elongation by the mutant enzymes, we used nucleic acid scaffolds with a 83 nt template strand (EC-T) hybridized to 9 nt of a 14 nt RNA (Figure 6A), and assembly protocols first described by Kashlev and coworkers (22,23). In a first set of experiments, we analyzed elongation of the RNA hybridized to a template single strand (Figure 6A, template 1a). To investigate the effect of downstream duplex DNA a scaffold was used that was similar to synthetic templates used for analysis of elongation by bacterial RNAP (24). This duplex template contained a 12 nt heteroduplex region basepaired in part with the 13 nt RNA primer (EC3, Figure 6A). A lower incubation temperature of 60°C was used to prevent DNA melting. The reconstituted wild-type polymerase was able to accumulate larger amounts of incomplete transcripts but required a downstream DNA duplex for synthesis of a full-length transcript (Figure 6B, lanes 1 and 8). This finding indicates that downstream duplex DNA is required for processive RNA synthesis by the WT enzyme. The cleft loop mutants generally showed defects in these elongation assays, with the exception of fork 1 (Figure 6B, lanes 5 and 12) and the mutant of residue K306 in switch 2 (data not shown). The Δlid, Δrudder and Δfork2 enzyme were unable to synthesize longer products on the single-stranded template a (Figure 6B, lanes 2–4), B-R445A synthesized a weak ∼40 nt product and A′-R313A a weak 20 nt product. These findings suggest that interaction of the cleft loops lid, rudder, fork2 and of R331A with non-template DNA are required for the formation of stable elongation complexes. In the presence of downstream duplex DNA the mutants rudder and lid retained the ability to synthesize RNA products up to a length of ∼40 nt (Figure 6B, lanes 9 and 10). Previous structural studies had suggested that downstream DNA strand separation may involve binding and distortion of the template strand by switch 2 (8) and interference of fork 2 with the path of the non-template strand (6,8). As predicted from this model, essentially no elongation activity was obtained for the ▵fork2 enzyme (Figure 6B, lane 11). Strikingly, even a point mutation in fork 2 that mutates the invariant arginine R445 to an alanine almost abolished elongation activity on a template containing downstream duplex DNA (Figure 6B, lanes 13), consistent with a role of this arginine in directing the non-template strand away from the active site. Also consistent with the proposed mechanism for downstream DNA separation, the switch 2 arginine mutation is totally inactive in elongation assays on template EC3 that contains a downstream DNA duplex (Figure 6B, lanes 14). Taken together, these results suggest that two invariant arginines, located in switch 2 and fork 2 on opposite sides of the cleft and incoming DNA play an important role in downstream DNA strand separation as suggested by structural studies. The role of the other adjacent amino acids still has to be investigated.
The ability of the archaeal RNAP to synthesize >40 nt transcripts on a single-stranded template (1a) was surprising since the bacterial enzyme forms only ∼20 nt transcripts on similar templates (11,12). To investigate whether the archaeal RNAP forms extended DNA–RNA hybrids on a single stranded template the products of reactions with template 1a were digested with RNase H (Figure 6C). To preclude post-transcriptional hybridization of RNA to the template during cooling down to 37°C, the transcription reaction carried out at 60°C were incubated at 70°C with a thermostable RNase H. This revealed that transcripts from template 1a were generally RNase H sensitive, indicating the formation of extended RNA–DNA hybrids. The shorter RNA products synthesized by E. coli polymerase on similar single-stranded templates are also RNase H sensitive (9,11).
DISCUSSION
Structural studies of RNAP II resulted in proposals for the functional roles of various polymerase elements that must be tested by mutational analysis. While such studies are now conveniently carried out in the bacterial system, only very few and non-lethal RNAP II mutations were so far introduced, because a reconstituted eukaryotic RNAP is not available (12). Thus, a detailed structure–function analysis of the RNAP II system is lacking. Here we used variants of a recombinant RNAP II-like archaeal RNA polymerase to investigate the functional role of the cleft loops rudder, lid, fork 1, fork 2 and the switch 2 element during various stages of transcription. We tested the polymerase variants in several assays, including promoter-dependent transcription (Figure 2), open complex formation (Figure 3), elongation of short RNA primers (Figure 4), de novo transcription from a pre-opened bubble (Figure 5) and transcription elongation with various nucleic acid scaffolds (Figure 6). The results of this study are summarized in Table 1. All mutants were impaired in these assays to various degrees, with the exception of fork 1 that is the only loop not present in the bacterial enzyme.
Table 1.
Comparison of the activities of the mutant enzymes relative to the reconstituted WT enzyme in various assays
| Run off transcription (Figure 2) | Open complex (Figure 3) | Abortive transcription (Figure 4) | Full-length transcript from open bubble (Figure 5B) | Elongation on double-stranded scaffold (EC3) (Figure 6B) | ||
|---|---|---|---|---|---|---|
| ≤40 nt | >40 nt | |||||
| Δ lid | – | + | – | – | + | – |
| Δ rudder | – | – | – | – | + | (+) |
| Δ fork2 | – | + | – | – | + | – |
| Δ fork1 | + | + | + | + | + | + |
| B -R445A | + | + | + | (+) | (+) | (+) |
| A′-R313A | – | + | (+) | – | (+) | – |
| A′-K306A | + | + | + | + | + (data not shown) | + (data not shown) |
(+) indicates strong impairment of activity, + good and – no activity.
Our analysis revealed that the rudder is important for stabilizing melted DNA in the open complex (Figure 3), consistent with formation of an inactive open complex by a bacterial rudder mutant (9). Initial transcription required the lid, rudder, fork 2 and switch 2 residue R313. The corresponding mutants were defective in extension of short RNA primers (Figure 4), and fork2 was incapable to synthesize longer RNAs from a pre-opened bubble (Figure 5). Our results are generally consistent with previous analysis of the bacterial rudder and lid, but some differences were also observed. During RNA elongation on the single-stranded template the archaeal enzyme was able to synthesize transcripts >40 nt (Figure 6C, lane 1) whereas the bacterial enzyme synthesized mainly 22–30 nt transcripts (11,10). The rudder was important for the maintenance of an actively elongating archaeal complex, whereas it seems less important in the bacterial system. The archaeal Δrudder enzyme was unable to reach the end of the template in preformed elongation complexes (Figure 6C), although the bacterial Δrudder enzyme is more processive and can produce full-length transcripts (9).
This work also provided insights into the crucial functions of fork loop 2. Fork 2 is strictly required for primed transcription and elongation, and a single-point mutation of its conserved arginine residue (R445A) showed a severe defect in elongation effectivity (Figure 6C, lane 13). This arginine is highly conserved in fork 2 (Figure 1A) and is located directly at the junction of the two DNA strands close to the point of downstream DNA separation (8,25). Several replacements in the eukaryotic fork 2, and mutations of sites interacting with fork 2 influence the elongation rate (12). Our data are consistent with an essential role of fork 2 in downstream DNA unwinding during elongation, most likely by interference with the path of the non-template strand as suggested by structural studies. However, the function of fork 2 is not restricted to downstream DNA unwinding. De novo RNA synthesis from a pre-melted bubble was totally impaired in the Δfork 2 variant (Figure 5B), suggesting that this loop is involved in stabilizing the NTPs during initial phosphodiester bond formation.
Unexpected insights into the transition from transcription initiation to elongation were obtained by the transcription assays with a pre-melted bubble (Figure 5). In the absence of initiation factors, run-off transcripts were synthesized in unprimed, de novo transcription. However, in the presence of initiation factors, RNA transcripts were generally limited to a length of 11–12 nt that were not displaced independent of the presence of a pre-formed bubble (Figure 5E). RNase H probing revealed that persistent DNA–RNA hybrids were formed under these conditions, and that the mismatch bubble prevented RNA displacement of the full-length transcript from the DNA template strand. Consistent with our findings, upstream re-closure of the transcription bubble begins when RNA has reached a length of 10–11 nt (21). The shorter RNA transcripts, however, were not induced by failure to extend the DNA–RNA hybrid past the lid, which normally stacks on the upstream end of the hybrid. Instead, they were due to the presence of the TBP–TFB complex. Synthesis of 11–12 nt RNAs in our assay must involve displacement of the TFB finger domain located in the hybrid site (26) but does apparently not entirely release TFB. TFB may remain bound to the dock with its N-terminal ribbon domain (26,27) but this alone cannot explain why RNA synthesis stops prematurely, since the ribbon domain would not interfere with the DNA–RNA hybrid emerging from the cleft between the polymerase protrusion, wall and clamp domains. Instead RNA synthesis may stop since the growing hybrid encounters the complex of TBP and the TFB core domain, situated above the cleft (27) when a length of 12–13 bp is reached. The existence of the 12–14 nt barrier on the duplex template (Figure 5E) that allows proper rewinding of upstream DNA indicates that the mismatch bubble is not the major cause of the first barrier. Furthermore, the ratio full-length to 11,12 nt transcripts is greatly increased when the length of duplex DNA downstream of the mismatch bubble was extended by 10 nt (Figure 5F). Taken together these results suggest that a minimal length of 27 nt of downstream duplex DNA are required for effective elongation past the barrier. Bubble reclosure at the upstream end of the open complex has been proposed as a key event in the promoter clearance transition and suggested to cause TFB-displacement (28) in the human RNAP II system, but is apparently not required for promoter escape and processive RNA synthesis in our system. The results with the preformed bubble described here are not specific for the archaeal enzyme. RNAP II from yeast shows similar properties on the pre-melted template analyzed in this study (C. Reich, S. Naji, J. Gerber, A. Küsser, H. Tschochner, P. Cramer, M. Thomm, manuscript in preparation) and both RNAP II (29) and the archaeal enzyme can transcribe ∼100 nt run-off transcripts with high effectivity when the pre-melted template contains longer segments of downstream duplex DNA (Figure 5F and Spitalny,P., Naji,S., Thomm,M. unpublished data).
Finally, we have uncovered the essential role of switch 2, and, in particular, an invariant arginine in switch 2, A′-R313, at various points during transcription. This arginine was required for chain elongation from the minimal nucleic acid scaffolds (Figure 6), for transcription from a pre-melted bubble (Figure 5) and for primed transcription (Figure 4). Thus, the interaction of this arginine with the DNA template strand backbone at position +2 is essential for correct template positioning in the active site. Further, these results provide insights into the structure of the open complex, since open complex formation apparently involves binding of the template in the active site at the location normally adopted during elongation, and thus leads to a complex prone to RNA chain initiation. Since mutation of R313 completely disabled RNA synthesis on templates containing downstream duplex DNA (Figure 6C), binding of the template strand to switch 2 is essential for DNA separation during elongation, most likely due to distortion of the incoming B-DNA duplex as suggested (8). Given the invariant nature of R313 in switch 2 in all three kingdoms of life, the same mechanisms and structural transitions will likely occur in all cellular RNA polymerases. The mutation of R313 directly reports on the function of this residue, and not on the function of switch 2 in general, since mutation of another switch 2 residue, K306, which is not conserved and more distant from the template and active site, has essentially no effects in most assays.
More generally, our analysis showed that the recombinant archaeal RNA polymerase can be used for a structure–function analysis of aspects of a eukaryote-like transcription mechanism, including initiation and the initiation–elongation transition. Many more mutant polymerases and different nucleic acid scaffolds, however, must be analyzed before a satisfactory mechanistic understanding of the dynamic transcription cycle will emerge.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants of the Deutsche Forschungsgemeinschaft to Michael Thomm and Patrick Cramer and the Leibniz award of the DFG to Patrick Cramer. We thank Bernd Goede for bioinformatic work. Funding to pay the Open Access publication charges for this article was provided by the University of Regensburg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cramer P, et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst S. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 4.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 5.Armache K-J, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 6.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 7.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 8.Kettenberger H, Armache K-J, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 9.Kuznedelov K, Korzheva N, Mustaev A, Severinov K. Structure-based analysis of RNA polymerase function: the largest subunit's rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 2002;21:1369–1378. doi: 10.1093/emboj/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 11.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 12.Trinh V, Langelier M-F, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol. Mol. Biol. Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner F, Weinzierl RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 14.Naji S, Grünberg S, Thomm M. The RPB/orthologue E′ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 2007;282:11047–11057. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 15.Pühler G, Leffers H, Gropp F, Palm P, Klenk HP, Lottspeich F, Garrett RA, Zillig W. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc. Natl Acad. Sci. USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goede B, Naji S, von Kampen O, Ilg K, Thomm M. Protein-protein interactions in the archaeal transcriptional machinery: binding studies of isolated RNA polymerase subunits and transcription factors. J. Biol. Chem. 2006;281:30581–30591. doi: 10.1074/jbc.M605209200. [DOI] [PubMed] [Google Scholar]
- 17.Kusser AG, Bertero MG, Naji S, Becker T, Thomm M, Beckmann R, Cramer P. Structure of an archaeal RNA polymerase. J. Mol. Biol. 2007 doi: 10.1016/j.jmb.2007.08.066. doi:10.1016/j.jmb.2007.08.066. [DOI] [PubMed] [Google Scholar]
- 18.Bell SD, Jackson SP. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 2001;4:208–213. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- 19.Thomm M, Hausner W. Transcriptional Mechanisms. In: Garrett R, Klenk H-P, editors. Archaea: Evolution, Physiology and Molecular Biology. Oxford, UK: Blackwell Publishing; 2006. pp. 185–198. [Google Scholar]
- 20.Hethke C, Geerling ACM, Hausner W, de Vos W, Thomm M. A cell-free transcription system for the hyperthermophilic Archaeon Pyrococcus furiosus. Nucleic Acids Res. 1996;24:2369–2376. doi: 10.1093/nar/24.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spitalny P, Thomm M. Analysis of the open region and of DNA-protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J. Biol. Chem. 2003;278:30497–30505. doi: 10.1074/jbc.M303633200. [DOI] [PubMed] [Google Scholar]
- 22.Kireeva ML, Komissarova N, Kashlev M. Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J. Mol. Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 23.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 24.Daube SS, von Hippel PH. Functional transcription elongation complexes from synthetic RNA-DNA bubble duplexes. Science. 1992;258:1320–1324. doi: 10.1126/science.1280856. [DOI] [PubMed] [Google Scholar]
- 25.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 26.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Ångstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 27.Chen H-T, Hahn S. Binding of TFIIB to RNA polymerase II: mapping the binding site fort he TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell. 2003;12:437–447. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 28.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 29.Holstege FCD, Tantin D, Carey M, van der Vliet PC, Timmers HThM. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.