Abstract
Chromatin immunoprecipitation (ChIP) is a powerful technique for studying protein–DNA interactions. Drawbacks of current ChIP assays however are a requirement for large cell numbers, which limits applicability of ChIP to rare cell samples, and/or lengthy procedures with limited applications. There are to date no protocols for fast and parallel ChIPs of post-translationally modified histones from small cell numbers or biopsies, and importantly, no protocol allowing for investigations of transcription factor binding in small cell numbers. We report here the development of a micro (μ) ChIP assay suitable for up to nine parallel quantitative ChIPs of modified histones or RNA polymerase II from a single batch of 1000 cells. μChIP can also be downscaled to monitor the association of one protein with multiple genomic sites in as few as 100 cells. μChIP is applicable to small fresh tissue biopsies, and a cross-link-while-thawing procedure makes the assay suitable for frozen biopsies. Using μChIP, we characterize transcriptionally permissive and repressive histone H3 modifications on developmentally regulated promoters in human embryonal carcinoma cells and in osteosarcoma biopsies. μChIP creates possibilities for multiple parallel and rapid transcription factor binding and epigenetic analyses of rare cell and tissue samples.
INTRODUCTION
Protein–DNA interactions are essential for genomic stability, DNA replication and repair, chromosome segregation, transcription and epigenetic silencing of gene expression. Chromatin immunoprecipitation (ChIP) is a powerful technique for studying interactions of DNA with proteins including transcription factors (1–3) and histones (4–6), whose post-translational modifications regulate gene expression (7–9). A drawback of current ChIP assays, however, is the requirement for large cell numbers (in the 106–107 range), which limits applicability of ChIP to rare cell samples such as small stem cell batches or tissue biopsies. A carrier ChIP protocol (CChIP) for a single histone immunoprecipitation from 100 cells under native conditions has been published, however it remains tedious and unsuitable for transcription factors (5). A recent fast ChIP protocol circumvents two cumbersome steps of a conventional ChIP assay (10,11), but it is suitable for large cell samples. We recently reported a quick and quantitative Q2ChIP assay suitable for up to 1000 histone immunoprecipitations or 100 transcription factor immunoprecipitations (including Oct4) from 100 000 cells (12).
Application of ChIP to stem cell and embryo biology requires a robust assay for immunoprecipitation of histones or transcription factors from small (hundreds or fewer) cell numbers. We report here a one-day micro (μ) ChIP assay suitable for up to nine parallel ChIPs of modified histones and/or RNA polymerase II (RNAPII) from a single batch of 1000 cells. The assay can be downscaled for monitoring association of one protein with multiple genomic sites in as few as 100 cells, and has been adapted for fresh or frozen tissue samples. The assay was validated by assessing several post-translational modifications of histone H3 and the binding of RNAPII in human embryonal carcinoma (EC) cells and in biopsies of human osteosarcoma xenografts, on the promoters of the developmentally regulated OCT4 and NANOG genes (13,14), the tissue-specific SLC10A6 gene (15) and the housekeeping gene GAPDH.
MATERIALS AND METHODS
Materials
Undifferentiated EC cells (NCCIT) were cultured as described (12). The osteosarcoma xenograft was established from a primary tumor of a male 7-year old. The tumor was evaluated as a Grade 4 Osteoblastic/Telangiectatic Osteosarcoma. Tumor tissue was grown subcutaneously on a nude mouse. The tumor was collected, washed in phosphate buffered saline (PBS) and dissected on ice into 1 mm3 pieces. The pieces were processed fresh or frozen in liquid nitrogen and stored at −80°C. Antibodies against H3K9ac (cat# 06-942), H3K9m2 (cat# 07-441), H3K9m3 (cat# 07-442) and H3K27m3 (cat# 05-851) were from Upstate (www.upstate.com). Anti-H3K9m3 antibodies used in biopsy ChIPs were from Diagenode (cat# pAb-056-050; www.diagenode.com). Antibodies to H3K4m2 (cat#Ab7766) and H3K4m3 (cat# Ab8580) were from Abcam (www.abcam.com). Antibodies to RNAPII were from Santa Cruz Biotechnology (cat# sc-899; www.scbt.com). Chelex-100 was from BioRad (cat# 142-1253; www.biorad.com). All other reagents were from Sigma-Aldrich (www.sigma-aldrich.com) unless otherwise stated.
μChIP assay
The basis of the μChIP assay was the Q2ChIP assay (12), with modifications. The LowCell# ChIP kit (Diagenode) was applied for the Q2ChIPs carried out here. Dynabeads Protein A (10 μl; Dynal Biotech; www.invitrogen.com) were added to 90 μl RIPA buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate, 100 mM NaCl) and 2.4 μg primary antibody in a 0.2 ml PCR tube and rotated at 40 rpm for 2 h at 4°C. PCR tubes were in the form of 8-tube strips handled in a magnetic rack (cat# kch-816-001; Diagenode) chilled on ice.
Chromatin preparation
Cells were harvested by trypsinization in presence of 20 mM butyrate and 1000 or 10 000 cells (depending on starting number; see Results) were re-suspended in 500 μl PBS/butyrate. The method described here is also applicable to cell numbers between 10 000 and 50 000.
We carried out an all-in-one tube preparation of chromatin in order to minimize exposure to plastic surfaces and sample handling, and reduce loss of material. Proteins and DNA were cross-linked with 1% formaldehyde for 8 min at room temperature and cross-linking was stopped with 125 mM glycine for 5 min. Cells were centrifuged at 470 g in a swing-out rotor with soft deceleration settings for 10 min at 4°C and washed twice in 0.5 ml ice-cold PBS/butyrate by gentle vortexing and centrifugation as above. Ten to twenty microliter of buffer was left with the pellet after removal of the last wash. Cells were lysed by addition of 115 µl room temperature lysis buffer [50 mM Tris–HCl, pH 8, 10 mM EDTA, 1% SDS, protease inhibitor cocktail (cat# P8340, Sigma-Aldrich), 1 mM PMSF, 20 mM butyrate] and subsequent vortexing. Cells were incubated on ice for 5 min, re-suspended by vortexing and sonicated for 3 × 30 s on ice with 30 s pauses, using a probe sonicator (Labsonic-M, 3-mm probe; cycle 0.5, 30% power; Sartorius AG; www.sartorius.com). This produced chromatin fragments of ∼500 bp. Alternatively, cells can be sonicated with a Bioruptor (Diagenode) (data not shown). The sonicated lysate was diluted 8-fold in RIPA buffer containing a protease inhibitor cocktail, 1 mM PMSF, 20 mM butyrate (RIPA ChIP buffer) to reduce SDS concentration to ∼0.1%. To this end, 480 µl RIPA ChIP buffer was mixed by vortexing with ∼120 μl lysate, the sample centrifuged at 12 000 g for 10 min at 4°C and the supernatant (chromatin) transferred to a chilled clean 1.5 ml tube. Care was taken to avoid aspiration of the sedimented material, thus ∼50 µl of the supernatant was left in the tube. RIPA ChIP buffer (330 μl) was added to the pellet (which is essentially invisible), mixed by vortexing and the sample spun as before. The supernatant was pooled with the first supernatant to yield a volume of chromatin suited for eight ChIPs and one input reference.
When starting from 100 cells, cells were cross-linked and washed as before and 20 µl PBS/butyrate was left with the washed pellet. Cells were lysed by mixing with 120 µl lysis buffer and a 3-min incubation, the nuclei were centrifuged at 860 g for 10 min and the supernatant discarded, leaving 20–30 µl lysis buffer in the tube (Note that leaving cells in lysis buffer for >3 min before centrifugation increases the chance of SDS precipitating. If this occurs, the lysis buffer is removed after centrifugation, 200 µl RIPA ChIP buffer are added, SDS is dissolved by vortexing and nuclei are pelleted as described). RIPA ChIP buffer (120 µl) was added, the tube vortexed thoroughly and cells sonicated as before but for 2 × 30 s. The resulting chromatin was pipetted several times with a siliconized tip before transfer into a 0.2 ml PCR tube containing beads pre-incubated with antibodies (see above) and from which the pre-incubation buffer (RIPA) was previously removed. A chromatin sample identical to that used in the ChIP was prepared to represent the input.
Immunoprecipitation and DNA recovery
Immunoprecipitation and washes of the ChIP material were as described (12). Chromatin was aliquoted into eight samples in 0.2 ml tubes containing antibody-bead complexes held to the wall by a magnet, and from which the pre-incubation buffer (RIPA) was removed, and one input sample (in a 1.5 ml tube for phenol-chloroform isoamylalcohol isolation, or 0.6 ml tube for Chelex-100-mediated DNA isolation; see subsequently). Beads were released into the suspension and rotated at 40 rpm for 2 h at 4°C. The ChIP material was washed three times by 4-min incubations in 100 μl of RIPA buffer and once in 100 μl of 10 mM Tris–HCl, pH 8.0, 10 mM EDTA (TE) buffer, and transferred into a new tube while in TE. DNA elution (1% SDS), cross-link reversal and proteinase K digestion (50 μg/ml) were carried out in a single step for 2 h at 68°C on a Thermomixer (Eppendorf; www.eppendorf.de). DNA was extracted with phenol-chloroform isoamylalcohol, ethanol-precipitated in presence of 10 μl acrylamide carrier (cat# A9099, Sigma-Aldrich) and dissolved in 100–300 μl TE (1000–100 000-cell ChIP) or 30–50 μl TE (100-cell ChIP). For ChIPs starting with 100 cells, DNA was purified using Chelex-100 (see subsequently).
Purification of ChIP DNA using Chelex-100
Chelex-100 mediated DNA purification was as described (10) with modifications for small samples and to speed up handling. Specifically, after transferring the washed ChIP material into a clean tube, the complex was centrifuged, TE was removed and 40 µl of 10% (wt/vol) Chelex-100 was added to the beads and vortexed for 10 s. Samples were boiled for 10 min in a PCR machine and cooled to room temperature before proteinase K (1 µl at 20 mg/ml) was added. Samples were vortexed, incubated at 55°C for 30 min at 1300 rpm on a Thermomixer, boiled for 10 min and quick-spun for 10 s. Tubes were then allowed to stand still for ∼1 min without a magnet to allow beads to settle. With a siliconized tip, 30 µl of the supernatant were transferred to a clean tube on ice. Ten microliter of MilliQ H2O were added to the remaining beads, the sample was vortexed and quick-spun for ∼3 s using a minifuge. After the beads settled, as much as possible of the supernatant (12–15 μl) was collected (volume must be consistent between tubes when ChIP samples are to be compared) and pooled with the first supernatant. The DNA sample was used for PCR or stored at −20°C. Input DNA from 100 cells was prepared using Chelex-100 as described earlier (10) except that 40 μl Chelex-100 beads were used and 10 μl acrylamide carrier was included in the ethanol precipitation. Purification of ChIP DNA with Chelex-100 can be applied to larger cell numbers (10) without affecting the ChIP PCR results compared to phenol-chloroform isoamyalcohol extraction (see Results).
Regardless of ChIP DNA isolation method, real-time PCR data were expressed as percent (±SD) precipitated (antibody-bound) DNA relative to input DNA, in two to nine independent replicate ChIPs.
μChIP from biopsies
Fresh and frozen osteosarcoma biopsies (∼1 mm3) were processed in a similar manner except that frozen biopsies were frozen without cross-linking and thawed in freshly prepared PBS containing 1% formaldehyde, 20 mM butyrate and protease inhibitors. Biopsies in PBS/formaldehyde/butyrate without (fresh biopsies) or with (frozen-thawed biopsies) protease inhibitors were vortexed and incubated for 10 min at room temperature. Fixation was terminated with 125 mM glycine for 5 min, the sample washed, and lysed by thorough resuspension by pipetting in 120 µl lysis buffer. The sample was incubated on ice and sonicated for 30 s. The lysate was centrifuged at 12 000 g for 10 min at 4°C and the supernatant transferred into a chilled 0.6 ml tube, leaving ∼30 μl of buffer with the pellet. This sedimentation step ensured removal of remaining tissue aggregates. Another 30 µl of lysis buffer was added and the tube vortexed. After centrifugation as before, ∼50 μl of the supernatant was pooled with the first supernatant and sonicated for another 2 × 30 s on ice. The sonicated chromatin sample was further processed for ChIP as described before (see Chromatin preparation and Immunoprecipitation and DNA recovery).
Polymerase chain reaction (PCR)
Immunoprecipitated DNA from 3–4 independent ChIPs was analyzed by duplicate quantitative (q)PCR as described (12). Reverse transcription (RT)–PCRs were performed from 500 ng total RNA using the Iscript cDNA synthesis kit (BioRad) and 30 cycles of 95°C 30 s, 60°C 30 s and 72°C 30 s. GAPDH, OCT4 and NANOG ChIP and RT–PCR primers were as described (12). SLC10A6 ChIP primers were 5′-TGGCTTTGGCTTTGTTGTTT-3′ and 5′-AGGCTTTCCACTTCTGTTCTT-3′ (228 bp amplicon) and RT–PCR primers were 5′-GGGTCCTCCTTCTGGTGGT-3′ and 5′-GCCAAGACTGGTGGGTAAAA-3′ (153 bp amplicon).
RESULTS
Set up and validation of μChIP
To test and validate the μChIP assay, we monitored the association of H3K9ac (a modification indicative of transcriptional activity), H3K4m2 and H3K4m3 [modifications associated with promoters but not necessarily indicative or predictive of transcriptional activity (16,17)] and of H3K9m2, H3K9m3 and H3K27m3 (three transcriptionally repressive marks) with the GAPDH, OCT4 and NANOG promoters in NCCIT cells (Figure 1A). RT–PCR analysis shows that GAPDH, OCT4 and NANOG were expressed (Figure 1B). Starting material was for each ChIP, either chromatin from 100 000 cells, chromatin from ∼1000 cells prepared from a starting 10 000 cells and divided into nine samples (‘1000-cell ChIP’), or chromatin from ∼100 cells prepared from 1000 cells and divided into nine samples (‘100-cell ChIP’). Among the nine samples, eight were dedicated to ChIP and one served as input chromatin reference.
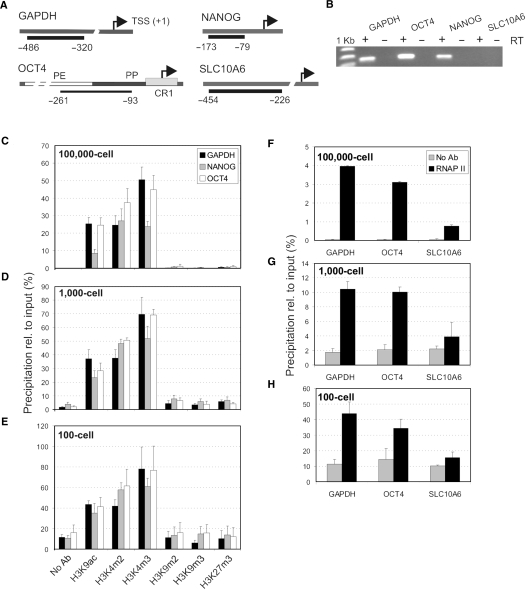
Figure 1.
Development and validation of μChIP. (A) Genomic regions examined. Bars and numbers indicate the position of ChIP amplicons relative to the transcription start site (TSS). PE, PP and CR1 delineate the proximal enhancer, proximal promoter and conserved region 1 of OCT4, respectively. (B) RT-PCR analysis of expression of GAPDH, OCT4, NANOG and SLC10A6 in NCCIT cells. Plus (+) and minus (−) indicate the presence or absence of reverse transcriptase (RT). (C–E) ChIP analysis of H3 modifications (x-axis) in NCCIT cells on promoters of indicated genes. (C) Analysis from chromatin of 100 000 cells, (D) 1000 cells and (E) 100 cells. (F–H) ChIP analysis of RNAPII binding to GAPDH, OCT4 and SLC10A6 promoters in the same chromatin samples as those examined for histones in (C–E). In (C–H), data are expressed as percent precipitation relative to input chromatin (mean ± SD; 2–9 independent experiments).
Quantitative PCR analysis of the ChIP DNA shows that with chromatin from 100 000 cells, H3K9ac, H3K4m2 and H3K4m3 were enriched on all promoters, whereas repressive histone marks were at background level (Figure 1C). This was in agreement with activation of the genes in NCCIT cells (Figure 1B) and with previously published Q2ChIP data (12). Remarkably, the 1000-cell ChIP (Figure 1D) and the 100-cell ChIP assays (Figure 1E) produced a similar histone enrichment profile as with 100 000 cells, indicating that scaling down by 1000-fold maintained specificity of the assay.
We consistently noted however, enhanced precipitation, relative to input, from small chromatin samples (compare y-axis scales in Figure 1C–E). This is suggested to be due to an increased antibody/bead-to-antigen ratio. Indeed, we showed that reducing antibody concentration to 40% relative to a standard Q2ChIP condition reduced the PCR signal by ∼50% while a 4-fold increase in antibody concentration enhanced the PCR signal by >30% (Supplementary Data, Figure 1). Nevertheless, while the ratio of antibody-to-chromatin increases 1000-fold between the 100 000-cell and the 100-cell ChIP assay, the ChIP efficiency (precipitation relative to input) increased by 1.5–2-fold. This indicates that the relationship between antibody concentration and ChIP efficiency is non-linear in the range examined and that with low amounts of chromatin, the antibody concentration is not limiting. The relative background signal also increased when scaling down ChIP (Figure 1D and E); however this did not affect the overall interpretation of the results (see Discussion). These results indicate therefore that μChIP maintains the profile of histone modifications detected in larger scale assays.
μChIP is suitable for detection of RNAPII binding to genomic sites
We also examined whether μChIP was applicable to assess transcription factor (RNAPII) binding to DNA. Engagement of RNAPII to target promoters has been used as an indicator of transcriptional activity or potential for activity in genome-wide epigenetic studies (3,18). RNAPII ChIPs were carried out in parallel to histone ChIPs presented before, from the same chromatin preparations (Figure 1F–H). RNAPII association with GAPDH and OCT4 promoters was clearly detected under each condition. As negative control for RNAPII binding, we monitored RNAPII association with the inactive SLC10A6 promoter (Figure 1B). Binding was dramatically weaker than with GAPDH and OCT4 and consistently near background levels (Figure 1F–H). We concluded that μChIP is suited for analysis of histone and RNAPII binding to genomic loci in parallel immunoprecipitations from chromatin from as few as 100 cells. Starting with chromatin from 10 cells did not yield consistent results (data not shown) indicating that the current reliable limit of μChIP for assessing RNAPII binding to promoters is chromatin from 100 cells.
μChIP is applicable to 100 cells as starting material
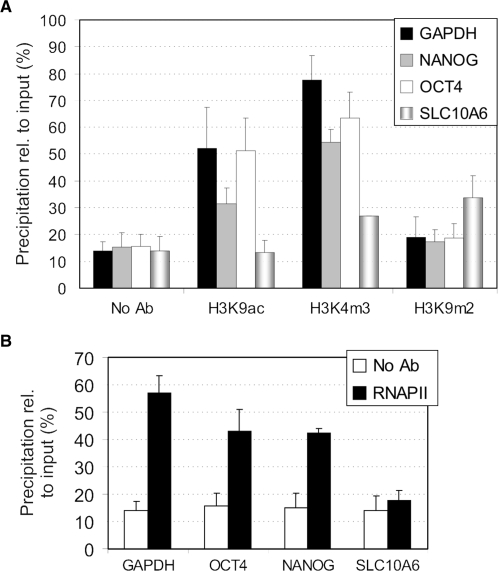
To determine whether μChIP was applicable to immunoprecipitation of histones and non-histone proteins from even smaller cell samples, chromatin was prepared from separate batches of 100 NCCIT cells. DNA was isolated from the ChIP material using Chelex-100 in order to minimize sample handling when very little material is present. We have shown that using Chelex-100 or phenol-chloroform isoamylalcohol to purify ChIP DNA does not affect ChIP results (Supplementary Data, Figure 2). Chelex-100-mediated DNA isolation also enhances DNA recovery (data not shown). Figure 2A shows that profiles of H3K9ac, H3K4m2 and H3K9m2 association with GAPDH, OCT4 and NANOG promoters in these samples were remarkably similar to those produced from those of a split chromatin sample from 1000 cells (compare with Figure 1E). In addition, μChIP enabled the detection of H3K9m2 enrichment on the SLC10A6 promoter, in agreement with its inactive state in NCCIT cells (Figure 2A). Moreover, RNAPII was successfully immunoprecipitated from 100-cell samples and found to be enriched on the GAPDH, OCT4 and NANOG promoters, but not on the silent SLC10A6 promoter (Figure 2B). Therefore, μChIP is compatible with immunoprecipitation of histones and RNAPII from as few as 100 cells as starting material.
Figure 2.
μChIP is suitable for analysis of histone or RNAPII binding in as few as 100 cells. (A) ChIP analysis of H3K9ac, H3K4m3 and H3K9m3 in separate 100-cell samples for each antibody, and for a no-antibody (No Ab) control. (B) ChIP analysis of RNAPII association with the GAPDH, NANOG, OCT4 and SLC10A6 promoters 100-cell samples. Two to four ChIPs were performed with each antibody. Data are expressed as in Figure 1.
Analysis of histone modifications and RNAPII binding in biopsies
Increasing evidence for a role of epigenetics in aging and in the development of cancer (19) reinforces the need for a method allowing detailed epigenetic and transcription factor binding studies in small tissue samples (20). Many tumor samples exist today in the form of stored frozen biopsies, not shown to date to be suited for ChIP analysis. Only limited ChIP studies have to date been reported using human lung, liver or post-mortem brain tissue (21–23), presumably due to the lack of an efficient ChIP protocol. In these and in mouse studies (24,25), tissue samples were relatively large, from >125 mm3 to processed large organ samples or organ slices.
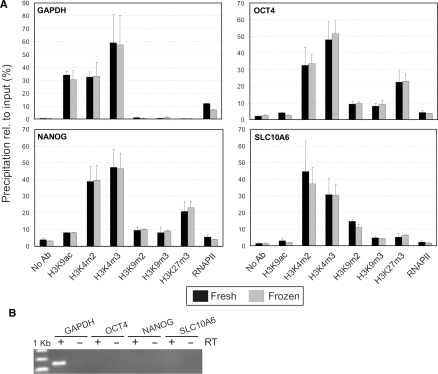
We determined whether μChIP was applicable to small tissue samples. We examined fresh and deep-frozen (−80°C) ∼1 mm3 biopsies (duplicates of each) from a human osteosarcoma xenograft. Chromatin from fresh biopsies was prepared as described under Materials and Methods. For frozen biopsies, a cross-link-while-thawing step in presence of 20 mM butyrate and protease inhibitors was introduced to preserve histone acetylation and intermolecular interactions in the dead tissue while thawing. The rest of the procedure was as for fresh samples. From each sample, six histone ChIPs, one RNAPII ChIP and one negative control ChIP were performed in parallel. PCR signal intensity from input chromatin for each gene examined indicated that total chromatin recovered from each biopsy was from ∼3000 cells (data not shown). Figure 3A shows histone H3 modifications and RNAPII binding patterns on the GAPDH, NANOG, OCT4 and SLC10A6 promoters, in relation to gene expression assessed by RT–PCR (Figure 3B). Identical results were obtained from fresh and frozen-thawed samples. Specifically, the active GAPDH promoter was clearly enriched in H3K9ac, H3K4m2 and K3K4m3, while none of the repressive marks was detected (Figure 3A). Consistent with these data, RNAPII also occupied the GAPDH promoter. The embryonic genes OCT4 and NANOG were not expressed in osteosarcomas and accordingly, promoters were enriched in H3K9m2, H3K9m3 and in particular H3K27m3. RNAPII was detected only at background levels on these promoters, corroborating their inactive state. The SLC10A6 promoter, also inactive in the osteosarcoma samples, did not show significant H3K9ac but was enriched in H3K9m2 and to a lesser extent H3K9m3 and H3K27m3 (Figure 3). All loci examined were enriched in di- and trimethylated H3K4 regardless of transcriptional status, in agreement with claims that H3K4m3 marks promoters regardless of activity (16,17).
Figure 3.
μChIP analysis of the association of modified histones and RNAPII to GAPDH, OCT4, NANOG and SLC10A6 promoters in human osteosarcoma biopsies. (A) ChIP analysis of fresh and frozen/thawed tissue samples (data from duplicate ChIPs from each sample). (B) RT-PCR analysis of expression of the respective genes (30 PCR cycles). Plus (+) and minus (−) indicate the presence or absence of reverse transcriptase (RT).
Altogether, these results indicate that seven different modified histones and RNAPII can be immunoprecipitated, along with a negative control ChIP, from ∼1 mm3 tissue samples, either fresh or stored frozen. The results provide an epigenetic profile on genomic loci that are compatible with transcription status. Under these conditions, the amount of DNA recovered from ChIP on biopsies is sufficient for examination of at least 20 genomic sites in duplicate qPCRs. Note that alternatively, chromatin prepared from tissue samples can be diluted further to enable more ChIPs, with however, fewer PCRs per ChIP.
DISCUSSION
We report here a μChIP assay suitable for up to nine parallel ChIPs from a single 1000-cell sample. Up to 50 μChIPs can be performed in 1 day. The assay is applicable to fresh or frozen tissue samples and is suitable for immunoprecipitation of both modified histones and of a transcription factor such as RNAPII. μChIP also enables the analysis of as few as 100 cells without a need for carrier chromatin as reported previously in the CChIP assay (5). Under these conditions, only one immunoprecipitation is possible per chromatin sample, as in CChIP. However, unlike CChIP, formaldehyde cross-linking makes μChIP suitable for the analysis of both histone and non-histone proteins. Although cross-linking is not required for immunoprecipitating histones (4,5), it makes the protocol more versatile than native ChIP and, as shown here, is compatible with efficient immunoprecipitation.
The specificity of larger scale ChIP assays is maintained with μChIP, as shown by the conservation of histone enrichment profiles on the promoters examined as the amount of input chromatin is decreased. Notably, μChIP precipitates more material (determined as a percentage of input) than standard ChIP, as illustrated by higher ChIP efficiency. A potential limitation, however, is that μChIP also results in enhanced background. This can be explained by the reduction in the amount of input chromatin while the total antibody-coupled bead surface area (the most likely source of unspecific chromatin binding) in the assay remains constant. An implication of increased background in low cell number ChIP is that the ratio of signals, e.g. H3K9ac over H3K9m2 decreases by ∼5-fold relative to 100 000-cell ChIP when the data are presented including background (compare, e.g. Figure 1C with E). Nevertheless, all modified histone binding profiles on the loci examined are maintained under each condition: H3K9 acetylated promoters remain clearly marked as acetylated, with no (or background) levels of H3K9 and H3K27 methylation. These arguments also hold true for RNAPII binding patterns on the various promoters examined (Figure F–H). It is likely that reducing cell numbers in ChIP assays systematically results in enhanced background [see also (12)], an issue simply dealt with recently in a miniChIP assay by subtracting background values (26). Clearly, a background subtraction in Figures 1 and 2 in the present article would produce histone enrichment profiles identical to those of 100 000-cell ChIP assays; however, we favor in this communication the presentation of all data to emphasize awareness of the background issue. Biological interpretation of the data remains unaffected.
Carrying out up to nine parallel ChIPs from a common chromatin preparation from 1000 cells enhances the consistency of the amount of input chromatin in each sample, making comparisons between ChIPs more robust than comparisons between single ChIPs from separate (100-cell) samples. With chromatin from 100 cells, four genomic sites can be examined in duplicate qPCRs on non-amplified DNA, and with 1 mm3 biopsies over 20 sites can be analyzed in duplicate qPCRs. The use of increasingly sensitive commercial DNA amplification kits should allow the analysis of more genomic sites and possibly of the whole genome with microarrays or by direct sequencing. This remains to be determined. One should nevertheless note that the amplitude of standard deviations from qPCR assays increases as cell numbers decrease. This is likely because the amount of target genomic sequence is closer to the detection limit by real-time PCR, so minor variations in the number of DNA template molecules for PCR translate into enhanced variability. The number of template molecules per PCR is so low that inaccessibility of one molecule in the first PCR cycle can affect final quantification. Increased apparent variation between replicates in 100-cell ChIP assays, therefore, reflects technical artifacts more than biological variation.
The demonstration of successful RNAPII immunoprecipitation with μChIP creates possibilities for examining other DNA-bound proteins. For example, detection of polycomb-group repressors on target genes, which correlates with H3K27 trimethylation on developmentally regulated genes in embryonic stem cells (3,27), should be possible from small amounts of human embryonic stem cells or even embryos. Additionally, the rapidity of the assay [see also (10)] may conceivably promote its implementation in routine epigenetic examinations of human material. However, because real-time PCR signals are weak, it remains to be determined how μChIP on 100 cells would perform in instances of precipitation of low abundance proteins or transiently bound proteins. We have previously shown that ChIP of the embryo-specific transcription factor Oct4 from chromatin from as a few as 1000 cells is possible (12), a number which at present constitutes the lowest reliable limit for this factor (our unpublished data).
The μChIP assay has been applied to small tumor samples. A limited number of studies have reported ChIP assays from human (21–23) or murine (23–25,28) tissue samples; however, these were significantly larger than the 1 mm3 samples used here. Furthermore, in these studies, ChIP protocols were conventional long protocols and no more than one histone modification or DNA binding protein was examined. μChIP allows parallel analysis of several histone modifications and RNAPII binding in single biopsies, and this from either fresh or frozen samples. Because μChIP can be applied to scarce biological material, it is anticipated to enable the long awaited clinical relevance of epigenetic assessments using precious cell or tissue samples (20).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Leif C. Lindeman for assistance with primer testing and Drs Leonardo Meza-Zepeda and Ola Myklebost (Rikshospitalet-Radiumhospitalet Medical Centre, Oslo) for providing osteosarcomas. This work was supported by the FUGE, YFF, STAMCELLE and STORFORSK programs of the Research Council of Norway. Funding to pay the Open Access publication charges for this article was provided by The Research Council of Norway.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Neill LP, Turner BM. Immunoprecipitation of chromatin. Methods Enzymol. 1996;274:189–197. doi: 10.1016/s0076-6879(96)74017-x. [DOI] [PubMed] [Google Scholar]
- 5.O’Neill LP, Vermilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 6.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 11.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2–e8. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- 13.Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 14.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 15.Geyer J, Doring B, Meerkamp K, Ugele B, Bakhiya N, Fernandes CF, Godoy JR, Glatt H, Petzinger E. Cloning and functional characterization of human sodium-dependent organic anion transporter (SLC10A6) J. Biol. Chem. 2007;282:19728–19741. doi: 10.1074/jbc.M702663200. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 19.Laird PW. Cancer epigenetics. Hum. Mol. Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 20.Kiermer V. Embryos and biopsies on the ChIP-ing forecast. Nat. Methods. 2006;3:583. doi: 10.1038/nmeth0806-583. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Ito M, Elliott WM, Cosio B, Caramori G, Kon OM, Barczyk A, Hayashi S, Adcock IM, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N. Engl. J. Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 22.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, MacDonald M, Fossale E, Zeitlin S, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy at its target genes in Huntington's disease. J. Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J. Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J. Biol. Chem. 2007;282:19071–19081. doi: 10.1074/jbc.M607486200. [DOI] [PubMed] [Google Scholar]
- 26.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl Acad. Sci USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 28.Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl Acad. Sci USA. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.