Abstract
All nuclear RNA polymerases are phosphoprotein complexes. Yeast RNA polymerase I (Pol I) contains approximately 15 phosphate groups, distributed to 5 of the 14 subunits. Information about the function of the single phosphosites and their position in the primary, secondary and tertiary structure is lacking. We used a rapid and efficient way to purify yeast RNA Pol I to determine 13 phosphoserines and –threonines. Seven of these phosphoresidues could be located in the 3D-homology model for Pol I, five of them are more at the surface. The single phosphorylated residues were systematically mutated and the resulting strains and Pol I preparations were analyzed in cellular growth, Pol I composition, stability and genetic interaction with non-essential components of the transcription machinery. Surprisingly, all Pol I phosphorylations analyzed were found to be non-essential post-translational modifications. However, one mutation (subunit A190 S685D) led to higher growth rates in the presence of 6AU or under environmental stress conditions, and was synthetically lethal with a deletion of the Pol I subunit A12.2, suggesting a role in RNA cleavage/elongation or termination. Our results suggest that individual major or constitutively phosphorylated residues contribute to non-essential Pol I-functions.
INTRODUCTION
Phosphoprotein complexes are involved in many biological processes. Although reversible phosphorylation at distinct sites and their consequences for complex function was extensively studied in many cases, a systematic functional and structural analysis of the predominant or stoichiometric phosphorylated residues within multiprotein complexes is rare. All three nuclear RNA polymerases are phosphoprotein complexes (1–3). Phosphorylation and function of the C-terminal domain of the largest Pol II subunit were analyzed in detail (4), but not much is known about the positions and roles of phosphorylation sites in other Pol I, Pol II and Pol III subunits. Yeast RNA Pol I consists of 14 subunits, 5 of which are phosphorylated in vivo (1–3): A190, the largest Pol I subunit, comprising part of the active site (5,6); A43, the interaction partner of the Pol I-specific transcription factor Rrn3p (7); the non-essential subunit A34.5, which interacts with the non-essential subunit A49 and was suggested to be involved in RNA elongation (8,9); ABC23 (= Rpb6), a subunit common to all three RNA polymerases, which is involved in enzyme assembly (10,11) and forms the main interaction interphase for the A43/A14 heterodimer (12,13); and AC19, one of the α-like subunits, shared by Pol I and Pol III, which is crucial for the assembly of the polymerase core (6,14,15). Although mammalian rRNA synthesis is mainly regulated by phosphorylation of Pol I-specific transcription factors (16,17), our previous analyses suggested that in yeast also the phosphorylation state of Pol I itself influences transcription of rRNA genes (18,19). Since its activity to synthesize rRNA is required for growth, Pol I represents an appropriate model both to investigate systematically in vivo the meaning of the major phosphorylated residues of a large phosphoprotein complex and to learn about their role in Pol I-activity. Furthermore, rRNA synthesis responds extremely sensitive to any slight changes in environmental conditions through signal transduction pathways. Thus, it should be possible to get insights whether and how the major phosphosites function in transcription regulation. Recent developments in mass spectrometry allowed to determine large numbers of phosphopeptides within complex protein mixtures (20,21). However, only few phosphopeptides of the nuclear RNA polymerases could be determined in these proteome-wide analyses (22–24). To obtain a more precise picture, we employed phosphopeptide mapping from purified polymerase fractions. We developed a new strategy to purify Pol I to homogeneity on a large scale. Using LC-MALDI-TOF/TOF mass spectrometry in combination with a chemical derivatization procedure (25,26), it was possible to determine the major phosphorylated Pol I residues, i.e. 13 phosphoserines and -threonines on the five subunits that are phosphorylated in vivo. Approximate or exact positions of the phosphorylated residues were located in either the homology model for the Pol I core enzyme or in the crystal structure of the Pol I subcomplex A14-A43 (9). After systematically replacing the phosphorylated amino acids by either alanine or aspartate to mimic a constitutively unphosphorylated or phosphorylated state, respectively, the resulting mutant strains were investigated in growth ability, Pol I subunit stability and —stoichiometry as well as in genetic interactions with non-essential components of the Pol I-transcription machinery. Surprisingly, our analyses demonstrate that most of the individual predominant or stoichiometrically phosphorylated residues of Pol I have no direct influence in Pol I assembly or essential steps of rRNA synthesis. Only one of the abundant Pol I-phosphorylation sites was found to play a detectable role in Pol I-activity. The same mutation was also the only one detected to genetically interact with another constituent of the Pol I machinery, the non-essential subunit A12.2. Our analyses suggest that reversible modification of A190 S685 is involved in RNA cleavage/elongation and/or termination.
MATERIALS AND METHODS
Pol I purification
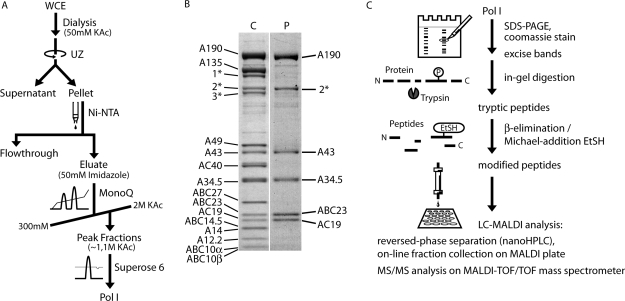
A purification scheme is shown in Figure 1A and Supplementary Fig. S1A. Strain GPY2, carrying the gene for an N-terminal His6/HA-tagged A43 on a plasmid was cultivated to OD600 = 5–6 at 30°C in YPD media in a 200 l fermenter (Infors ABEC). The cells were harvested by continuous flow centrifugation (Padberg) and lysed by bead beating in ice-cold buffer A (400 mM (NH4)2SO4, 60 mM MgCl2, 150 mM HEPES pH 7.8, 30% glycerol, 5 mM DTT, 1 mM PMSF, 1 mM benzamidine, 200 μM pepstatin, 60 μM leupeptin). All subsequent steps were performed at 4°C. Glass beads were separated by filtration prior to clearing the lysate by centrifugation (30 min, 8000g, Sorvall SLA-1500). The final whole cell extract (WCE) was obtained after centrifugation at 100 000g for 90 min (Beckman SW28) by separating the clear middle phase from the turbid lower phase. The WCE was dialyzed against buffer B (50 mM KAc, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 mM benzamidine) over night, using Spectra/Por 2 dialysis tubing (MWCO 12–14 kDa) (Spectrum Laboratories). After centrifugation at 30 000g for 1 h (Beckman Ti45), the pellet was resuspended in buffer C (1.5 M KAc, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10% glycerol, 1 mM PMSF, 1 mM benzamidine) and applied to 4 ml Ni-NTA Agarose (Qiagen) per 150 g of cells. Binding was performed in batch for 4 h on a turning wheel, before pouring the suspension into disposable Econo-Pac™ columns (Bio-Rad) for subsequent wash-steps with buffer C and buffer D (300 mM KAc, 20 mM HEPES pH 7.8, 1 mM MgCl2, 10% glycerol) and elution with buffer E (buffer D + 100 mM imidazole). The eluting proteins were loaded onto a Mono Q anion exchange column (Mono Q™ 10/100 GL, GE Healthcare) and fractionated by applying a salt gradient from 300 mM to 2 M KAc with buffers F (20 mM HEPES pH 7.8, 1 mM MgCl2, 10% glycerol) and G (buffer F + 2 M KAc). The pooled Pol I containing fractions (eluting at 1.1 M KAc) were either directly applied to the gelfiltration chromatography step (Figure 1A and B) or, to obtain the highest purity, subjected to cation exchange chromatography (Supplementary Fig. S1). To enable binding to the Mono S column (Mono S™ 5/50 GL, GE Healthcare), the Mono Q eluate was diluted to a salt concentration of 200 mM KAc with buffer F, loaded onto the column and fractionated by applying a gradient from 200 mM to 2 M KAc using buffers F and G, with Pol I eluting at 490 mM KAc. Finally the combined Pol I peak fractions from either the Mono Q or the additional Mono S chromatography were concentrated to a volume of 500 µl using an Amicon™ Ultra–4 centrifugal filter unit (MWCO 100 kDa, Millipore) and applied to gelfiltration chromatography on a Superose 6 column (Superose™ 6 10/300 GL, GE Healthcare) with buffer H [60 mM (NH4)2SO4, 5 mM HEPES pH 7.8, 1 mM MgCl2, 10 µM ZnCl2, 5 mM DTT].
Figure 1.
RNA polymerase I purification and identification of phosphorylation sites. (A) Pol I purification scheme. See Supplementary Figure S1 for the extended scheme, including an additional cation exchange chromatography step. (B) Coomassie staining of purified Pol I [C] and phosphoprotein staining of the same gel [P] using phosphostain ProQ Diamond (Invitrogen). Three proteins of an eIF3 subcomplex co-purified with Pol I, namely Rpg1p (1*), Nip1p (2*) and Prt1p (3*). (C) Scheme of the strategy used for the identification of phosphorylation sites from purified Pol I.
Phosphoprotein staining
SDS–polyacrylamide gels were stained with Pro–Q™ Diamond Phosphoprotein stain (Invitrogen) according to the manufacturer's instructions and visualized on a 315 nm UV transilluminator. The same gel was then stained with SimplyBlue™ SafeStain (Invitrogen) for total protein staining.
Identification of phosphorylation sites
Following in-gel digestion, ∼37.5 pmol of the tryptic peptides of any phosphorylated Pol I subunit were used for the chemical derivatization at a time. The sulfhydryl groups of the cysteine residues were protected by reduction with 10 mM DTT at 37°C and carboxymethylation with 35 mM Iodoacetic acid at 24°C for 1 h each. The β–elimination and Michael-addition reactions were performed in a single step by incubation of the alkylated peptides in the presence of 64.5 mM Ba(OH)2 [taken from a freshly prepared saturated Ba(OH)2 solution] with either 450 mM ethanethiol or 400 mM pentanethiol in 30% acetonitrile at 50°C for 90 min. The reactions were stopped by precipitation of BaCO3 after the addition of NH4HCO3 to the final concentration of 100 mM. Residual alkanethiols, NH4HCO3 and acetonitrile were removed by lyophilization. The modified peptides were either analyzed after desalting with ZipTip™ C18 pipette tips (Millipore) as described for protein identification (Supplementary Methods) or fractionated via C18 reversed phase liquid chromatography. The latter was performed using a Dionex UltiMate™ nanoHPLC applying a gradient from 15 to 60% acetonitrile/0.05% TFA at a flow rate of 300 nl/min within 90 min. Fractions were mixed with CHCA matrix and collected directly on the MALDI-target using a Dionex Probot™ system. All samples were analyzed on an Applied Biosystems 4700 Proteomics Analyzer™ MALDI-TOF/TOF mass spectrometer. Identification of the originally phosphorylated residues is based on the unique mass shift of +44.0085 Da (for ethanethiol) or +86.0554 Da (for pentanethiol) compared to the expected mass of an unmodified serine or threonine. A customized Pol I sequence database (including trypsin and all co-purifying proteins) was used to allow more variables in the search parameters and the results were confirmed by searching against the NCBInr protein sequence database.
RESULTS AND DISCUSSION
Identification and 3D-localization of major Pol I-phosphosites
To generate homogenous Pol I on large scale we modified a previously developed purification protocol for active Pol I initiation complexes. We took advantage of the fact that soluble Pol I complexes in WCEs can be precipitated by dialysis against buffers with low ionic strength (27,28). This precipitation step reduces significantly the volume of the Pol I containing fraction and, thus, facilitates large-scale purification. Subsequent purification of His-tagged Pol I (N-terminal His-tag on subunit A43) on Ni-Agarose, chromatography on MonoQ and MonoS, and gelfiltration on Superose 6 yielded an overall recovery of 5 mg Pol I from 200 l yeast culture (Figure 1A and B and Supplementary Fig. S1). The purity of this Pol I fraction (Supplementary Fig. S1B) enabled structure determination at 12 Å by cryo electron microscopy (9). The quality of the purified Pol I was also appropriate to determine Pol I-specific phosphopeptides. Staining of purified Pol I fractions using the phosphostain Pro-Q Diamond (Invitrogen) revealed that all five in vivo phosphorylated subunits (1–3,29) remained phosphorylated after purification (Figure 1B). The purified Pol I was tested to be active in non-specific transcription assays (data not shown).
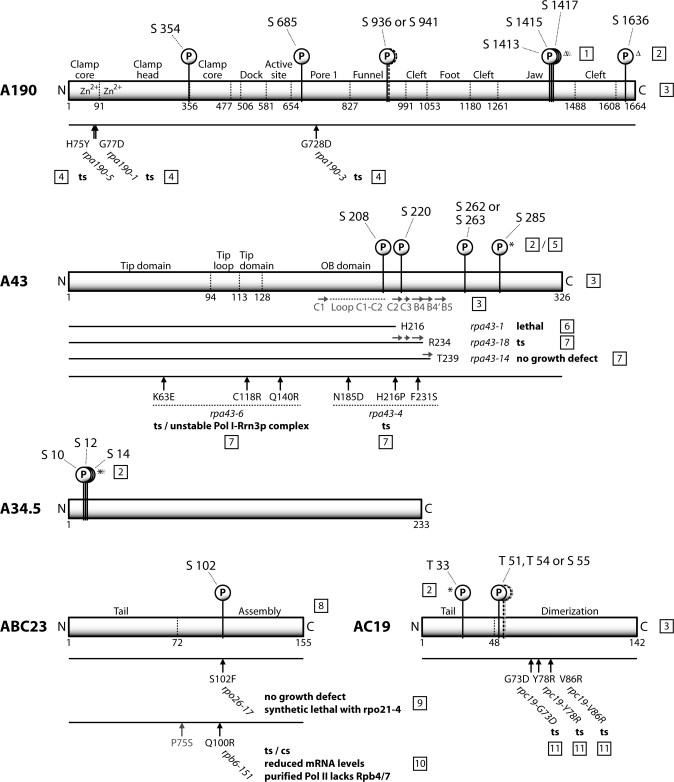
To identify phosphorylated peptides after tryptic digestion of the single Pol I subunits, we used a chemical derivatization procedure to replace the phosphogroups by either ethanethiol (S-EtSH) or pentanethiol (S-PeSH) (25,26). In combination with C18 reversed phase liquid chromatography, this method overcomes some of the problems associated with the analysis of phosphorylated peptides in MALDI–MS, like the neutral loss of H3PO4 and suppression effects (30), while increasing the ionization efficiency of the modified peptides (26,31). The originally phosphorylated serines or threonines could then be identified by their additional mass of +44.0085 Da (S-EtSH) or of +86.0554 Da (S-PeSH) using MALDI-TOF/TOF-MS (Figure 1C). We identified 11 phosphopeptides (Table 1), including a total of 13 phosphoserines and –threonines; four on subunit A43, three on each of the subunits A190 and A34.5, two on subunit AC19, which is shared in Pol III, and one on subunit ABC23 (= Rpb6), which is common to all three RNA polymerases. Our results complement the identification of phosphoserines S1413/S1415/S1417 (22) and S1636 on subunit A190 (24) in proteome-wide large-scale approaches. These recent analyses also confirmed one of the four herein described phosphoserines on subunit A43 (S285), the triple cluster on A34.5 (S10/S12/S14) and one of two phosphothreonines found in subunit AC19 (T33), using a completely different methodology (23,24). The positions of the phosphosites in the primary structure with regard to the domain organization of the subunits and in the context of published mutations that lead to a growth phenotype are indicated in Figure 2. Overall, the number of the so far identified phosphosites approximately matches the calculated 15 ± 3 phosphate groups per enzyme and the average number of in vivo phosphorylated residues (6 for A190, 4 for A43, 2 for A34.5, 1 or 2 for ABC23, 1 or 2 for AC19, respectively) (29), indicating that most of the constitutively or strongly phosphorylated sites could be determined. However, it is possible that additional phosphosites exist, which have not been detected yet, either due to methodological constraints or due to limited cellular amounts of the respective Pol I population, like the initiation active Pol I-Rrn3p complex.
Table 1.
Identified Pol I phosphopeptides and –sites and corresponding amino acids in the Pol I homology model
| Subunit | Phosphopeptide | 3D-localization | ||
|---|---|---|---|---|
| Position | Sequencea | Phosphosite | Amino acid in homology modelb | |
| A190 | 352–366 | ADSFFMDVLVVPPTR | S354 | Rpb1 W234 |
| 684–689 | DSFFTR | S685 | Rpb1 T539 | |
| 935–955 | GSNVNVSQIMCLLGQQALEGR | S936 or S941c | Rpb1 S754/A759 | |
| A43 | 207–212 | FSFGNR | S208 | N/D |
| 213–228 | SLGHWVDSNGEPIDGK | S220 | A43 S220 | |
| 242–264 | VVSVDGTLISDADEEGNGYNSSR | S262 or S263c | N/D | |
| 278–289 | IVFDDEVSIENK | S285 | N/D | |
| A34.5 | 7–31 | DYVSDSDSDDEVISNEFSIPDGFKK | S10/S12/S14 | N/D |
| ABC23 | 98–119 | ALQISMNAPVFVDLEGETDPLR | S102 | ABC23 S102 |
| AC19 | 21–46 | HIQEEEEQDVDMTGDEEQEEEPDREK | T33 | Rpb11 A3 |
| 49–77 | LLTQATSEDGTSASFQIVEEDHTLGNALR | T51 or T54 or S55c | Rpb11 I21/D24/T25 | |
aPhosphoserines/-threonines are presented as bold, underlined characters.
bSome phosphosites could not be localized in the Pol I homology model due to weak homology of the respective protein regions to Pol II, missing homologous subunits or deletions in the A43/14 crystal structure.
cThree phosphopeptides could be identified due to the specific mass shift after chemical derivatization and a partial MS/MS spectrum, but the phosphorylation site could not be unambiguously assigned. The remaining possible serines and threonines are shown in bold, italic letters in the peptide sequence.
Figure 2.
Phosphorylation sites of Pol I. Positions of the identified Pol I phosphorylation sites in the primary structure of the five phosphorylated subunits. Phosphorylation sites marked with an asterisk were confirmed by proteome-wide, large-scale phosphorylation analyses (see text). Sites identified exclusively by proteome-wide analyses are marked by a triangle. The domain organization of the subunits and the positions of amino acid exchanges of known Pol I mutants are shown, as well as a part of the secondary structure of A43. References: [1] (22); [2] (24); [3] (9); [4] (53); [5] (23); [6] (54); [7] (7); [8] (55); [9] (10); [10] (45); [11] (15).
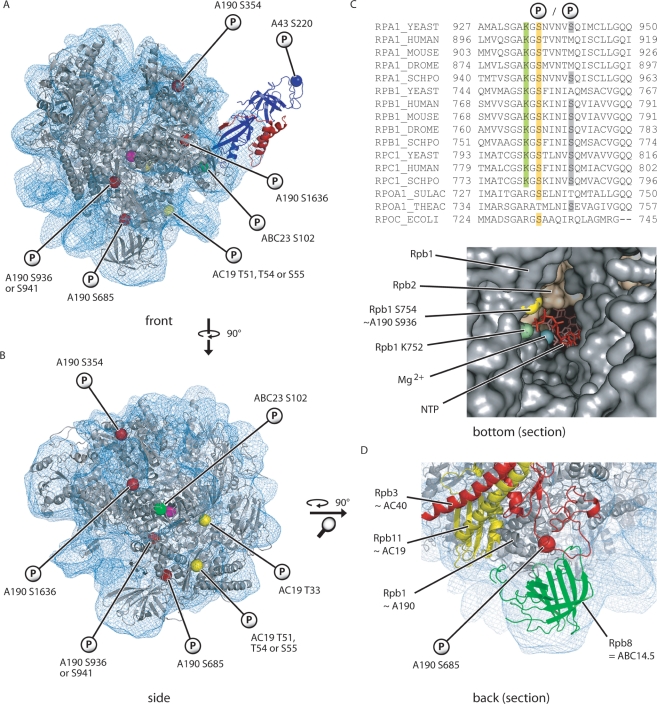
Using the complete Pol II structure (32), a recently established Pol I-cryo-EM structure and a new crystal structure of the Pol I subcomplex A43-A14, a model of a 12-subunit form of Pol I has been constructed (9). Based on this model and the sequence conservation between Pol I and Pol II subunits, the 3D-positions of seven phosphorylated residues were allocated (Figure 3A and B). Interestingly, all phosphosites except two (A190 S936/941 and ABC23 S102) are located more at the surface of the enzyme indicating that they could be accessible by modifying enzymes. The most striking position is that of S936/941 in the funnel domain of A190 (Figure 3C). The side chain of S936 points towards the pore/funnel through which the nucleotides are presumed to enter (33) and into which the 3′ end of the RNA chain is apparently extruded during backtracking of the enzyme. Many residues (including S936/941) in the funnel- and pore 1-domains are highly conserved among species as well as among nuclear RNA polymerases and are apparently important for the mechanism of NTP-binding, -selection and -incorporation [Figure 3C and (34)]. For instance, yeast Rpb1 K752, which is identical to K934 in A190, is crucial for NTP binding in the A-site (34–36). Thus, it was surprising that neither replacement of S936 nor of S941 by alanine, aspartate or even glutamate resulted in a severe growth phenotype (see subsequently). Apparently, the nucleotides can still pass through the funnel without constraint despite the obvious steric and electrochemical hindrance of a negatively charged side chain that occupies a significant part of the pore/funnel. To allow efficient passage of NTPs, it is possible that the funnel region is more flexible than anticipated from its crystal structure. Furthermore, the movement of the trigger loop during NTP incorporation (35) is apparently not influenced by changing the negative charge and the amino acid side chain at the positions S936/941.
Figure 3.
Three-dimensional localization of the identified Pol I phosphorylation sites. Localization of the phosphorylated amino acids in the Pol I homology model (9). The polymerase core and the A43/A14 heterodimer are shown in gray, blue and red, respectively. The fitted Pol I EM-density is depicted by the blue mesh. The catalytic Mg2+-ion was added for better orientation (magenta sphere in the middle of the structure). (A) Front view. (B) Side view without A43/A14. (C) Sequence alignment of the largest subunits of several eukaryotic RNA polymerases, as well as the homologous subunits from two Archaea species and Escherichia coli. Highlighted amino acids are shown in the structure of the pore/funnel-region below. A section of the bottom view of a Pol II elongation complex (PDB 1Y77) is presented with the downstream DNA on the right hand side. Rpb1 A759 is not located directly on the surface and thus not shown. (D) Localization of A190 S685 on the backside of the polymerase. A magnification of the lower part of the homology model is shown.
A190 S685 is another phosphorylation site of the pore/funnel region. In contrast to S936/941, this residue is not located inside the pore but on the part of the pore 1-domain which contributes to the backside of the polymerase (Figure 3D). A190 S685 lies in the vicinity of ABC14.5, the AC40/AC19 heterodimer and a conserved loop of the hybrid binding-domain of A135.
Only one phosphorylation site in subunit A43 falls within the ordered part of the new crystal structure, whereas the others are located to its mobile C-terminal region or within a disordered loop that are not revealed by crystallography (9). The phosphorylated S220 localizes between the C2 and C3 β-sheets which form together with the β-sheet C1 the outermost part of the A43-OB domain (9). The exposed position of S220 on the OB domain, the fact that A43 plays a crucial role in transcription initiation (7) and the findings that mutants with truncated β-sheets in this region are severely affected in growth ability (Figure 2), makes this region an appropriate candidate for important interactions with further components of the initiation machinery.
In vivo phenotypes after mutation of the major Pol I-phosphosites
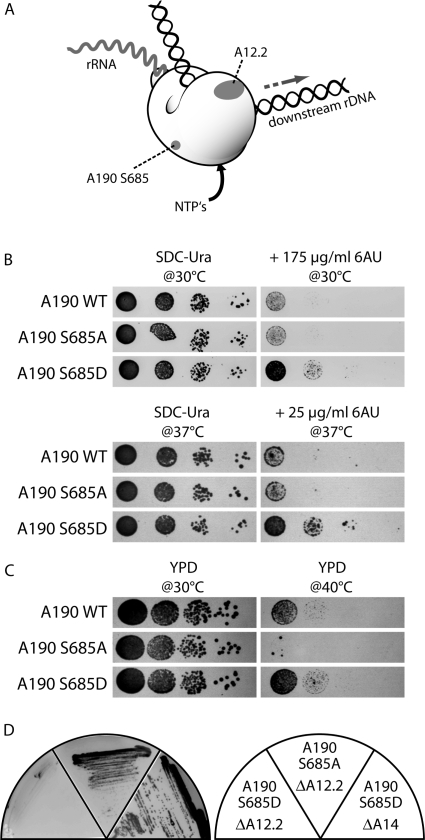
To study functional consequences of states that mimic either the non-phosphorylated or constitutively phosphorylated state, each of the single phosphorylated residues was replaced by either an alanine or aspartate, respectively. The clustered phosphoserines in A190 and A34.5 were each mutated in combination. We replaced each wild-type subunit by the corresponding mutant copies and investigated growth complementation at 16, 24, 30 and 37°C as well as the resulting growth behavior in the presence of 6-azauracil (6AU) and rapamycin. 6AU helps to detect elongation defects by indirectly lowering the pool of the nucleotides GTP and UTP (37,38), whereas rapamycin inhibits the TOR pathway through which cells sense nutrient availability. In total 27 different mutants were generated and analyzed (Supplementary Fig. S3). None of the mutations showed a significant growth phenotype (data not shown) arguing that crucial steps in rDNA transcription are not absolutely dependent on the single phosphosites investigated. Accordingly, for instance a single phosphorylated residue in subunit A43 or any other subunits tested cannot be crucial for interaction with transcription factor Rrn3p, which was previously shown to be growth-dependent (7). Since we found previously that in vitro interaction between Rrn3p and Pol I depends on the phosphorylation status of Pol I (19), it is possible that either combinations of several phosphosites are required for Rrn3p-binding or that the crucial site(s) was yet not identified. Also, no growth phenotype was detected if all serines in the motif present both in A190 (S1413/S1415/S1417) and A34.5 (S10/S12/S14) were exchanged as well as if both serines S208 and S220 were replaced in subunit A43. However, a change in growth behavior in the A190 S685D mutant was observed in the presence of 6AU. Replacement of S685 by an aspartate resisted 6AU concentrations under which growth of both wild-type cells and A190 S685A mutants is significantly reduced (Figure 4B). One possible explanation for this phenotype is that A190 S685D mutants do not respond properly if environmental conditions produce cellular stress, which in wild-type cells leads to growth inhibition and down-regulation of rRNA synthesis. This observation is supported by the growth behavior of different A190 S685-mutants at elevated temperatures. While the exchange to an alanine results in almost complete growth inhibition, wild-type cells can slowly grow at 40°C and replacement of S685 by an aspartate allows even better growth at this temperature (Figure 4C).
Figure 4.
Synthetic lethal effects and growth analysis of A190 S685 mutants. (A) Scheme of Pol I, showing the localization of A190 S685 and A12.2. (B) A yeast strain carrying the mutation A190 S685D is less sensitive to the NTP-pool depleting drug 6–azauracil (6AU) than the isogenic wild-type or a strain with the analogous mutation to alanine. (C) The same mutant (A190 S685D) grows better on YPD at elevated temperatures (40°C) than the corresponding wild-type strain, while the alanine variant is growth inhibited. (D) Mutation of A190 S685 to aspartate (left), but not to alanine (middle) is synthetic lethal with a deletion of RPA12. No genetic interaction with other non-essential Pol I subunits was observed. The deletion of RPA14 is shown as an example (right).
The finding that many major phosphosites are not involved in essential functions of Pol I-activity is reminiscent to the non-essential subunits in Pol I and Pol II which are not required for growth although their evolutionary conservation suggests that they participate in important steps of transcription. We investigated systematically genetic interactions between most of the mutated phosphosites and non-essential components of the Pol I transcription machinery like the Pol I subunits A12.2, A14, A34.5, A49, the transcription factors Uaf30, Hmo1 and other factors that interact with rDNA chromatin like the topoisomerases Top1 and Top3 or the histone assembly factor Asf1 by synthetic lethality assays. We replaced each phosphorylated Pol I subunit-gene by the corresponding phospho-mutant allele in the knock out strain of the candidate interaction partner (Supplementary Fig. S4). Remarkably, only one of the tested 180 combinations between a mutated phosphoserine/threonine and a gene deletion revealed a significant growth reduction. This is the same mutation leading to an altered growth behavior on 6AU-plates: replacement of S685 in the subunit A190 by an aspartate, but not by alanine resulted in synthetic lethality, if RPA12 was lacking, which encodes the non-essential Pol I subunit A12.2 (Figure 4D). In contrast yeast strains containing this phosphomutation, but lacking other non-essential Pol I subunits were not affected. It is possible that A190 S685 phosphorylation supports a non-essential function of subunit A12.2, which was discussed to be involved in elongation and/or stabilization of Pol I (39), transcription termination (40) and/or rRNA cleavage (9). If A12.2 is absent, the switch to phosphorylation might lead Pol I into a dead end situation, which then also impairs essential functions of the enzyme and which could only be avoided if S685 stays dephosphorylated. Accordingly, the viability of a Δrpa12 strain depends on the non-phosphorylated state of A190 S685. Thus, we conclude that this residue is reversibly phosphorylated in wild-type cells. Interestingly, A190 S685 is located well apart from A12.2 on the 3D structure of the enzyme [Figure 3A, B and D, Figure 4A, (9)] suggesting a functional co-operation of two spatially separated entities. It is possible that binding of A12.2 induces changes in the structure of Pol I (39) which, finally, ensures a high processivity of the enzyme in context with the reversible phosphorylation of A190 S685. Since subunit A12.2 was reported to be involved in transcription elongation (39), transcription termination (40) and rRNA cleavage (9) and, furthermore, the A190 S685D mutation caused growth resistance in the presence of 6AU, it is likely that the phosphorylation of this residue is involved in rRNA cleavage/elongation or termination. We are presently investigating which and how these steps are modulated by A190 S685 phosphorylation.
Phosphorylation and assembly/stability of Pol I
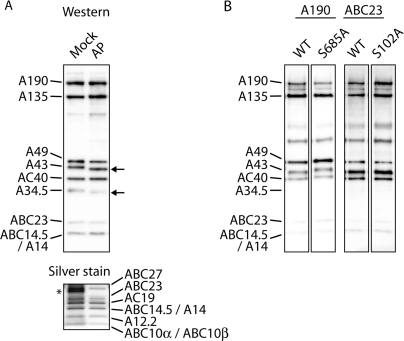
In many cases stability of protein–protein interaction or degradation of polypeptides depends on a distinct phosphorylation pattern (41–44). In particular, subunits ABC23 and AC19 are the ω-like and one of the α-like subunits of the Pol I complex, respectively, which are involved in enzyme-assembly (6,10,11,14,15). Therefore, we analyzed whether dephosphorylation of Pol I results in a loss of interactions between Pol I subunits. Such a reduced stability of Pol I could explain the loss of transcriptional activity which was previously found if Pol I-containing fractions were treated with alkaline phosphatase (18,19). Purified Pol I was dephosphorylated using alkaline phosphatase and affinity purified via its His-tagged A43 subunit under high ionic strength (1.5 M KAc) (Figure 5A). The dephosphorylation reaction resulted in a shift of subunit A34.5 and A43, respectively to a lower molecular weight on SDS-gels. No significant dissociation of any subunit could be detected, suggesting that the phosphosites analyzed are not required for complex stability as determined by subsequent western analysis and silver staining. The loss of transcriptional in vitro activity after Pol I-dephosphorylation (18,19) can, therefore, not be due to an instability of the core-enzyme at the promoter or during the transcription reaction. However, analysis by phosphostaining demonstrated that it was not possible to achieve a complete dephosphorylation of subunits A43, A190 and AC19 (data not shown) even, if extended incubation times or increased amounts of phosphatase were used. This suggests that some phosphorylation sites are not accessible to alkaline phosphatase [see also (29)]. To find out whether one of the phosphomutants is affected in either complex stability or in vivo assembly we systematically immunopurified Pol I from phosphomutant extracts using high salt conditions (1 M NaCl). None of the mutated Pol I molecules tested differed significantly in their substoichiometric composition suggesting that phosphorylation of the residues investigated in this study play neither a role for assembly nor for the stability of the mutated subunits under the conditions analyzed (Figure 5B and Supplementary Fig. S2). In particular, this means that A34.5 phosphorylation is neither required in formation of the A49/A34.5 dimer (9) nor in the association of the heterodimer to the core polymerase (8).
Figure 5.
Stability of phosphatase-treated and of mutated Pol I complexes. (A) Purified Pol I was treated with heat-inactivated (Mock) or active alkaline phosphatase (AP), affinity purified and analyzed by western blotting (upper panel; anti-Pol I antiserum was used) or silverstaining (lower panel). No significant changes in the complex stoichiometry were visible. Shifts of dephosphorylated A43 and A34.5 are indicated by arrows. The asterisk indicates an additional proteinband, which is due to heat-treated alkaline phosphatase fraction. (B) Pol I variants and the corresponding wild-types were immunoprecipitated from whole cell extracts and analyzed by western blotting. Two examples are shown. No differences in complex stability were detectable.
S102 of subunit ABC23 is one of the few phosphosites within interfaces between Pol I subunits. It is located in close proximity of amino acid Q100 (Figure 2) which is required for binding of the Pol II-specific subunits Rpb4 and Rpb7 (45). Exchange of Q100 with an arginine results in a temperature sensitive phenotype and a reduction in mRNA- and tRNA- however not in rRNA-synthesis. It is possible that the modulation of the close-by serine helps the common ABC23 subunit to distinguish between the different contact sites required for interaction with the distantly related Pol I, Pol II and Pol III-specific subcomplexes A14/A43, Rpb4/Rpb7 and C17/C25, respectively (13,46–48). However, since S102 mutations exhibit no significant growth phenotype, subunit and enzyme selection cannot exclusively be triggered by phosphorylation at this position. It is more likely that S102 phosphorylation is involved in a distinct function related to the largest Pol subunits, since genetic interactions between ABC23 S102F and an insertion mutation (W954(LELE)P) positioned in the foot domain of Rpb1 were reported (10,49). This insertion belongs to the domains which differ significantly between the largest Pol I and Pol II subunits (9). Further investigations will show whether the phosphorylation of ABC23 (2,3,50) appears also in Pol II and Pol III at the same position and whether this modification is either crucial for the assembly in a specific polymerase complex (10,32) or whether it is involved in a regulatory role as suggested by the 3D-neighborhood of ABC23 to the clamp (51). Accordingly, similar considerations are possible for the incorporation of the shared subunit AC19 into Pol I or Pol III, respectively.
In summary, our systematic studies about the Pol I-phosphoprotein complex show that, most residues that are phosphorylated at a molar ratio make no exclusive contribution to control cellular growth and, thus, to regulate ribosome synthesis. Furthermore, they are not essential for subunit stability or enzyme assembly. In this regard they are reminiscent of yeast Pol I subunits, which are not essential for growth though they are evolutionary conserved constituents. Apparently, they fulfill important functions for Pol I which are not detectable through the growth rate of the respective deletion strain. Possible roles of these non-essential subunits are emerging with the application of assays which allow to resolve single steps of transcription or processes related to nuclear transcription (8,9,40,52). Our phosphomutant strains are suitable to perform similar studies on the function of the major phosphorylated residues in Pol I subunits. On the other hand, cooperation between two or more phosphosites might be required to accomplish a significant impact on Pol I assembly or -activity or, for instance, to allow nuclear import of (preassembled) Pol I subunits. Investigations of such combinatorial effects can now be started.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Prof. Deutzmann, Regensburg for his support using LC-MALDI-TOF/TOF and Drs Ch. Carles and M. Riva for antibodies. This work was supported by grants of the Deutsche Forschungsgemeinschaft and of the Fonds der Chemischen Industrie. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft (grants to H.T. and P.C.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell GI, Valenzuela P, Rutter WJ. Phosphorylation of yeast RNA polymerases. Nature. 1976;261:429–431. doi: 10.1038/261429a0. [DOI] [PubMed] [Google Scholar]
- 2.Bell GI, Valenzuela P, Rutter WJ. Phosphorylation of yeast DNA-dependent RNA polymerases in vivo and in vitro. Isolation of enzymes and identification of phosphorylated subunits. J. Biol. Chem. 1977;252:3082–3091. [PubMed] [Google Scholar]
- 3.Buhler J-M, Iborra F, Sentenac A, Fromageot P. The presence of phosphorylated subunits in yeast RNA polymerases A and B. FEBS Lett. 1976;71:37–41. doi: 10.1016/0014-5793(76)80893-9. [DOI] [PubMed] [Google Scholar]
- 4.Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 5.Memet S, Gouy M, Marck C, Sentenac A, Buhler JM. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J. Biol. Chem. 1988;263:2830–2839. [PubMed] [Google Scholar]
- 6.Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 7.Peyroche G, Milkereit P, Bischler N, Tschochner H, Schultz P, Sentenac A, Carles C, Riva M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo. J. 2000;19:5473–5482. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadal O, Mariotte-Labarre S, Chedin S, Quemeneur E, Carles C, Sentenac A, Thuriaux P. A34.5, a nonessential component of yeast RNA polymerase I, cooperates with subunit A14 and DNA topoisomerase I to produce a functional rRNA synthesis machine. Mol. Cell Biol. 1997;17:1787–1795. doi: 10.1128/mcb.17.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn C-D, Geiger S, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P. Cell. 2007. Functional architecture of RNA polymerase I. in press. [DOI] [PubMed] [Google Scholar]
- 10.Nouraini S, Archambault J, Friesen JD. Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerases I and II and for the stability of the largest subunits of these enzymes. Mol. Cell Biol. 1996;16:5985–5996. doi: 10.1128/mcb.16.11.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanzendorfer M, Smid A, Klinger C, Schultz P, Sentenac A, Carles C, Riva M. A shared subunit belongs to the eukaryotic core RNA polymerase. Genes Dev. 1997;11:1037–1047. doi: 10.1101/gad.11.8.1037. [DOI] [PubMed] [Google Scholar]
- 12.Bischler N, Brino L, Carles C, Riva M, Tschochner H, Mallouh V, Schultz P. Localization of the yeast RNA polymerase I-specific subunits. Embo. J. 2002;21:4136–4144. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peyroche G, Levillain E, Siaut M, Callebaut I, Schultz P, Sentenac A, Riva M, Carles C. The A14–A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl Acad. Sci. USA. 2002;99:14670–14675. doi: 10.1073/pnas.232580799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dequard Chablat M, Riva M, Carles C, Sentenac A. RPC19, the gene for a subunit common to yeast RNA polymerases A (I) and C (III) J. Biol. Chem. 1991;266:15300–15307. [PubMed] [Google Scholar]
- 15.Lalo D, Carles C, Sentenac A, Thuriaux P. Interactions between three common subunits of yeast RNA polymerases I and III. Proc. Natl Acad. Sci. USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 17.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fath S, Kobor MS, Philippi A, Greenblatt J, Tschochner H. Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J. Biol. Chem. 2004;279:25251–25259. doi: 10.1074/jbc.M401867200. [DOI] [PubMed] [Google Scholar]
- 19.Fath S, Milkereit P, Peyroche G, Riva M, Carles C, Tschochner H. Differential roles of phosphorylation in the formation of transcriptional active RNA polymerase I. Proc. Natl Acad. Sci. USA. 2001;98:14334–14339. doi: 10.1073/pnas.231181398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters EC, Brock A, Ficarro SB. Exploring the phosphoproteome with mass spectrometry. Mini Rev. Med. Chem. 2004;4:313–324. doi: 10.2174/1389557043487330. [DOI] [PubMed] [Google Scholar]
- 21.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 23.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J. Proteome. Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 25.Byford MF. Rapid and selective modification of phosphoserine residues catalysed by Ba2+ ions for their detection during peptide microsequencing. Biochem. J. 1991;280(Pt 1):261–265. doi: 10.1042/bj2800261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molloy MP, Andrews PC. Phosphopeptide derivatization signatures to identify serine and threonine phosphorylated peptides by mass spectrometry. Anal. Chem. 2001;73:5387–5394. doi: 10.1021/ac0104227. [DOI] [PubMed] [Google Scholar]
- 27.Tschochner H. A novel RNA polymerase-I dependent RNase activity that shortens nascent transcripts from the 3′end. Proc. Natl Acad. Sci. USA. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milkereit P, Schultz P, Tschochner H. Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol. Chem. 1997;378:1433–1443. doi: 10.1515/bchm.1997.378.12.1433. [DOI] [PubMed] [Google Scholar]
- 29.Breant B, Buhler JM, Sentenac A, Fromageot P. On the phosphorylation of yeast RNA polymerases A and B. Eur. J. Biochem. 1983;130:247–251. doi: 10.1111/j.1432-1033.1983.tb07143.x. [DOI] [PubMed] [Google Scholar]
- 30.Areces LB, Matafora V, Bachi A. Analysis of protein phosphorylation by mass spectrometry. Eur. J. Mass. Spectrom. (Chichester, Eng) 2004;10:383–392. doi: 10.1255/ejms.601. [DOI] [PubMed] [Google Scholar]
- 31.Klemm C, Schroder S, Gluckmann M, Beyermann M, Krause E. Derivatization of phosphorylated peptides with S- and N-nucleophiles for enhanced ionization efficiency in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:2697–2705. doi: 10.1002/rcm.1676. [DOI] [PubMed] [Google Scholar]
- 32.Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 33.Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc. Natl Acad. Sci. USA. 2004;101:17361–17364. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 35.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Archambault J, Lacroute F, Ruet A, Friesen JD. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Van Mullem V, Landrieux E, Vandenhaute J, Thuriaux P. Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol. Microbiol. 2002;43:1105–1113. doi: 10.1046/j.1365-2958.2002.02824.x. [DOI] [PubMed] [Google Scholar]
- 40.Prescott EM, Osheim YN, Jones HS, Alen CM, Roan JG, Reeder RH, Beyer AL, Proudfoot NJ. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl Acad. Sci. USA. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang CN, Hayen A, Wilkins MR. Surface accessibility of protein post-translational modifications. J. Proteome. Res. 2007;6:1833–1845. doi: 10.1021/pr060674u. [DOI] [PubMed] [Google Scholar]
- 42.Edmunds JW, Mahadevan LC. MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J. Cell Sci. 2004;117:3715–3723. doi: 10.1242/jcs.01346. [DOI] [PubMed] [Google Scholar]
- 43.Yaffe MB, Cantley LC. Signal transduction. Grabbing phosphoproteins. Nature. 1999;402:30–31. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 44.Gutierrez GJ, Ronai Z. Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem. Sci. 2006;31:324–332. doi: 10.1016/j.tibs.2006.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Q, Prysak MH, Woychik NA. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III. Mol. Cell Biol. 2003;23:3329–3338. doi: 10.1128/MCB.23.9.3329-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadhale PP, Woychik NA. C25, an essential RNA polymerase III subunit related to the RNA polymerase II subunit RPB7. Mol. Cell Biol. 1994;14:6164–6170. doi: 10.1128/mcb.14.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol. Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jasiak AJ, Armache KJ, Martens B, Jansen RP, Cramer P. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol. Cell. 2006;23:71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Archambault J, Schappert KT, Friesen JD. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell Biol. 1990;10:6123–6131. doi: 10.1128/mcb.10.12.6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolodziej PA, Woychik N, Liao SM, Young RA. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell Biol. 1990;10:1915–1920. doi: 10.1128/mcb.10.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 52.Gadal O, Labarre S, Boschiero C, Thuriaux P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. Embo. J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittekind M, Dodd J, Vu L, Kolb JM, Buhler JM, Sentenac A, Nomura M. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae. Mol. Cell Biol. 1988;8:3997–4008. doi: 10.1128/mcb.8.10.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thuriaux P, Mariotte S, Buhler JM, Sentenac A, Vu L, Lee BS, Nomura M. Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA polymerase I. J. Biol. Chem. 1995;270:24252–24257. doi: 10.1074/jbc.270.41.24252. [DOI] [PubMed] [Google Scholar]
- 55.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.