Abstract
Activation of pre-messenger RNA (pre-mRNA) splicing requires 5′ splice site recognition by U1 small nuclear RNA (snRNA), which is replaced by U5 and U6 snRNA. Here we use crosslinking to investigate snRNA interactions with the 5′ exon adjacent to the 5′ splice site, prior to the first step of splicing. U1 snRNA was found to interact with four different 5′ exon positions using one specific sequence adjacent to U1 snRNA helix 1. This novel interaction of U1 we propose occurs before U1-5′ splice site base pairing. In contrast, U5 snRNA interactions with the 5′ exon of the pre-mRNA progressively shift towards the 5′ end of U5 loop 1 as the crosslinking group is placed further from the 5′ splice site, with only interactions closest to the 5′ splice site persisting to the 5′ exon intermediate and the second step of splicing. A novel yeast U2 snRNA interaction with the 5′ exon was also identified, which is ATP dependent and requires U2-branchpoint interaction. This study provides insight into the nature and timing of snRNA interactions required for 5′ splice site recognition prior to the first step of pre-mRNA splicing.
INTRODUCTION
Many eukaryotic pre-messenger RNAs (pre-mRNAs) contain introns between coding exon sequences that must be removed by pre-mRNA splicing. Splicing occurs via sequential transesterification reactions involving conserved sequences within the intron at the 5′ splice site, 3′ splice site and branchpoint (1). The first step of splicing produces 5′ exon and lariat intron-3′ exon splicing intermediates and the second step releases the intron and ligates the exons. A large ribonucleoprotein complex called the spliceosome catalyzes these reactions. The spliceosome is composed of five small nuclear RNAs (snRNAs) U1, U2, U4, U5 and U6 assembled into small nuclear ribonucleoprotein particles (snRNPs) and numerous associated non-snRNP proteins (2,3).
Sequential interactions of snRNA and protein components of the snRNPs with the pre-mRNA correctly orient the pre-mRNA for the two steps of splicing (4). During spliceosome assembly a commitment complex is followed by a prespliceosome, a complete spliceosome then an active spliceosome (5). Yeast commitment complex (CC) forms in two stages, CC1 and CC2 (6). Initially the 5′ end of U1 snRNA base pairs with the 5′ splice site to form CC1 (7). U1 base pairing with the 5′ splice site is well established in both yeast and mammalian splicing systems (8–11). Interactions of U1 at 5′ exon positions (−1) and (−2) in yeast and mammalian spliceosomes were identified by crosslinking with 4-thiouridine (4-thioU) (12–14). These interactions were not mapped within U1 to determine if they represented base-pairing interactions of U1 with the 5′ splice site but were assumed to represent the base-pairing interaction of U1 as genetic analysis has indicated base pairing between 5′ exon position (−1) and (−2) and U1 positions 9 and 10 (8). Following U1-5′ splice site base pairing, the branchpoint and 3′ splice site are recognized by branchpoint bridging protein (BBP) and Mud2, respectively, which contact U1 snRNP protein Prp40 bringing the two ends of the intron together to form CC2 (15). The mammalian equivalent to the commitment complex, E complex, contains U2 snRNP independent of ATP and branchpoint sequence (16). In fact, E complex formation is dependent on the presence of U2 snRNP (16,17). Within mammalian E complexes the 5′ end of U2 is close to the 5′ exon of the pre-mRNA near the splice site, further upstream within the 5′ exon and also several regions within the intron (18). U2 is also close to U1 snRNA within E complexes (18). U2 may help in bridging the ends of the intron in E complex in addition to protein interactions described above. Stable U2 association with the branchpoint then requires ATP to form prespliceosomes. Within minor U12 prespliceosomes, U12 snRNA also interacts with the 5′ exon of pre-mRNA at (−2) position in an ATP-dependent manner (19). It is not known whether U2 association with the 5′ exon is conserved in the yeast system.
The U4/U6.U5 tri-snRNP next joins the spliceosome and U1 at the 5′ splice site is exchanged for U5 snRNA loop 1 on the exon side of the splice site (20). Interaction of U5 loop 1 encompasses 5′ exon nucleotides (−1), (−2) and (−3) (12–14,21–23). U5 loop 1 based crosslinking indicates that loop 1 contacts the 5′ splice site from (−5) in the exon to (+2) in the intron (24,25). U5 loop 1 maintains its contact with (−1), (−2) or (−3) of the 5′ exon intermediate after the first step of splicing when loop 1 then contacts the 3′ exon to align the exons for ligation during the second step of splicing (12,13,21,26,27). U5 also crosslinks to 5′ exon position (−8) but, unlike its interactions adjacent to the 5′ splice site, this interaction does not persist after the first step of splicing and involves U5 nucleotides at the base of loop 1 (12). This suggests that the (−8) interaction is part of an earlier distinct interaction of U5 snRNP with pre-mRNA before it is finally positioned at the 5′ splice site prior to the first step of splicing.
When tri-snRNP joins the spliceosome U6 snRNA is also exchanged for U1 snRNA but on the intron side of the 5′ splice site (13,28–30). U6 base pairs with U2 adjacent to the U2-branchpoint interaction site (31) bringing branchpoint and 5′ splice site into close proximity for the first step of splicing. Prp19-associated complex (NTC) joins the spliceosome after U4 snRNP has dissociated and is required for stable U5 and U6 snRNP association in active spliceosomes (32). U5 loop 1 crosslinks to pre-mRNA are confined to (−1) and (−2) in the 5′ exon in NTC containing spliceosomes (33). The NTC may act as a specificity factor defining interactions of U5 and U6 with the 5′ splice site (33).
The snRNA interactions with the 5′ exon of pre-mRNA have an important role in spliceosome assembly and 5′ splice site definition. Although interactions of U1 and U5 with the 5′ exon have been investigated, the extent and nature of these interactions within the exon and each snRNA is not fully understood. We set out to precisely define interactions of U1 and U5 snRNAs with the 5′ exon and search for possible interactions of other snRNAs with the 5′ exon. Here we use site-specific crosslinking to reveal that both U1 and U5 interact with each 5′ exon position in pre-mRNA from (−3) to (−7). Only interactions of U5 with 5′ exon positions (−3) and (−4) continue with the 5′ exon intermediate after the first step of splicing, whereas interactions from (−5) to (−7) occur only with pre-mRNA. A novel crosslink of U2 to the 5′ exon at position (−6) was identified that was reliant on ATP and U2-branchpoint interaction. This interaction occurs in prespliceosomes independent of tri-snRNP addition. Crosslink mapping has defined a specific site in U1 that interacts with the 5′ exon before the known base-pairing interaction of U1. In contrast, the position of 5′ exon crosslinks to U5 progressively shift along U5 loop 1 as the crosslinking group is placed further from the 5′ splice site. Finally, U2 was found to interact with position (−6) of the 5′ exon revealing parallels in 5′ exon interactions between yeast and mammalian spliceosomes.
MATERIALS AND METHODS
Pre-mRNA production
Body labeled CYH2 pre-mRNA was produced as previously described (24). CYH2 pre-mRNAs containing 4-thioU were produced by RNA ligation of a chemically synthesized 5′ RNA (Dharmacon) to an in vitro transcribed 3′ RNA (21) by published procedures (34). The 3′ RNA was made by in vitro transcription from PCR product containing a T7 promoter and primed with UpG (Sigma) to allow end labeling with 32P. The RPS10B sequence was amplified from genomic DNA then inserted into pBluescript II KS+ (Stratagene) to produce plasmid pBS-RPS10B. PCR was carried out with this plasmid to produce a full-length RPS10B pre-mRNA T7 transcription template. Body labeled RPS10B was produced by in vitro transcription of the PCR product as previously described (27). RPS10B pre-mRNA containing the crosslinking group 4-thioU and control pre-mRNA was made by RNA ligation according to published methods (34). This ligation procedure produced an RPS10B pre-mRNA with a 4-thioU at position (−6) and a 32P label between positions (−2) and (−3).
In vitro splicing and UV crosslinking
Splicing extract was made from strain BJ2168 as described (24). Splicing was carried out at 23°C with 40% extract in 1× splicing buffer (60 mM potassium phosphate pH 7.0, 3% PEG 8000, 2.5 mM MgCl2, 2 mM ATP) with approximately 2 nM body labeled or ligated CYH2 pre-mRNA. Splicing reactions ranged from 5 to 1500 μl. UV crosslinking and precipitation of RNA from reactions was carried out according to published procedures (12). RNA was then analyzed by 4% or 5% polyacrylamide/8 M urea gel electrophoresis. For analysis with the Prp2p dominant negative protein a final concentration of 8.5 ng/μl wild type or Prp2pLAT was incubated with extract in splicing buffer at 23°C for 25 min. Body labeled or ligated CYH2 was added to give final concentrations of 40% extract in 1× splicing buffer then spliced for 15 min at 23°C before crosslinking. ATP was depleted from extract in splicing buffer without ATP by incubation with a final concentration of 1 mM glucose for 5 min at 23°C. Pre-mRNA was added to give 40% extract in 1× splicing buffer then spliced for 15 min before UV crosslinking. U6 snRNA was depleted from extracts by oligonucleotide targeted RNaseH degradation (35). Reactions containing 50% extract in 1 × splicing buffer were incubated with 1 μM oligonucleotide U6KO36−50 5′-CTGTATTGTTTCAAA for 30 min at 30°C. Ligated pre-mRNA was then added in 1 × splicing buffer to give 40% extract and spliced for 15 min before UV crosslinking. 2′-O-methyl RNA oligonucleotide complementary to the CYH2 branchpoint, 5′-UUAAAUCGUUAGUAAAACAG, was annealed to body labeled CYH2 to give final concentrations of 300, 500 or 700 ng total oligonucleotide in the reaction. Five hundred nanogram of oligonucleotide was annealed to CYH2 pre-mRNA for analysis of the U2 snRNA crosslink. The RNAs were annealed by heating to 95°C followed by slow cooling to 30°C in splicing buffer. Extract was then added to give final concentrations of 40% extract in 1 × splicing buffer and reactions were spliced for 15 min before UV crosslinking.
RNaseH analysis, crosslink selection with biotinylated oligonucleotides and primer extension mapping
RNaseH analysis was carried out as described (21). Specific crosslink species were selected from RNA isolated from splicing reactions using 5′-biotinylated oligonucleotides and streptavidin magnetic beads. Selected crosslinks were gel purified and used for U1 primer extension mapping as described (26). For U2 crosslink mapping extract was depleted of U6 snRNA as described above to inhibit tri-snRNP formation. For U2 and U5 primer extension mapping pre-mRNA with 4-thioU at position (−5), (−6) or pre-mRNA without 4-thioU was added to the extract and spliced for 30 min followed by UV irradiation. RNA isolated from the reactions was hybridized with a biotin containing oligonucleotide complementary to the CYH2 pre-mRNA and CYH2-containing RNA species were selected with streptavidin magnetic beads. Selected RNA on the magnetic beads was subjected to U5 or U2 primer extension mapping.
Prp2p purification
The pGEX-6P-2 plasmids C458-PRP2 and C451-PRP2 SAT > LAT (a gift from Andy Newman) carrying PRP2 genes with both GST and 6His tags were transformed into Escherichia coli strain BL21 (DE3) (Novagen). Two litres of LB-culture for each was grown to an OD600 of 0.5 before transfer to 18°C for 30 min. To induce protein expression, IPTG was added to a concentration of 25 μM and cells were incubated at 18°C overnight. Cells were then pelleted and frozen at −80°C overnight. Pellets were resuspended in 30 ml ice-cold lysis buffer (50 mM NaH2PO4 at pH 8, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 1% Triton X-100) and sonicated. Lysate was cleared at 35 000g for 15 min then loaded onto a 3 ml Ni2+-NTA (Sigma) column. Unbound protein was washed away with 10 ml wash buffer (50 mM NaH2PO4 at pH 8, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 1% Triton X-100) then Prp2p was eluted with 6 ml elution buffer (50 mM NaH2PO4 at pH 8, 300 mM NaCl, 250 mM imidazole, 10% glycerol, 1% Triton X-100). All eluate was loaded onto a 2 ml Glutathione Sepharose 4B (Amersham Biosciences) column and washed with 1 × phosphate buffered saline containing 10% glycerol and 1% Triton X-100. A second wash carried out with cleavage buffer (50 mM Tris–HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1% Triton X-100). Prp2p was eluted by cleavage of the GST tag with 160 U of PreScission Protease (GE Healthcare) at 4°C overnight and eluted with cleavage buffer then dialyzed against buffer containing 50 mM NaH2PO4 at pH 8, 300 mM NaCl, 1 mM DTT and 20% glycerol overnight. Protein was then visualized by 10% SDS–PAGE and the protein concentration determined using the BioRad dye binding reagent with bovine serum albumin as the standard.
Oligonucleotides and synthetic RNAs
The sequences of the chemically synthesized CYH2 5′ RNAs, with 4-thioU represented as X, are:
(−3) 5′-GACUAGAAAGCACAGAGGUCACGUCUXA,
(−4) 5′-GACUAGAAAGCACAGAGGUCACGUCXUA,
(−5) 5′-GACUAGAAAGCACAGAGGUCACGUXUUA,
(−6) 5′-GACUAGAAAGCACAGAGGUCACGXCUUA,
(−7) 5′-GACUAGAAAGCACAGAGGUCACXUCUUA.
Control RNA for positions (−3), (−4) and (−6) contained uridine in place of 4-thioU. Control RNAs for positions (−5) and (−7) contained a uridine in place of cytidine and guanosine, respectively.
PCR primers for cloning RPS10B gene are:
5′-GCTCTAGAGAAACAAAATTCACCAATACTTG
5′-CGGGATCCTGGTATCAATTTCTTCGTGCTTAG
PCR primers for T7 transcription template of full length RPS10B pre-mRNA are:
5′-CGTAATACGACTCACTATAGAAACAAAATTCACCAATAC,
5′-TGGTATCAATTTCTTCGTGC
The sequence of the chemically synthesized RPS10B 5′ RNAs, with 4-thioU represented as X, are: 5′-GAAACAAAAUUCACCAAUACUUGUXUCA,
control RNA 5′-GAAACAAAAUUCACCAAUACUUGUUUCA. The bridge oligonucleotide for RPS10B ligation was:
5′-GCATATCACTGCATATTTCACCTAAAAAATGCAAACATACCTTGAAACAAGTATTG GTGAATTTTGTTTC
PCR primers for T7 transcription template of RPS10B 3′ RNA are:
5′-CGTAATACGACTCACTATAGGTATGTTTGCATTTTTTAGG, and the back primer described above for the full length RPS10B. The 3′ RNA transcription was primed with ApG (Sigma) to allow end labeling with 32P.
Biotin oligonucleotides used to specifically select RNAs with streptavidin paramagnetic beads:
(CYH2 BioTEG) 5′Biotin TEG—CAGCGATAATTAGTGCGTTCGCAATC
(U5 BioTEG) 5′Biotin TEG—ATGGCAAGCCCACAGTAACGGACAGC
(U2 BioTEG) 5′Biotin TEG—AAGGTAATGAGCCTCATTGAGGTCAT
(U1 Bio2TEG) 5′Biotin TEG—CGATGTTTGATCAGTAGGACTTCTTGATC
RESULTS
U1, U2 and U5 snRNAs crosslink to the 5′ exon of the pre-mRNA
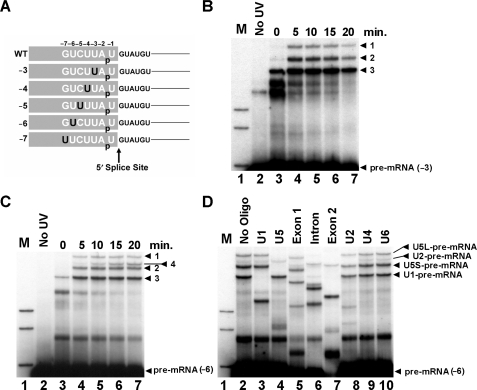
Crosslinking has demonstrated interactions of U1 and U5 with pre-mRNA at 5′ exon positions (−1) and (−2) as well as U5 at (−3) and (−8) (12−14, 21). We sought to define snRNA interactions with the 5′ exon in more detail during spliceosome assembly and 5′ splice site recognition. Five CYH2 pre-mRNAs with a single crosslinkable 4-thiouridine (4-thioU) at each 5′ exon position from (−3) to (−7) were produced by RNA ligation (Figure 1A). Each pre-mRNA also contained a single 32P label between positions (−1) and (−2) so only pre-mRNA, 5′ exon intermediate and spliced mRNA, but not lariat-3′ exon intermediate and released lariat intron, could be visualized by autoradiography when these pre-mRNAs were spliced in vitro (Supplementary Figure S1). These pre-mRNAs were added to yeast in vitro splicing reactions and irradiated with long wave UV at different time points to activate the 4-thioU. 4-thioU at position (−3) produced three major crosslink bands migrating above the pre-mRNA that appeared over time (Figure 1B, labeled 1, 2 and 3). 4-thioU at (−4), (−5) and (−7) produced an identical pattern of crosslinked bands as observed at (−3) (data not shown). Interestingly, a fourth additional band was observed with 4-thioU at (−6) (Figure 1C, labeled 4). All crosslinks were UV dependent (Figure 1B and C, lane 2) and specific for 4-thioU (Supplementary Figure S1).
Figure 1.
snRNA crosslinking to the 5′ exon of the pre-mRNA. (A) Six CYH2 pre-mRNAs were produced by RNA ligation. The sequence of the CYH2 pre-mRNA around the 5′ splice site is shown with 5′ exon positions represented by negative numbers. The five pre-mRNAs are named according to the 4-thioU positions shown in black. A single 32P label is shown between positions (−1) and (−2). (B) In vitro splicing time course of ligated CYH2 pre-mRNA with 4-thioU at (−3). Zero time point with no UV irradiation (lane 2). Reactions were incubated at 23°C and samples withdrawn at 5 min intervals then UV irradiated (lanes 3–7). RNA isolated from the reactions was analyzed by 5% denaturing PAGE. The pre-mRNA and three major crosslinked species are indicated on the right. Markers (lane 1), 32P-labeled pBR322 Msp1-digested DNA. (C) In vitro splicing and crosslinking time course of ligated CYH2 pre-mRNA with 4-thioU at (−6). Lanes as in (B). The pre-mRNA and four major crosslinked species are indicated on the right. (D) Oligonucleotide targeted RNaseH cleavage of crosslinks formed with CYH2 pre-mRNA with 4-thioU at (−6). RNA recovered from a UV irradiated splicing reaction carried out for 20 min was incubated with the oligonucleotides specified above each lane. The pre-mRNA and four major crosslinked species are indicated on the right. The long and short forms of U5 are indicated with L and S, respectively. Markers (lane 1), as in (B).
To identify crosslinks they were hybridized with oligonucleotides complementary to pre-mRNA or snRNAs then incubated with ribonuclease H (RNaseH). A shift in migration of crosslink following incubation with RNaseH indicates the presence of the specific RNA complementary to the oligonucleotide. All crosslinks produced with 4-thioU at (−6) were targeted by 5′ exon, intron and 3′ exon oligonucleotides indicating all crosslinks contained the pre-mRNA (Figure 1D, lanes 5, 6 and 7). Crosslinks 1 and 2 were also targeted by the U5 oligonucleotide identifying these crosslinks as the long and short forms of U5 (36) crosslinked to the pre-mRNA (Figure 1D, lane 4). Crosslink 3 was also targeted by the U1 oligonucleotide identifying this crosslink as U1 crosslinked to the pre-mRNA (Figure 1D, lane 3). Oligonucleotides complementary to U2, U4 and U6, were utilized to identify crosslink 4 as U2 crosslinked to pre-mRNA (Figure 1D, lanes 8–10). RNaseH analysis carried out on crosslinks at positions (−3), (−4), (−5) and (−7) confirmed crosslinks 1, 2 and 3 were U5- and U1-pre-mRNA crosslinks (data not shown). Therefore, U1 and U5 interactions with the 5′ exon of pre-mRNA encompass every nucleotide from (−3) to (−7) and U2 contacts pre-mRNA at position (−6). A time course of splicing and crosslinking reveals that the U1-pre-mRNA crosslinks occur before the U5-pre-mRNA and U2-pre-mRNA crosslinks first appear at the 5 min time point (Figure 1C).
Not all U5-pre-mRNA crosslinks are maintained with the 5′ exon intermediate
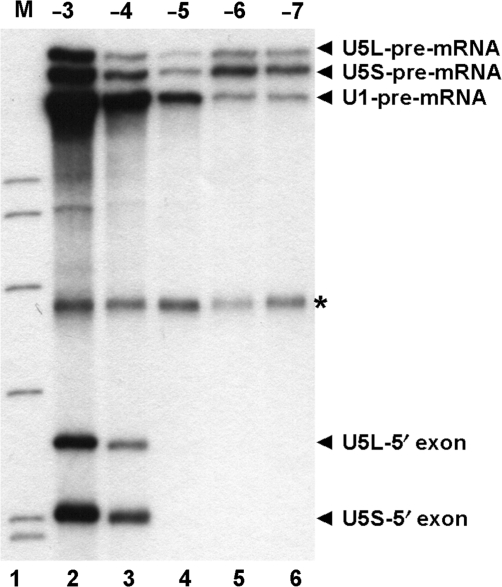
Following the first step of splicing U5 loop 1 holds the 5′ exon intermediate and aligns it with the 3′ exon for the second step of splicing (26,27). U5-pre-mRNA crosslinks at (−1) and (−3) continue with the 5′ exon intermediate (12,13,21). U5-pre-mRNA crosslinks at (−8) do not continue with the 5′ exon intermediate suggesting this interaction of U5 is distinct from interactions adjacent to the 5′ splice site (12). We were interested in the extent of 5′ exon interactions of U5. As U5-5′ exon intermediate crosslinks migrate faster than unspliced pre-mRNA they are obscured by pre-mRNA degradation products. To visualize U5-5′ exon crosslinks U5 containing species were captured from UV irradiated splicing reactions using biotinylated olionucleotides complementary to U5 and streptavidin magnetic beads. Crosslinks were eluted from the beads, run on a gel and visualized by autoradiography. U1 containing crosslinks were also selected from the same reactions. A band is visualized in the gel only when unlabeled snRNAs are crosslinked to labeled pre-mRNA or 5′ exon intermediate. U1 and U5 crosslinks were selected from reactions with each 4-thioU containing pre-mRNA (Figure 2). U1- and U5-pre-mRNA crosslinks were observed from (−3) to (−7) (Figure 2, lanes 2–6). U5-5′ exon intermediate crosslinks produced following the first step of splicing were only selected when 4-thioU was at (−3) and (−4) (Figure 2, lanes 2 and 3). Therefore, although U5 interacts with (−1) to (−8), the interactions from (−5) to (−8) only occur before 5′ splice site cleavage. As previously suggested, this indicates two distinct interactions of U5 with pre-mRNA (12), interactions upstream of (−4) that occur before the first step and interactions from (−1) to (−4) that are maintained with the 5′ exon intermediate after the first step of splicing.
Figure 2.
Isolated crosslinks of U1 and U5 snRNA with the pre-mRNA and 5′ exon intermediate. In vitro splicing was carried out for 20 min followed by crosslinking using CYH2 pre-mRNA with 4-thioU at (−3), (−4), (−5), (−6) or (−7) (lanes 2–6). RNA recovered from reactions was hybridized to biotin containing oligonucleotides complementary to U1 and U5 then U1- and U5-containing RNA species were captured with streptavidin magnetic beads. RNA eluted from magnetic beads was analyzed by denaturing PAGE. The U1-pre-mRNA, the U5-pre-mRNA and the U5-5′ exon crosslinks are indicated on the right. The long and short forms of U5 are indicated with L and S, respectively. The asterisk indicates background pre-mRNA captured with streptavidin magnetic beads. Markers (lane 1), 32P-labeled pBR322 Msp1-digested DNA.
The U2- and U5-pre-mRNA crosslinks at position (−6) are ATP dependent
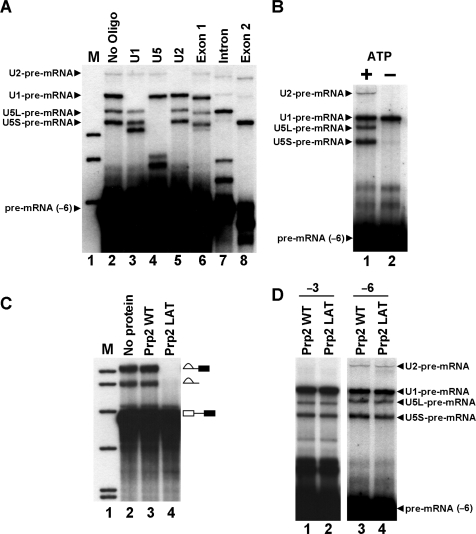
Intron recognition by U1 to form commitment complex occurs independently of ATP (10,11). A U2 containing E complex (mammalian commitment complex) has been isolated that contains U2 independent of ATP (16). Subsequent steps in spliceosome assembly require ATP (4). We, therefore, decided to investigate the ATP dependence of pre-mRNA crosslinks at (−6). U2-pre-mRNA crosslinks at (−6) migrated between the long and short crosslinked forms of U5 in the 5% gel used in previous experiments. To allow better visualization of U2-pre-mRNA crosslinks reactions were separated on 4% gels. RNaseH analysis of (−6) crosslinks separated on 4% gel reveals that U2–pre-mRNA crosslinks migrate above the other crosslinks (Figure 3A). U1-pre-mRNA crosslinks run below U2-pre-mRNA crosslinks and U5-pre-mRNA crosslinks run below U1-pre-mRNA crosslinks (Figure 3A). All subsequent analysis of crosslinks was carried out using 4% gels.
Figure 3.
The ATP and Prp2 dependency of snRNA-pre-mRNA crosslinks. (A) Oligonucleotide directed RNaseH cleavage analysis of crosslinks by 4% denaturing PAGE. RNA recovered from a UV irradiated splicing reaction carried out for 20 min using CYH2 pre-mRNA with 4-thioU at the (−6) position was incubated with oligonucleotides specified above each lane (lanes 2–8). The pre-mRNA and four major crosslinked species are indicated on the left. Markers (lane 1), 32P-labeled pBR322 Msp1-digested DNA. (B) Formation of U2-pre-mRNA crosslink requires ATP. Extract was pre-incubated with normal splicing conditions (lane 1) or in buffer lacking ATP with addition of glucose to deplete endogenous ATP from the extract (lane 2). The CYH2 pre-mRNA with 4-thioU at (–6) was then added and spliced for 20 min before UV irradiation. Isolated RNA was analyzed by 4% denaturing PAGE. The pre-mRNA and four major crosslinked species are indicated on the left. (C) Recombinant Prp2LAT inhibits the first step of splicing. Extracts were pre-incubated without protein (lane 2), wild-type recombinant Prp2 (lane 3) or recombinant Prp2LAT mutant (lane 4), before addition of CYH2 pre-mRNA and splicing for 20 min. Isolated RNA was analyzed by denaturing PAGE. The splicing intermediates and products indicated on the right. Markers (lane 1), as in (A). (D) The U1- and U5- pre-mRNA crosslinks at (−3) and (−6) and U2-pre-mRNA crosslink at (−6) occur before the Prp2-dependent step of spliceosome assembly. Extracts were pre-incubated with wild-type Prp2 (lanes 1 and 3) or Prp2LAT (lanes 2 and 4) before addition of CYH2 pre-mRNA with 4-thioU at (−3) (lanes 1 and 2) or (−6) (lanes 3 and 4) followed by splicing for 20 min then UV irradiation. Isolated RNA was analyzed by 4% denaturing PAGE. The pre-mRNA and four major crosslinked species are indicated on the left.
To investigate ATP dependence of U2- and U5-pre-mRNA crosslinks ATP was depleted from extracts. Incubation of (−6) pre-mRNA was then carried out without ATP before UV irradiation. Only U1-pre-mRNA crosslinks appear without ATP (Figure 3B, lane 2). U2- and U5-pre-mRNA crosslinks at (−6) require ATP. This suggests that U2-pre-mRNA crosslinks occur in splicing complexes following 5′ splice site recognition by U1. U5–pre-mRNA interactions at (−6), which do not continue after the first step of splicing, also occur following initial 5′ splice site recognition.
The U2- and U5-pre-mRNA crosslinks occur before the Prp2-dependent step of spliceosome assembly
The RNA-dependent ATPase Prp2 functions prior to the first step of splicing inducing a structural change that forms fully assembled spliceosomes (37). A dominant negative mutant of Prp2 containing a serine to leucine mutation in the conserved SAT motif III, Prp2LAT, competes with wild-type Prp2 when added to extracts. This results in stalled, unactivated, spliceosomes and inhibition of the first step of splicing (38). Extracts incubated with Prp2LAT do not carry out the first step of splicing (Figure 3C, lane 4). Addition of wild-type Prp2 had no effect on splicing (Figure 3C, lane 3). To determine if crosslinks occur before or after the Prp2-dependent step of spliceosome assembly extracts were pre-incubated with Prp2LAT before splicing and UV irradiation with 4-thioU containing pre-mRNA. Pre-mRNAs with 4-thioU at (−3) and (−6) were employed to analyze the timing of snRNA-pre-mRNA crosslinks at these positions. The U1- and U5-pre-mRNA crosslinks at (−3) (Figure 3D, lanes 1 and 2) and U1-, U2- and U5-pre-mRNA crosslinks at (−6) (Figure 3D, lanes 3 and 4) appear in the presence of wild-type or mutant Prp2p. Since all crosslinks are present with Prp2LAT mutant they must occur before the Prp2p-dependent step of spliceosome assembly.
The U2-pre-mRNA crosslink occurs in prespliceosomes and with another pre-mRNA
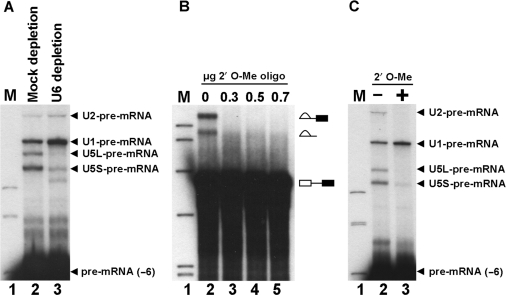
U2-pre-mRNA crosslinks occur following the first ATP-dependent step in spliceosome assembly but before the Prp2-dependent step of spliceosome assembly. To determine the precise point in spliceosome assembly when U2-pre-mRNA crosslinks occur we investigated whether crosslinks could form before or after tri-snRNP joins the spliceosome. U6 was specifically depleted from extracts using an oligonucleotide complementary to U6 that prevents formation of tri-snRNP during spliceosome assembly (35). Pre-mRNA with 4-thioU at (−6) was added to extract depleted of U6 and incubated under splicing conditions followed by UV irradiation to allow crosslink formation. All four previously identified snRNA-pre-mRNA crosslinks appear in a mock depletion (Figure 4A, lane 2). In U6 depleted extract, both U1- and U2-pre-mRNA crosslinks increased in intensity (Figure 4A, lane 3) suggesting they occur before tri-snRNP joins the spliceosome. As expected U5-pre-mRNA crosslinks are greatly reduced upon U6 depletion (Figure 4A, lane 3) as U5 joins the spliceosome as part of the tri-snRNP. As U2-pre-mRNA crosslinks occur before tri-snRNP addition this interaction could be in prespliceosomes, when U2 has formed its branchpoint interaction, or earlier, before branchpoint interaction is established. To determine whether U2-branchpoint interaction is required for U2-pre-mRNA crosslinks at (−6), a 2′-O-methyl (2′O-Me) RNA oligonucleotide complementary to the branchpoint was utilized to block U2-branchpoint interaction. 2′O-Me oligonucleotide effectively blocked U2-branchpoint interaction since the first step of pre-mRNA splicing was prevented (Figure 4B). When (−6) pre-mRNA was spliced and crosslinked in the presence of 2′O-Me oligonucleotide U2- and U5-pre-mRNA crosslinks no longer appeared (Figure 4C, lane 3). This suggests that U2- and U5-pre-mRNA crosslinks at (−6) require prior U2-branchpoint contact. We conclude that U2-pre-mRNA crosslinks at (−6) form in prespliceosomes after U2 is base paired at the branchpoint but before tri-snRNP joins and completes spliceosome assembly.
Figure 4.
The U2-pre-mRNA crosslink occurs in prespliceosomes. (A) The U2-pre-mRNA crosslink at (−6) occurs before tri-snRNP addition to the spliceosome. Extracts were depleted of U6 by pre-treating with an oligonucleotide complementary to U6 (lane 2) or water (lane 1) before the addition of CYH2 pre-mRNA with 4-thioU at (−6), followed by splicing for 20 min then UV irradiation. Isolated RNA was analyzed by 4% denaturing PAGE. The pre-mRNA and four major crosslinked species are indicated on the right. Markers (lane 1), 32P-labeled pBR322 Msp1-digested DNA. (B) A 2′-O-Me RNA oligonucleotide complementary to the CYH2 branchpoint inhibits the first step of splicing. Body labeled CYH2 was hybridized in splicing buffer with 0, 0.3, 0.5 or 0.7 μg 2′ O-Me RNA oligonucleotide complementary to the branchpoint (lanes 2–5). Splicing was carried out for 20 min and isolated RNA was analyzed by denaturing PAGE. The splicing intermediates and products indicated on the right. Markers (lane 1), as in (A). (C) The U2-pre-mRNA crosslink at (−6) requires prior U2-branchpoint interaction. CYH2 pre-mRNA with 4-thioU at position (−6) was hybridized to 0.5 μg of 2′-O-Me RNA oligonucleotide complementary to the branchpoint (lane 3) or water (lane 2). Extract was added and splicing carried out for 20 min before UV irradiation. Isolated RNA was analyzed by 4% denaturing PAGE. The pre-mRNA and four major crosslinked species are indicated on the right. Markers (lane 1), as in (A).
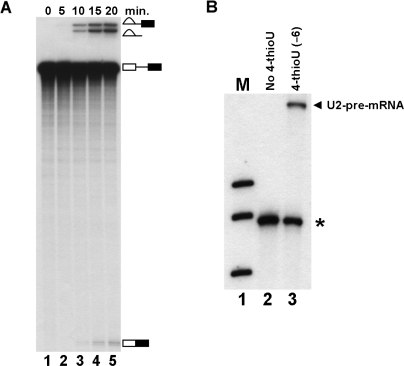
To confirm U2-pre-mRNA crosslinks at (−6) occurred in all spliceosomes and were not unique to CYH2 pre-mRNA an alternative pre-mRNA was utilized for splicing and crosslinking. RPS10B was chosen because it contained a relatively small 5′ exon with a U at (−6). A body labeled RPS10B pre-mRNA was competent for splicing in vitro (Figure 5A). RPS10B pre-mRNAs with wild-type 5′ exon or 4-thioU at (−6) and a single 32P label between (−2) and (−3) were produced by RNA ligation then utilized for splicing and crosslinking. RNA species containing U2 were selected from UV-irradiated splicing reactions with a biotin containing oligonucleotide complementary to U2 and streptavidin magnetic beads. A radioactive band is selected only when unlabeled U2 is crosslinked to labeled pre-mRNA. A U2-pre-mRNA crosslink is observed when RPS10B pre-mRNA contained 4-thioU at (−6) but not in pre-mRNA lacking 4-thioU (Figure 5B). Since U2-pre-mRNA crosslinks occur with both CYH2 and RPS10B pre-mRNA it is more than likely to represent a real interaction with all pre-mRNAs.
Figure 5.
The U2 snRNA crosslinks to RPS10B pre-mRNA. (A) Splicing of RPS10B pre-mRNA. Splicing was carried out and RNA isolated at 0, 5, 10, 15 and 20 min (lanes 1–5) then analyzed by denaturing PAGE. The splicing intermediates and products indicated on the right. The 5′ exon intermediate runs below the mRNA product and is not shown. (B) U2 crosslinks to the (−6) position of RPS10B pre-mRNA. Splicing was carried out for 20 min then the reactions were crosslinked using RPS10B pre-mRNA without 4-thioU (lane 2) and with 4-thioU at position (−6) (lane 3). RNA recovered from reactions was hybridized with a biotin containing oligonucleotide complementary to U2 then U2-containing RNA species were captured with streptavidin magnetic beads. RNA eluted from magnetic beads was analyzed by denaturing PAGE. The U2-pre-mRNA crosslink is indicated on the right. The asterisk indicates background pre-mRNA captured with streptavidin magnetic beads. Markers (lane 1), 32P-labeled pBR322 Msp1-digested DNA.
Mapping of snRNA-pre-mRNA crosslinks
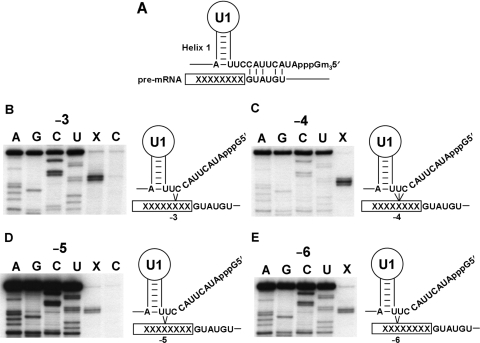
To identify specific sites in U1, U2 and U5 that crosslink to the 5′ exon of pre-mRNA a selection of crosslinks were mapped. Prior to the first step of splicing the 5′ end of U1 base pairs with the 5′ splice site (Figure 6A). Four U1-pre-mRNA crosslinks were mapped from (−3) to (−6) (Figure 6B–E). The four U1-pre-mRNA crosslinks mapped to one specific site in the 5′ end of U1 at nucleotides 9, 10 and 11. Interestingly, these interactions detected by crosslinking are not compatible with base pairing of U1 with the 5′ splice site region (5) and may represent an alternative conformation of U1 that occurs before base pairing.
Figure 6.
Mapping of the U1-pre-mRNA crosslinks. (A) Diagram of base-pairing interaction of the 5′ end of the yeast U1 with the 5′ splice site. (B, C, D, E) Four positions in the pre-mRNA crosslink to a specific site in the 5′ end of the U1. Crosslinks selected from UV-irradiated splicing reactions by hybridization with a biotin oligonucleotide complementary to U1 were separated by denaturing PAGE. Crosslinks were cut from the gel and isolated by electroelution for use as primer extension templates with an end-labeled U1-specific oligonucleotide. U1-pre-mRNA crosslinks were mapped (lanes marked X) from UV irradiated splicing reactions containing CYH2 pre-mRNA with 4-thioU at positions (−3), (−4), (−5) and (−6). Control reactions (lanes marked C) were identical except that pre-mRNA without a 4-thioU cross-linking group was utilized and an identical region of gel cut out for electroelution. The control mapping for (−3) applies to positions (−4) and (−6) as these control RNAs are identical. U1 sequencing tracks are displayed as reference for the primer extension mapping. During the primer extension the reverse transcriptase stops one nucleotide before the site of the crosslink. A diagrammatic representation of crosslinking at each pre-mRNA position analyzed is displayed to the right of each panel.
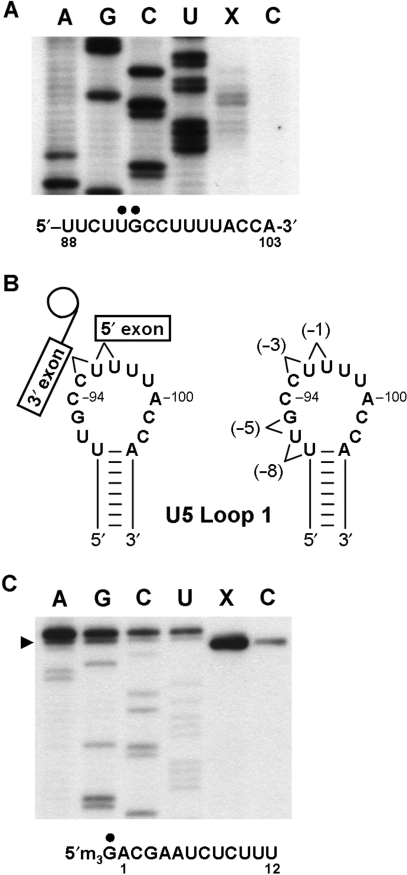
One U5-pre-mRNA crosslink was mapped at (−5) as mapping of U5-pre-mRNA crosslinks at (−1), (−3) and (−8) had been previously undertaken (12,21). U5-pre-mRNA crosslinks at (−5) mapped to nucleotides 92 and 93 on the 5′ side of U5 loop 1 (Figure 7A). As U5-pre-mRNA crosslinks at (−3) mapped to nucleotides 95 and 96 in U5 loop 1 (21) and (−8) to nucleotides 91 and 92 in U5 loop 1 (12) this indicates that as the crosslinking group is moved further upstream in the 5′ exon of pre-mRNA, U5-pre-mRNA crosslinks progressively shift along U5 loop 1 in the 5′ direction (Figure 7B).
Figure 7.
Mapping of U2- and U5-pre-mRNA crosslinks. (A) The (−5) position of the pre-mRNA crosslinks to positions 92 and 93 in U5 snRNA loop 1. U5 sequencing tracks are displayed as reference for the primer extension mapping. Crosslinks were selected from UV-irradiated splicing reactions containing CYH2 pre-mRNA with or without 4-thioU at position (−5) of the pre-mRNA by hybridization with a biotin oligonucleotide complementary to CYH2 and streptavidin magnetic beads. Primer extension was carried out directly from RNA captured on the streptavidin magnetic beads with an end-labeled U5-specific oligonucleotide. Primer extension reactions were separated by denaturing PAGE next to U5 sequencing tracks. The site of the pre-mRNA (−5) position crosslink to U5 is indicated with a black dot above the U5 sequence. (B) Pre-mRNA crosslinks to U5 loop 1 shift towards the 5′ end of loop 1 as crosslinking groups are placed further from the 5′ splice site. Diagrammatic representation of U5–exon interations prior to the second step of splicing. U5 snRNA loop 1 indicating the positions in loop 1 that crosslink to the (−1) (12), (−3) (21), (−5) and (−8) (12) positions of the pre-mRNA. (C) The 5′ end of U2 crosslinks to the (−6) position in the pre-mRNA. Crosslink selection as in (A). Primer extension was carried out with an end-labeled U2-specific oligonucleotide. Primer extension reactions were separated by denaturing PAGE next to U2 sequencing tracks of a U2 template with an extra nucleotide at the 5′ end. The authentic 5′ end of U2 is indicated by an arrow on the left. The site of the pre-mRNA (−6) position crosslink to the 5′ end of the U2 sequence is indicated with a black dot above the U2 sequence. Lane (X) is the primer extension reaction of RNA selected from UV-irradiated splicing reactions containing CYH2 pre-mRNA with 4-thioU at position (−6) and lane (C) is the control primer extension of RNA selected from UV-irradiated splicing reactions containing CYH2 pre-mRNA without 4-thioU at position (−6).
Given the lower abundance of U2-pre-mRNA crosslinks we undertook an alternative approach to their mapping. To enrich for U2-pre-mRNA crosslinks we depleted U6 from splicing extract as previously described (Figure 4A). Splicing and UV irradiation of U6 depleted extract containing pre-mRNA with, and without, 4-thioU at (−6) was carried out. RNA species that contained CYH2 pre-mRNA were selected with a biotin containing oligonucleotide complementary to the CYH2 intron and streptavidin magnetic beads. U2 will only be selected if directly crosslinked to CYH2 pre-mRNA. Beads were then washed and any selected U2 was subjected to primer extension mapping directly from the beads. No primer extension stops were found within the first 100 nucleotides of U2 from reactions with, or without, 4-thioU at (−6). However, a significant amount of primer extension product was detected corresponding to the 5′ end of U2 in reactions containing 4-thioU at (−6) whereas only background levels of U2 were detected in control reactions (Figure 7C). This indicates that significant quantities of U2 were crosslinked to pre-mRNA containing 4-thioU at (−6) compared to the control and suggests that this crosslink occurs at the 5′ end of U2. In support of this conclusion is the fact that mobility of U2-pre-mRNA crosslinks in a 5% denaturing gel was between the long and short forms of U5 (182 and 214 nt) crosslinked to pre-mRNA (Figure 1C). While it is hard to predict exactly how crosslinked molecules will run in a denaturing gel the fact that U2 (1175 nts) crosslinked to pre-mRNA migrates with U5-pre-mRNA crosslinks suggests that (−6) crosslinks at, or near, the end of U2. Therefore, we conclude that the 5′ end of U2 most likely crosslinks to position (−6) in the pre-mRNA.
DISCUSSION
Incorporation of a single crosslinking group from positions (−3) to (−7) upstream of the 5′ splice site has allowed us to determine the nature and timing of snRNA interactions before the first step of splicing. We found a specific interaction between the (−6) position of pre-mRNA and the 5′ end of U2 snRNA that brings to light parallels between yeast and mammalian spliceosomes (18,19). U5 snRNA was found to interact with every position within the pre-mRNA from (−3) to (−7) encompassing the whole 5′ side of U5 loop 1. In contrast, only a specific site in U1 interacts with each position examined. Mapped U1–pre-mRNA interactions are not compatible with base pairing of U1 with the 5′ splice site and suggest an alternative conformation of U1 before the base-pairing interaction. Overall, we provide new insight into snRNA interactions with the pre-mRNA that occur prior to the first step of pre-mRNA splicing.
A revised model for U1–pre-mRNA interactions prior to the first step of splicing
A model where U1 base pairs with the 5′ splice site began with recognition that the 5′ end of U1 was complementary to the 5′ splice site (39,40). Later biochemical and genetic studies proved U1 snRNP bound to the 5′ splice site, the 5′ end of U1 was required for splicing and base pairing between the 5′ end of U1 and the 5′ splice site was required for splicing (9–11,41,42). Crosslinking analysis provided evidence for direct interaction between U1 and 5′ splice site region (12–14). However, the nature of these crosslink interactions has never been investigated in detail and was assumed to reflect a base-pairing interaction of U1 with the 5′ splice site.
Our analysis of U1 crosslinking to pre-mRNA has identified a previously undiscovered interaction. Mapping of U1-pre-mRNA crosslinks at four positions within the pre-mRNA has identified a specific site in U1, adjacent to helix 1, which interacts with each position in the pre-mRNA. The site of crosslinks in U1 are not compatible with a conformation of U1 that is base pairing at the 5′ splice site, therefore, we believe this interaction is distinct from the classical base-pairing interaction of U1 with the 5′ splice site. It is not surprising that we have identified non-base-pairing interactions of U1 as RNA crosslinks generated with 4-thioU are known to favor non-Watson–Crick interactions (43). In addition, if the 5′ end of U1 was base paired to the 5′ splice site, as in the base-pairing model (Figure 6A), then base pairing would only extend to the (−2) position of the pre-mRNA as this is the limit of available single-stranded 5′ end of U1. This is not the case, as our 4-thioU crosslinking has detected U1 interactions to at least (−7) in the pre-mRNA. We propose that previous crosslinking studies using 4-thioU detected non-base-pairing interactions of U1 (12–14). A prediction would be that mapping of U1-pre-mRNA crosslinks at positions in the region of the 5′ splice site (−1 or +2 from 5′ splice site) would also map to the site our crosslinks have mapped to.
Base pairing of U1 to the 5′ splice site is not required for interaction of U1 snRNP with pre-mRNA in yeast or 5′ splice site selection in mammalian extracts (44,45). U1 snRNP protein U1C recognizes and binds the 5′ splice site consensus sequence when the 5′ end of U1 is removed and this protein–RNA interaction may precede U1-pre-mRNA base-pairing interaction during 5′ splice site recognition (46,47). In fact, no direct interaction of U1C with U1-pre-mRNA base-paired duplex was found and it was proposed that the first interaction of U1 snRNP with the 5′ splice site is via U1C, which prevents base pairing between the 5′ end of U1 and 5′ splice site (48). This initial commitment complex, termed CC0, is proposed to be an unstable, non-base-paired, conformation of U1 snRNP that then undergoes a conformational change to canonical base-paired configuration facilitated by U1C (48). In vivo, chromatin immunoprecipitation analysis of snRNP association with nascent pre-mRNA also supports an initial non-base-paired interaction of U1 snRNP with nascent pre-mRNA (49). We believe that the interaction we detect by 4-thioU crosslinking may be this initial CC0 conformation. We propose a model where U1 snRNP scans the 5′ splice site region with a site in U1 adjacent to helix 1 in a non-base-paired conformation mediated by U1C. This scanning leads to a temperature-dependent conformational change, mediated by U1C, to the canonical base-paired conformation of U1 snRNP with the 5′ splice site as previously proposed (48).
Extensive interaction of U5 snRNA loop 1 with the 5′ exon region
U5 loop 1 is required for aligning the two exon ends during the second step of pre-mRNA splicing (12,13,23,26,27). While U5–exon interactions are not required for the first catalytic step to take place in vitro they are required for holding the 5′ exon intermediate for ligation to the 3′ exon during the second step of splicing in yeast (26). We have defined U5–5′ exon interactions before and after the first step of splicing. U5 extensively interacts with pre-mRNA at all positions we tested from (−3) to (−7). Only interactions at (−3) and (−4) continue to the 5′ exon intermediate produced following the first step of splicing. Mapping of U5–pre-mRNA interactions with different positions in the 5′ exon in this work and in previous work (12,21) reveals that as the crosslinking group is placed further from the 5′ splice site the location of U5-pre-mRNA crosslink shifts along the 5′ portion of U5 loop 1 (Figure 7B). U5–pre-mRNA interactions at (−3) and (−4) that persist with the 5′ exon intermediate are predicted to overlap with the interactions of U5 loop 1 and lariat-3′ exon intermediate (Figure 7B). Therefore, we propose a conformational change must occur immediately before, or following, the first step of splicing that allows alignment of the 5′ exon and lariat-3′ exon intermediates with U5 loop 1 for the second step of splicing. A likely candidate for regulating this conformation change before the first step of splicing is the NTC complex as it has been shown to be required for defining specificity of U5 interaction with the 5′ splice site (33). A candidate for regulating conformational change in alignment of the 5′ exon with U5 loop 1 after the first step of splicing is Prp8. Prp8 is proposed to modulate conformational changes within the spliceosome (50) and crosslinks to both the 5′ exon as well as U5 loop 1 (51,52). Another candidate is Prp18 that has recently been implicated in stabilizing U5–exon interactions prior to the second step of splicing (53).
Conservation of snRNA–pre-mRNA interactions between yeast and mammalian spliceosomes
We identified an interaction of U2 with the 5′ exon of pre-mRNA that takes place in prespliceosomes before addition of tri-snRNP to form active spliceosomes. This U2–pre-mRNA interaction is ATP dependent, requires prior interaction of U2 with the branchpoint and occurs with more than one pre-mRNA. While our data point to the 5′ end of U2 interacting with the 5′ exon of the pre-mRNA there is still the possibility that a region of U2 outside the first 100 nts may interact with the 5′ exon or that the nature of the crosslink has not allowed us to map it by reverse transcription blockage. An interaction of the 5′ end of U2 with the 5′ exon of pre-mRNA has been identified in mammalian major spliceosomes and an interaction of the 5′ end of U12 snRNA with the 5′ exon of pre-mRNA in mammalian minor spliceosomes (18,19). Mammalian U2-pre-mRNA interaction takes place in ATP independent E complexes and ATP dependent A complexes (18). In addition, as a wide-ranging hydroxyl-radical production approach was utilized, the 5′ end of U2 was also found to be in proximity to 5′ splice site, branchpoint, polypyrimidine tract and 3′ splice site of pre-mRNA as well as to U1 (18). In mammalian minor spliceosomes U12 snRNA was found to interact with (−2) of pre-mRNA and required ATP and prior branchpoint interaction (19). This U12 interaction is similar to our findings in yeast and may reflect the fact that both studies utilized short-range crosslinking group 4-thioU. As it is the 5′ end of U2 that is involved in the interaction with the 5′ exon of the pre-mRNA in each spliceosome this brings up the question of whether the 5′ cap structure might be important. At least in the mammalian major spliceosome, this does not appear to be the case as the crosslinking group utilized was attached directly to the 5′ end of U2 without a cap structure (18). Overall, our data indicate that there is conservation of snRNA–pre-mRNA interactions between yeast and mammalian spliceosomes. It will be interesting to determine whether U1–pre-mRNA and U5–pre-mRNA interactions we have identified in yeast are also conserved in the mammalian system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Andy Newman for the Prp2 expression plasmids, Joseph Burhani for cloning RPS10B and members of the O’Keefe lab for helpful discussions and advice on the manuscript. This work was supported by a grant from the BBSRC. Funding to pay the Open Access publication charges for this article was provided by the BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNA by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA World. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 2.Jurica MS, Moore MJ. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 3.Lührmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim. Biophys. Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- 4.Nilsen TW. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons RW, Grunberg-Manago M, editors. RNA Structure and Function. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1998. pp. 279–307. [Google Scholar]
- 5.Brow DA. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 6.Séraphin B, Rosbash M. Identification of functional U1 snRNA-premRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 7.Séraphin B, Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNP-pre-mRNA complex and is recognised in the absence of U2 snRNA. EMBO J. 1991;10:1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Séraphin B, Kandels-Lewis S. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993;73:803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- 9.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA-pre-mRNA base-pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siliciano PG, Guthrie C. 5′ splice site selection in yeast: Genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang Y, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 12.Newman AJ, Teigelkamp S, Beggs JD. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 13.Sontheimer EJ, Steitz JA. The U5 and U6 small nuclear RNAs as the active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 14.Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 15.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 16.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 17.Donmez G, Hartmuth K, Lührmann R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10:1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donmez G, Hartmuth K, Kastner B, Will CL, Lührmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol. Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Frilander MJ, Meng X. Proximity of the U12 snRNA with both the 5′ splice site and the branch point during early stages of spliceosome assembly. Mol. Cell Biol. 2005;25:4813–4825. doi: 10.1128/MCB.25.12.4813-4825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman AJ. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997;16:5797–5800. doi: 10.1093/emboj/16.19.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrail JC, Tatum EM, O’Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J. 2006;25:3813–3822. doi: 10.1038/sj.emboj.7601258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 23.Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 24.Alvi RK, Lund M, O’Keefe RT. ATP-dependent interaction of yeast U5 snRNA loop 1 with the 5′ splice site. RNA. 2001;7:1013–1023. doi: 10.1017/s135583820101041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConnell TS, Steitz JA. Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J. 2001;20:3577–3586. doi: 10.1093/emboj/20.13.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Keefe RT, Newman AJ. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell. 1996;86:679–689. doi: 10.1016/s0092-8674(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 28.Kandels-Lewis S, Séraphin B. Role of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 29.Kim CH, Abelson J. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- 30.Lesser CA, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 31.Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 32.Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 33.Chan S-P, Cheng S-C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 34.Moore MJ, Sharp PA. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 35.Fabrizio P, McPheeters DS, Abelson J. In vitro assembly of yeast U6 snRNP: A functional assay. Genes Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 36.Patterson B, Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- 37.Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plumpton M, McGarvey M, Beggs JD. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. Are snRNPs involved in splicing? Nature. 1980;283:220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- 40.Rogers J, Wall R. A mechanism for RNA splicing. Proc. Nat Acad. Sci. USA. 1980;77:1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krämer A, Keller W, Appel B, Lührmann R. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984;38:299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- 42.Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell. 1983;33:509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- 43.Sontheimer EJ. Site-specific RNA crosslinking with 4-thiouridine. Mol. Biol. Rep. 1994;20:35–44. doi: 10.1007/BF00999853. [DOI] [PubMed] [Google Scholar]
- 44.Du H, Rosbash M. Yeast U1 snRNP-pre-mRNA complex formation without U1 snRNA-pre-mRNA base pairing. RNA. 2001;7:133–142. doi: 10.1017/s1355838201001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lund M, Kjems J. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA. 2002;8:166–179. doi: 10.1017/s1355838202010786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du H, Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D, Rosbash M. Identification of eight proteins that crosslink to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du H, Tardiff DF, Moore MJ, Rosbash M. Effects of the U1C L13 mutation and temperature regulation of yeast commitment complex formation. Proc. Natl Acad. Sci. USA. 2004;101:14841–14846. doi: 10.1073/pnas.0406319101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA: 5′ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat. Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 51.Dix I, Russell CS, O’Keefe RT, Newman AJ, Beggs JD. Protein-RNA interactions in the U5 snRNP of Saccharomyces cerevisiae. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teigelkamp S, Newman AJ, Beggs JD. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggests a role in stabilization of exon alignment by U5 snRNA. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21:1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.