Abstract
Myocyte enhancer factor 2 (MEF2) proteins play a key role in promoting the expression of muscle-specific genes in differentiated muscle cells. MEF2 activity is regulated by the association with several transcriptional co-factors and by post-translational modifications. In the present report, we provide evidence for a novel regulatory mechanism of MEF2C activity, which occurs at the onset of skeletal muscle differentiation and is based on Lys4 acetylation. This covalent modification results in the enhancement of MEF2C binding to DNA and chromatin. In particular, we report that the kinetic parameters of MEF2/DNA association change substantially upon induction of differentiation to give a more stable complex and that this effect is mediated by Lys4 acetylation. We also show that Lys4 acetylation plays a prominent role in the p300-dependent activation of MEF2C.
INTRODUCTION
The myocyte enhancer factor 2 (MEF2) transcriptional activators are members of the MADS box (MCM1, Agamous and Deficiens, SRF) family of proteins. There are four vertebrate MEF2 proteins, MEF2A, B, C and D, encoded by distinct genes. They share high amino acid identity (95%) throughout the highly conserved amino-terminal MADS box (aa 1–57) and the adjacent MEF2-specific domain (aa 58–86); these domains mediate the DNA-binding affinity and specificity and the homo and hetero dimerization (1). The C-terminal region of MEF2 proteins is more divergent and acts as a transcriptional activation domain. MEF2 factors were originally identified in skeletal muscle cells, they lack myogenic activity, but strengthen the activity of myogenic bHLH proteins. Several lines of genetic and biochemical evidence underscore the central role played by MEF2 proteins in promoting skeletal muscle differentiation. Loss-of-function mutations in the single Drosophila MEF2 gene prevent myoblast differentiation (2–4) and dominant-negative MEF2 mutants inhibit myoblast differentiation in vitro (5). The pivotal role played by MEF2 proteins in skeletal myogenesis has been reinforced by two recent papers where the role of MEF2 proteins in vertebrate skeletal muscle in vivo has been clarified. The combined knock down of mef2c and mef2d in zebrafish revealed the essential role of MEF2 proteins for thick filament formation after terminal differentiation (6). Similar results were obtained in mice where skeletal muscle-specific mef2c deletion results in sarcomere disorganization and myofibres deterioration after birth (7).
The transcriptional activity of MEF2 is tightly regulated during skeletal muscle differentiation. MEF2 proteins are expressed in proliferating C2C12 myoblasts but they fail to activate MEF2-dependent transcription of endogenous or transiently transfected genes unless the cells are induced to differentiate (8,9). Multiple pathways exist to ensure the repression of these transcription factors in dividing myoblasts. For example, Cdk4/Cyclin D represses the activity of MEF2 proteins by blocking their interactions with GRIP1 (10). In C2C12 myoblasts repression of MEF2 activity depends on its association with class II HDACs-4-5-7-9 (11). This interaction does not affect MEF2 DNA-binding activity and implies the recruitment of HDACs to MEF2-containing transcriptional complexes.
During muscle differentiation, class II HDACs are sequestered in the cytoplasm. As a result, transcriptional repression by HDACs is relieved, leading to up-regulation of MEF2 target genes, such as ‘Muscle Creatine Kinase (MCK)’. Release of class II HDACs from MEF2 may occur by phosphorylation of conserved serine residues in the HDAC N-terminal region, resulting from the activation of number of Ser/Thr kinases such as CaMK, PKCδ, PKD, MARK2, Mirk/dyrk1B and SIK1 serine/threonine kinases (12–14). The released MEF2 is then able to associate with the acetyltransferase co-activator p300 and stimulate MEF2-dependent genes. Importantly, MEF2 activity is regulated by several post-translational modifications in the C-terminal region: MEF2C is acetylated by the histone acetyltransferase (HAT) p300 selectively in differentiated muscle cells and this post-translational modification enhances MEF2 activity (15). Moreover, MEF2 proteins are sumoylated in vitro and in vivo on a C-terminal lysine residue and this post-translational modification inhibits their transcriptional activity, likely through the recruitment of transcriptional repressors other than class II HDACs (16–19).
In the present report, we contribute to the understanding of the mechanisms underlying the activation of MEF2 proteins in differentiating muscle cells, by providing evidence of a regulation of the DNA-binding properties of MEF2C. In particular, we describe for the first time a differentiation-dependent post-translational modification that occurs in the MADS box, consisting of lysine 4 acetylation. This modification results in the enhancement of the binding of MEF2C to its cognate DNA site, as well as of its transcriptional activity. In addition, we find evidences for p300 playing a role in this regulatory mechanism: p300 enhances the DNA-binding activity of MEF2 and acetylates it on Lys4, furthermore, Lys4 acetylation contributes substantially to the p300-mediated activation of MEF2-dependent transcription.
MATERIALS AND METHODS
Plasmids
The bacterial expression vector pET32b/MEF2C was generated by subcloning a PCR product of the mouse MEF2C cDNA in the pET32b(+) vector (Novagen), resulting in fusion of His6-thioredoxin with MEF2C (His6MEF2C). Briefly, the cDNA encoding MEF2C [gi:293728] was amplified from pcDNAI/Amp/MEF2C, using the following primers:
TA_FW 5′-GGGAAGCTTGGGAGAAAAAAGAT-3′
TA_REV 5′-TCTAGATCATGTTGCCCATCCTTC-3′.
The PCR product was inserted in the pCR3.1 vector using the TA cloning kit (Invitrogen). The insert was excised from pCR3.1 by cutting with HindIII/NotI restriction enzymes and inserted in the same restriction sites in the pET32b(+) plasmid vector.
The expression vector pET32b/MEF2C1-92 was obtained by deleting the DNA sequence encoding the amino acids 93–466 of MEF2C from the pET32b/MEF2C plasmid by site-directed mutagenesis with the Quick Change Site-Directed Mutagenesis Kit (Stratagene). The deleted plasmid was obtained by PCR amplification of pET32b/MEF2C with the following oligonucleotides:
FW 5′-GAGAAAGAAGGGCCTCGCGGCCGCACTCGAGC-3′
RV 5′-GCTCGAGTGCGGCCGCGAGGCCCTTCTTTCTC-3′
pcDNAI/Amp/MEF2C_K4R, pcDNAI/Amp/MEF2C_K4Q, pGFP/MEF2C_K4R, pGFP/MEF2C_R3T and pFLAG/MEF2C_K4R, pET32b/MEF2C1-92_K4R were obtained by mutagenesis respectively of pcDNAI/Amp/MEF2C (20), pGFP/MEF2C, pFLAG/MEF2C (21) and pet32b/MEF2C1-92. The mutagenesis reactions were performed using the Quick Change Site-Directed Mutagenesis Kit (Stratagene). The plasmid encoding the HA-tagged p300 (22) was a generous gift of Pier Lorenzo Puri (Dulbecco Telethon Institute at Fondazione A. Cesalpino, Rome, Italy). The reporter plasmid for MEF2-dependent transcription, pGL3(desMEF2)3, contains three tandem MEF2 sites from the mouse Desmin enhancer (23) inserted in the pGL3 vector (Promega) and was a kind gift of S. Schiaffino (University of Padova, Italy).
Cell cultures and transfections
C2C7 is a skeletal muscle cell line subcloned (24) from the original C2 cell line derived from C3H mice (25). This cell line was kindly provided by M. Buckingham (Pasteur Institute, Paris, France). C2C7 cells were grown in Dulbecco's Modified Eagle's Medium (DMEM, Euroclone) containing 20% fetal calf serum (FCS, Life Technologies) (Growth Medium, GM) at low density and, when approaching confluence, induced to differentiate with DMEM–2% Horse Serum (HS, Hyclone) (Differentiation Medium DM). COS1 simian kidney cells (26) and C3H10T1/2 mouse fibroblasts were maintained in DMEM containing 10% FCS. Cells, grown at 80% confluence, were transfected using the lipid-based Lipofectamine Plus Reagent (Invitrogen) according to the manufacturer's instructions. For confocal imaging, cells were grown in glass-bottom Petri dishes (WillCo).
C3H10T1/2 myogenic conversion assay
C3H10T1/2 cells were grown to 60% confluence in 60-mm dishes and transfected with Lipofectamine-Plus reagent (Invitrogen) with 3 μg of pEMSV-MyoD (27) in association with 1 μg of pcDNAI/Amp (empty vector), pcDNAI/Amp/MEF2C and pcDNAI/Amp/MEF2C_K4R. After 24 h, cells were split in 40-mm dishes and mantained in growth medium for 24 h and then transferred to differentiation medium changing media every 2 days. Myogenic conversion was assessed by western blot analysis of Myosin heavy chain (MyHC) expression. Cells were harvested at different time points and lysed in 100 μl of single detergent buffer [50 mM Tris–HCl pH 8, 150 mM NaCl, 1% Triton X-100 supplemented with protease inhibitors (Roche)]. The extracts were analysed by western blotting, using the antibodies against actin and MyHC. The results were quantitated by densitometry.
Transcription reporter assays
COS1 cells were co-transfected with pGL3(desMEF2)3, pRSVβ-gal, the expression vectors for MEF2C wild-type (WT) or MEF2C mutant (K4R) and for the HA-tagged p300 enzyme. Thirty-six hours after transfection, cell lysates were obtained by three freeze-and-thaw cycles in 250 mM Tris–HCl pH 7.5, 15% glycerol and 10 mM Dithiothreitol (DTT); 30% of the extract was used for the β-galactosidase assay and up to 30% for the luciferase assay. For the luciferase assay, samples were diluted to a 350 µl volume with LUC buffer (25 mM Glycil–glycine pH 7.8, 2 mM ATP, 10 mM MgSO4) and incubated with 40-nm Luciferin (Sigma). Luciferase activity was measured in the luminometer LUMAT LB 9501; Berthold. For the β-galactosidase assay samples were processed as described previously (28). Luciferase activity was normalized to the β-galactosidase activity.
Protein analysis
Samples were diluted in 1× SDS gel-loading buffer (50 mM Tris–HCl pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol) and separated by SDS-polyacrylamide gel electrophoresis (SDS–PAGE). The separated proteins were subsequently analysed by western blot as previously described (21). The following antibodies were used: mouse M2 monoclonal anti-FLAG (1:1000, F3165, Sigma), rabbit polyclonal anti-Acetylated-Lysine (1:1000, #9441, Cell Signaling), rabbit polyclonal anti-MEF2 (1:5000, C21 sc-313X, Santa Cruz), mouse monoclonal anti-His6 (1:500, #11922416001, Roche), rabbit polyclonal anti-actin (1:1500, #MAB 1501, Chemicon) and mouse monoclonal anti MyHC (1:200, MF20 Developmental Studies Hybridoma Bank, IA, USA).
Chromatin immunoprecipitation (ChIP) and PCR amplification
The ChIP protocol was performed on C2C7 cells as described (29). C2C7 cells were grown in GM. At 90% of confluence (0 h), they were induced to differentiate in DM for 24 h. The following antibodies were used: rabbit polyclonal anti-MEF2 (C21 sc-313X, 200 mg/0.1 ml, Santa Cruz) and, as a control, goat polyclonal anti-enolase (C-19 sc-7455, 200 mg/m1, Santa Cruz).
Precipitated DNA fragments were analysed by semi-quantitative PCR with BD Advantage TM 189 2 PCR Kit (BD Biosciences). Immunoprecipitated DNA was subjected to 28 cycles of PCR with primers specific for the Myogenin promoter (sense 5′-TTTCTGTGGCGTTGGCTATATATTTATC-3′; antisense 5′-TGCTGGGTGCCATTTAAAC-3′) or for an enhancer region of the liver-specific Phenylalanine Hydroxylase (PAH) gene (30) (sense 5′-CAAAATGGTGCTGTATCTCTGATATTC-3′, antisense 5′-GGCACCAACTTCCTCTTTGAGT-3′). As a control for DNA content, PCR reactions were also performed on chromatin samples prior to immunoprecipitation (input), undiluted or diluted by a factor of 50, 100 and 200 to assess whether the assay is being performed in the linear range. The resulting PCR products were resolved through a 10% native acrylamide gel and DNA visualized by Ethidium Bromide staining. The immunoprecipitated chromatin was quantified by real-time quantitative PCR with an ABI 7900 PRISM™ DNA Sequence Detector (PE Applied Biosystems) using a SYBR GREEN PCR Core Reagent Kit (PE Applied Biosystems). The chromatin was subjected to 40 amplification cycles. Known quantities of total input were used as standards. Fold enrichment was estimated relative to the control antibody sample.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed on a Leica TCS SP2 confocal laser scanner microscope using a 63X/1,4 N.A. oil immersion objective. Transfected C2C7 cells, expressing the fluorescent fusion proteins GFP-MEF2C_WT, K4R or R3T, were excited with an Argon laser at 488 nm. Images were collected every 0.064 s at a resolution of 512 × 64 pixels as follows: 5 images for pre-bleaching, 5 images for bleaching, 150 images for post-bleaching. Bleaching was performed using the 488 nm and 514 nm lines of an Ar laser at ∼100% laser power (100 mW total nominal output) on a circular region of 3-μm diameter; recovery was observed with low laser intensity. In order to correct for the loss in fluorescence during the acquisition, data were doubly normalized as described previously (31,32). For each experiment, at least 10 datasets were collected, which were used to calculate the half-life of maximal recovery. This is defined as the time point after bleaching when the normalized fluorescence has increased to half the value of maximal recovery. Each experiment was repeated at least twice.
Nuclear extracts and electrophoretic mobility shift assays (EMSAs)
The nuclear extracts of COS1 cells ectopically expressing MEF2C_WT or K4R and C2C7 cells were obtained as described previously (21). The double-stranded probe, representing the MCK MEF2-binding site, was obtained by in vitro annealing of complementary oligonucleotides. Terminal labelling was obtained by incubation with polynucleotide kinase (PNK) in the presence of [γ-32P]ATP. The sequences of the sense strands of the probes, wt or mutant, were as described previously (33).
EMSAs were performed as described (34). The binding reaction was performed in a total volume of 15 µl containing 4 µg of nuclear extract proteins and 1 ng of labelled DNA probe. Alternatively, the labelled MEF2-binding site was incubated with 4 μl of the in vitro translated proteins MEF2C_WT or MEF2C_K4R and MEF2C_K4Q. Where indicated the C2C7 cells nuclear extracts were added with 0.5 µl of p300 enzyme obtained in the baculovirus system (35). In vitro translated proteins were obtained with the TnT Quick Coupled Transcription/translation System (Promega). p300 was a generous gift of Roberto Mantovani (University of Milano). For the supershift experiments, 0.2 μl of MEF2 antibody (C21 sc-313X, Santa Cruz) was used. Competition experiments were performed by diluting the labelled probe with a 100-fold molar excess of cold wt or mutant competitor probe.
Purification of bacterially expressed recombinant proteins
The recombinant proteins His6MEF2C1-92_WT or K4R were purified from transformed BL21 bacterial cells. Expression of the recombinant proteins was induced with 1mM isopropyl-b-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Recombinant proteins were affinity purified with a Ni-NTA Agarose (Qiagen) according to the manufacturer's instructions.
In vitro acetylation assay
One microgram of bacterially expressed protein was incubated with 0.5 µl of baculovirus-purified p300 for 1 h at 30°C in a 30-μl reaction mixture containing acetylation buffer (50 mM Tris–HCl pH 8.5, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, 10 mM sodium butyrate, 1 mM PMSF) and 1 mM acetyl coenzyme A (AcCoA). Mock acetylation reactions were performed using AcCoA in the absence of p300 or incubating the complete reaction mix with the p300-specific inhibitor LysCoA (100 nM). The p300 selective inhibitor LysCoA (36) was a generous gift of Phil Cole (Johns Hopkins University, Baltimore).
Acetylated proteins were analysed in western blot with an antibody that recognizes acetylated lysine residues and successively with an antibody directed against the His6 protein tag.
Protein purification and MALDI-TOF MS analysis
One day prior to transfection, C2C7 myoblasts were plated at a density of 106 cells/dish. Four micrograms of plasmid expressing FLAG-MEF2C were used to transfect 10-cm diameter dishes. Cells were maintained in growth medium for 36 h or alternatively in differentiation medium for 72 h. Cells were washed twice with PBS and incubated for 30 min at 4°C in 1 ml of lysis buffer [50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Igepal, supplemented with 1 mM PMSF, 330 nM TSA, 10 mM sodium orthovanadate and protease inhibitors tablet (Complete; Roche)]. FLAG-MEF2C was purified by affinity gel chromatography with the anti-FLAG M2-affinity gel (Sigma), according to the manufacturer's instructions. Proteins recovered from the affinity columns were separated by SDS–PAGE and then visualized by Coomassie brilliant blue staining (Sigma). The band corresponding to FLAG-MEF2C was excised and cut into small pieces of ∼1 mm3. Gel pieces were washed, reduced with DTT, S-alkylated with iodoacetamide, and in-gel digested with trypsin (from bovine pancreas, unmodified, sequencing grade, Roche) as described elsewhere (37). The digestion was allowed to proceed overnight at 37°C. Digested aliquots were analysed directly or after a desalting/concentration step on μZipTipC18 (Millipore) before MALDI-MS analysis. One microlitre of the sample was mixed (1:1, v/v) with matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% TFA), spotted onto the MALDI target and analysed by using a Voyager-DE STR mass spectrometer (Applied Biosystems). The search program ProFound, developed by The Rockefeller Mass Spectrometry Laboratory and the New York University (New York, NY), was used for database searches (38). Peptides were selected in the mass range of 700–4500 kDa. Spectra were calibrated using a matrix and tryptic autodigestion ion peaks as internal standards.
Multiple sequence alignment
Amino acid sequences of mouse MEF2C [gi:293728], human MEF2C [gi:2500875], mouse MEF2A [gi:2500873], human MEF2A [gi:1170908], mouse MEF2D [gi:19526812], human MEF2D [gi:2500876], mouse MEF2B [gi:6678852], human MEF2B [gi:5174543], Drosophila MEF2 [gi:25453445], DEFICIENS [gi:118426], AGAMOUS [gi:399096], Saccharomyces cerevisiae MCM1 [gi:6323686], SRF_human [gi:134876] were aligned with Clustal W.
RESULTS
MEF2 binds the regulatory region of the Myogenin gene and increases its residence time on chromatin in C2C7 cells upon induction of muscle differentiation
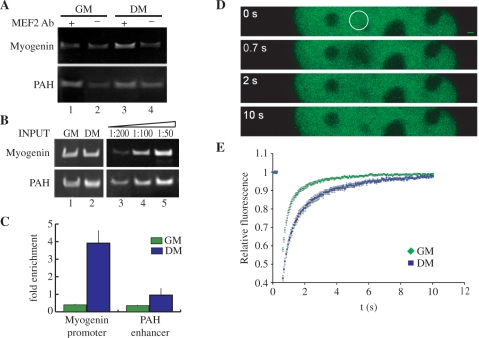
In proliferating C2C7 myoblasts, MEF2 proteins are expressed but are transcriptionally silent (8,9). To investigate whether MEF2 proteins change their binding properties to DNA during muscle cell differentiation, we performed ChIP experiments (Figure 1A–C) on the chromatin extracted from proliferating C2C7 cells cultured in high serum (GM) or cells stimulated to differentiate in low serum for 24 h (DM). Specifically, we verified the enrichment of the regulatory region of the Myogenin gene in the chromatin immunoprecipitated by the MEF2 antibody versus the control antibody, by PCR amplification of the DNA region encompassing nucleotides from −82 to −14 (Figure 1A, upper panel). The enhancer region of the liver-specific PAH gene (Figure 1A, lower panel) was used as a control. Equivalent amounts of input chromatin were used for each sample as shown in Figure 1B (upper and lower left panels). PCR reactions were conducted in semi-quantitative conditions (Figure 1B, upper and lower right panels). The MEF2 antibody immunoprecipitates a complex where the Myogenin sequence is enriched, in comparison with the background level seen with control antibody, only in differentiated muscle cells. (Figure 1A, upper panel, lane 3 compared to control lane 4). The presence of MEF2 proteins in the immunoprecipitates of C2C7 cells at both stages was confirmed by western blot analysis (data not shown).
Figure 1.
Analysis of MEF2 association to its cognate sites on chromatin in proliferating and differentiating C2C7 muscle cells. (A) Soluble chromatin prepared from proliferating myoblasts cultured in growth medium (GM) or confluent cells cultured in differentiation medium for 24 h (DM), was processed for ChIP assays with the antibody directed against MEF2 (MEF2 Ab, lanes 1 and 3) or anti-enolase as a control antibody (lanes 2 and 4). PCR products, obtained with primers spanning the MEF2 site in the Myogenin regulatory region (nt −82 to −14, upper panel), or a region of the promoter of the liver-specific gene PAH (lower panel), were analysed by gel electrophoresis. (B) Equivalent amounts of INPUT chromatin were used as shown by direct PCR of the chromatin samples (lanes 1 and 2). Dilutions (1:50, 1:100; 1:200) of the input DNA from proliferating muscle cells were subjected to PCR in order to verify if the reaction is being performed in semi-quantitative conditions. (C) Quantification of the Myogenin promoter and PAH enhancer immunoprecipitated in the ChIP assays described above using SYBR green and real-time PCR. The quantities of DNA fragments immunoprecipitated with the MEF2 antibody are shown as fold the quantity precipitated with the control antibody, this quantity was arbitrarily taken as 1. The results shown are the average ± SE of at least three estimations. (D, E) FRAP analysis of GFP-MEF2C. C2C7 cells were transfected with GFP-MEF2C and maintained in growth medium (GM) or switched to low serum for 24 h (DM). (D) Time laps imaging of a nucleus, maximal laser power was applied to a selected nuclear area (circle). The recovery of fluorescence in the bleached area was imaged at the indicated time points (0.7, 2 and 10 s; 0 s is the image taken before bleaching) (Bar: 1 µm). (E) Fluorescence intensities in the bleached regions were quantified and corrected for the change in total fluorescence. The obtained relative fluorescence values were plotted against time. Bleaching was at t = 0.
No significant binding of MEF2 to the Myogenin control region could be detected at the myoblast stage (Figure 1A, upper panel, lane 1 compared to control lane 2). A similar enrichment for the PAH control sequence was not observed (Figure 1A, lower panel). The above results were confirmed by a quantitative real-time PCR amplification of the Myogenin and PAH regulatory regions from the immunoprecipitated chromatin. Results in Figure 1C show a significant higher recruitment of MEF2 proteins to the promoter of the Myogenin gene in differentiated muscle cells than in myoblasts; in contrast, the level of MEF2 proteins recruitment to the enhancer of the PAH gene is negligible both in proliferating and in differentiating C2C7 muscle cells. These data show that MEF2 proteins bind the Myogenin promoter in myotubes but not in myoblasts.
Myogenin is an early muscle-specific gene, whose mRNA starts to be detected 24 h after induction of muscle differentiation in C2C7 cells. In addition, the expression of the Myogenin gene is entirely dependent, in vitro, on the integrity of the MEF2 site in its proximal regulatory region (9).
The ability of a transcription factor to activate the transcription of its target genes depends in part on its DNA-binding affinity. ChIP experiments enable the analysis of a fixed binding situation. In fact, due to formaldehyde cross-linking, changes in key kinetic parameters of DNA–protein interactions, like the ‘off-rate’, are evened up. Therefore, we explored the dynamic and kinetic properties of the MEF2–DNA interactions in proliferating and differentiating C2C7 cells ectopically expressing the GFP-MEF2C fusion protein, using the technique of FRAP (Figure 1D and E). The FRAP technique allows the measurement of the mobility of a protein in a living cell, this parameter contains information about the in vivo binding properties of a protein, the main determinant of the intranuclear mobility of transcription factors is their ability to bind DNA (32,39).
First, we verified that the tagged protein GFP-MEF2C binds DNA and activates transcription of a MEF2-driven reporter gene with an efficiency which is comparable to wt MEF2C (data not shown). The representative image of a FRAP experiment shown in Figure 1C, indicates that GFP-MEF2C localizes homogenously throughout the nucleus and is excluded from the nucleolus. Fluorescence bleaching was obtained by applying maximal laser power to a selected circular region of the nucleus (Figure 1D, 0 s, pre-bleaching); fluorescence recovery in the bleached area was then determined at the indicated time intervals (Figure 1E). The relative fluorescence of the bleached area (fluorescence in the bleached area corrected for the change in total nuclear fluorescence) in myoblasts and in differentiating myotubes as a function of time is reported in Figure 1E. It is evident that recovery after bleaching is slower in differentiating muscle cells than in myoblasts. The plot was also used to determine the t1/2 of recovery (protein fluorescence half-life). Since the recovery of fluorescence in the bleached area is due to the migration of unbleached molecules from the surroundings of the bleached area, the protein fluorescence half-life can be considered a measure of the mobility of the protein and therefore of its association to chromatin. As it is shown in Figure 5D, the t1/2 of GFP-MEF2C in myoblasts is 0.34 s (±0.03 s SE) versus a t1/2 of 0.64 s (±0.05 s SE) measured in cells induced to differentiate. The ChIP and the FRAP results strongly suggest an increase of the affinity of MEF2C for its chromatin sites in response to a differentiation stimulus, concomitant with the activation of MEF2-dependent gene expression.
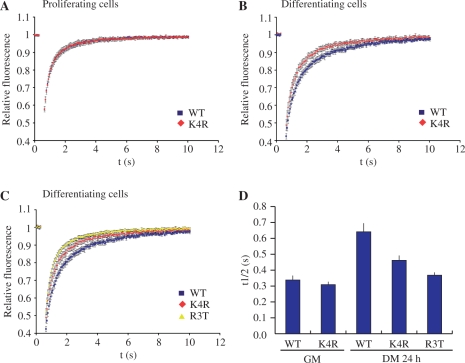
Figure 5.
FRAP analysis of GFP-MEF2C mutants. (A and B) Fluorescence recovery curves obtained with GFP-MEF2C_K4R (red) and wild-type GFP-MEF2C (blue) in proliferating (A) and differentiating (B) C2C7 cells. (C) Fluorescence recovery curves obtained with GFP-MEF2C_R3T (yellow), GFP-MEF2C_K4R (red) and wild-type GFP-MEF2C (blue) in differentiating C2C7 cells. (D) Half-life values (time in seconds when fluorescence intensity in bleached area reaches half of the maximal value) in proliferating (GM) or in differentiating (DM 24 h) C2C7 cells transfected with wild-type GFP-MEF2C (WT), GFP-MEF2C_K4R (K4R), GFP-MEF2C_R3T (R3T). Reported values are average values ± SE, obtained from at least 10 cells in two independent experiments.
MEF2C is acetylated in the MADS box at Lys4 in differentiated muscle cells
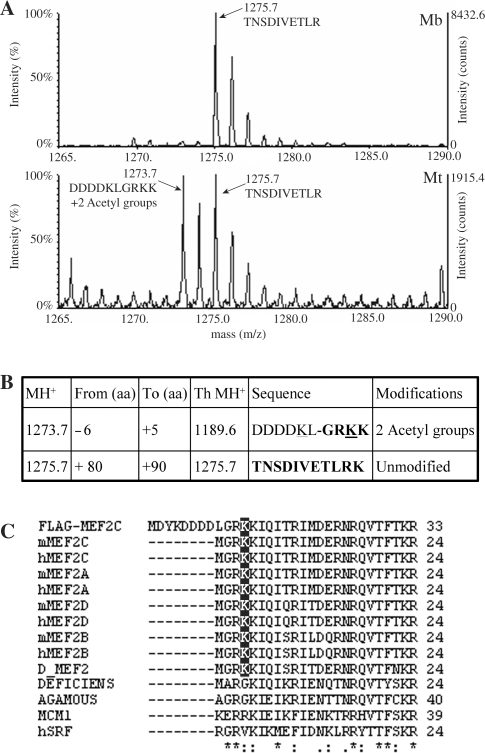
The results of the ChIP and the FRAP experiments indicate that the stability of MEF2–DNA interaction increases considerably at the transition from proliferating to differentiated C2C7 muscle cells, suggesting the existence of a regulatory mechanism that modulates the DNA-binding activity of MEF2 in the course of myogenesis. Since MEF2 proteins are subjected to extensive post-translational modifications that in some cases were shown to regulate their activity, we undertook a systematic analysis of MEF2C purified from proliferating or differentiated C2C7 muscle cells, with the aim of identifying differentiation-specific covalent modifications that could account for the activation of MEF2C upon induction of differentiation. The mass spectrometry analysis of the tryptic digest of FLAG-MEF2C, purified from transfected C2C7 myoblasts and myotubes, revealed an ion peak (1273.7 kDa), which is present only in differentiated cells (Figure 2A, compare upper and lower panels). The mass of this ion does not correspond to any of the theoretical peptides and is coherent with the addition of two acetyl groups to the N-terminal part of the recombinant protein, which includes the six C-terminal amino acids of the FLAG peptide and the amino acids from 2 to 5 in the native MEF2C sequence (Figure 2, panel B; see also the alignment between the N-terminal sequences of FLAG-MEF2C and of native MEF2C in Figure 2C). This N-terminal fragment contains three hypothetical acetyl acceptor lysine residues; one in the FLAG peptide and two in the MEF2C sequence, corresponding to Lys4 and Lys5. Since the acetylated peptide observed in the mass spectrum results from the cut after Lys5 and trypsin cuts proteins in correspondence of unmodified lysine residues, we can exclude the acetylation of Lys5 and infer that Lys4 in MEF2C and the lysine residue in the C-terminus of the FLAG epitope are the acetylated amino acids in the peptide. The peak corresponding to the unmodified peptide was not revealed, neither in the tryptic digest of the protein purified from myoblasts. A simple explanation is that this region is highly enriched of basic amino acids, which are recognized by trypsin if not covalently modified; thus, a complete tryptic digestion would produce peptides too small to be reliably detected by mass spectrometry. The described 1273.7-kDa peptide could not be subjected to MS/MS sequence analysis due to its very low relative concentration. As shown in Figure 2C, Lys4 is conserved in all MEF2 proteins, including D-MEF2, but is absent from all the other members of the MADS box family of transcription factors. Although we did identify many other modified peptides (i.e. phosphopeptides), all of them were present in both proliferating and differentiated cells (data not shown). Finally, we could not reliably detect acetylated peptides other than the one containing Lys4.
Figure 2.
Identification of a novel acetylation site in MEF2C. (A) MALDI spectra (range 1265–1290 kDa) of FLAG-MEF2C tryptic digests obtained from transfected C2C7 myoblasts (Mb, upper panel) and myotubes (Mt, lower panel). The monoisotopic peaks at 1275.7 and 1273.7 kDa are indicated. (B) A table is shown indicating the mass of the monoisotopic ions (MH+), the residues range (from aa to aa), the theoretical molecular weight (Th MH+), the sequence of unmodified peptides and compatible modifications. The residues amenable to the indicated modifications are underlined. (C) Sequence comparison between the MADS box of MEF2 proteins and of other members of the MADS box family. The lysine 4 residue is highlighted.
Lysine 4 in MEF2C is acetylated both in vitro and in vivo
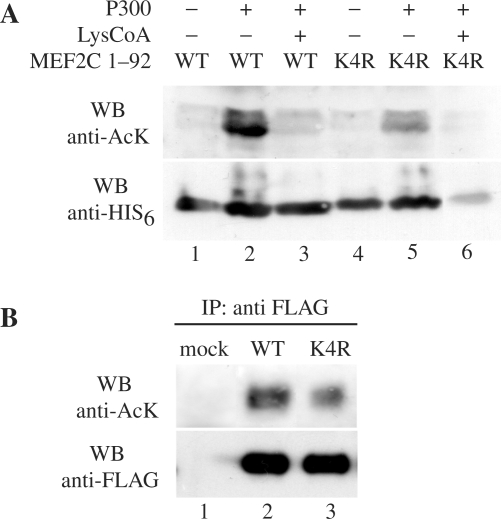
A previous report (15) described several acetylated lysine residues in the C-terminal domain of MEF2C, notably only in differentiated C2C7 muscle cells. Our mass spectrometry results indicate an additional acetylation event that takes place on Lys4. With the aim to get more details about the acetylation of this residue, we performed in vitro acetylation assays. p300 purified from baculovirus was incubated with a purified recombinant His6-thioredoxin-fusion protein containing the N-terminal MADS box and MEF2 domain of MEF2C WT or a mutant where Lys4 was changed to arginine (K4R), an amino acid that cannot be acetylated. The incorporation of AcCoA was monitored with the antibody that recognizes acetylated lysine residues. As shown in Figure 3A, the acetylation of MEF2C depends on the presence of the enzyme (compare lanes 2 and 1) and on its catalytic activity, since the p300-specific inhibitor LysCoA abolishes the reaction (compare lanes 2 and 3). The extent of acetyl incorporation in the mutant protein is negligible in comparison with the wt protein (compare lanes 5 and 2) indicating that Lys4 is indeed a target of acetylation by p300 in vitro. Comparable amounts of protein substrates were used, as shown in Figure 3A, lower panel. Importantly, we found that Lys4 is acetylated also in cultured cells. To show this, we ectopically expressed FLAG-MEF2C_WT or FLAG-MEF2C_K4R together with GFP-p300 in COS1 cells. The FLAG fusion proteins were purified by affinity gel chromatography from cell lysates 48 h after transfection and then detected with the antibodies directed to the FLAG peptide and to acetylated lysine. The results shown in Figure 3B demonstrate that MEF2C is acetylated in vivo in COS1 transfected cells that overexpress p300 and that Lys4 is one of the prominent acetylated residues, since the signal of FLAG-MEF2C_K4R is lower than FLAG-MEF2C_WT (at least 30% lower as evaluated by densitometric analysis, data not shown). The signal associated to the K4R mutant is not abolished, consistently with the observation that other acetylated residues are present in the full-length protein (15).
Figure 3.
MEF2C is acetylated in vitro and in vivo. (A) Wild-type (WT) His6MEF2C1-92, or the mutant where the Lys4 residue is substituted with an arginine (K4R) fusion proteins consisting of the MADS box and the MEF2 domains of MEF2C fused to the His6 tag, were analysed by gel electrophoresis after in vitro acetylation in the reaction buffer alone (lanes 1 and 4), containing the p300 enzyme purified from baculovirus (lanes 2 and 5) or containing the p300 enzyme and the p300-specific inhibitor LysCoA (lanes 3 and 6, respectively). Equivalent amounts of the acetylation products were loaded in two distinct gels and analysed respectively with the antisera that recognizes acetylated lysine residues (anti-AcK) or the His6 tag (anti His6), (upper and lower panels, respectively). (B) Western blot analysis of FLAG-MEF2C WT and FLAG-MEF2C_K4R purified from transfected COS1 cells by immunoprecipitation with the anti-FLAG antibody (IP anti-FLAG). Immunoprecipitated proteins were revealed with the antibodies directed to acetyl lysine (anti-AcK, upper panel) and to the FLAG epitope (anti-FLAG). Mock-transfected cells were used as a control (lane 1).
Acetylation of Lys4 enhances the DNA-binding activity of MEF2C
EMSA assay
The crystal and the solution structures of MEF2 dimers bound to DNA revealed that Lys4 is involved in contacting DNA in the minor groove (40,41). Since acetylation neutralizes the positive charge of Lys ɛ-amino groups, we sought to determine how the acetylation of Lys4 affects the interaction of MEF2 with DNA. To investigate this, we performed EMSA experiments with a 32P-radiolabelled oligonucleotide representing the MEF2 site from the enhancer of the MCK gene. The binding reactions were performed by incubating the labelled DNA probe with in vitro translated MEF2C proteins (Figure 4A) or the nuclear extracts of cells ectopically expressing MEF2 proteins (Figure 4B). We examined the DNA-binding properties of wt MEF2C and of the mutant proteins MEF2C_K4R and MEF2C_K4Q, where Lys4 is substituted with arginine or glutamine. The K4R mutation prevents acetylation and retains the positive charge of the lateral chain, thus mimicking a permanent deacetylated status. On the contrary, the substitution with a glutamine residue (K4Q) approximates the physical changes associated with acetylation. We observed that the K4R mutant exhibits substantially the same activity as the wt protein (Figure 4A, lane 6 versus lane 2). This is not surprising since MEF2C from reticulocyte lysates is probably not acetylated. On the contrary, as shown in lane 7, the K4Q mutant exhibits a strongly enhanced DNA-binding activity compared to the wt protein (at least 7-fold higher as evaluated by a PhosphorImager analysis, data not shown). We next investigated if Lys4 acetylation alters the DNA-binding properties of the MEF2C protein ectopically expressed in transfected cells in culture. COS1 cells were transfected with expression vectors encoding wt MEF2C or the K4R mutant. As shown in Figure 4B, the K4R mutant binds DNA less efficiently than wt MEF2C (compare lanes 3 and 2).
Figure 4.
Effect of Lys4 acetylation on MEF2 binding to DNA. (A) An oligonucleotide encompassing the MEF2 site of the MCK enhancer was used as probe in EMSA assay performed with in vitro translated proteins (IVT). The EMSA assay was performed with the following in vitro translated proteins: wild-type MEF2C (WT, lanes 2–5), MEF2C_K4R (K4R, lane 6) and MEF2C_K4Q (K4Q, lane 7); the unprogrammed reticulocyte lysate was used as a control (ctrl, lane 1). Lane 3 shows the effect of adding the anti-MEF2 antibody to the binding reaction. For competition assays, a 100-fold molar excess of unlabelled native (lane 4) or mutant probe (lane 5) was added to the reaction. Arrows indicate the MEF2–DNA complex (lower) and the complex supershifted by the MEF2 antibody (upper) (B) EMSA assay performed with nuclear extracts obtained from COS1 cells transfected with: empty expression vector (ctrl, lane 1), wild-type MEF2C (WT, lanes 2, 4, 5 and 6) and MEF2C_K4R (K4R, lane 3). Lane 4 shows the effect of adding the anti-MEF2 antibody to the binding reaction. Lanes 5 and 6 show respectively the results of competition assays with a 100-fold molar excess of unlabelled native probe (lane 5) or mutant probe (lane 6). (C) Immunoblot analysis of the in vitro translated proteins used in the EMSA experiment shown in panel A. (D) Immunoblot analysis of the nuclear extracts used in the EMSA experiment shown in panel B.
FRAP assay
The effect of Lys4 acetylation on the kinetic properties of MEF2–DNA interaction in the chromatin context of a living cell, were then explored by the FRAP technique. FRAP experiments were performed in C2C7 cells, proliferating or stimulated to differentiate for 24 h, transiently transfected with wt or Lys4 mutant GFP-MEF2C. To investigate the contribution of Lys4 acetylation to the observed decreased mobility of GFP-MEF2C in myotubes, we compared GFP-MEF2C_WT and GFP-MEF2C_K4R. GFP-MEF2C_WT and GFP-MEF2C_K4R display the same mobility in the nucleus of proliferating cells (Figure 5A), the t1/2 of the two proteins being 0.34 s (±0.03 s SE) and 0.31 s (±0.02 s SE), respectively (Figure 5D). Upon induction of differentiation, the mobility of both wt and mutant MEF2C is decreased, but at a different extent: the mobility of the mutant protein is higher than that of the wt protein (Figure 5B). In a parallel set of experiments FRAP was used to determine the mobility of GFP-MEF2C_R3T, a mutant that was shown to be unable to bind the MEF2 responsive elements, albeit retaining the ability of homo- and heterodimerizing with other MEF2 members (42). In differentiated C2C7 cells, the mobility of GFP-MEF2C_R3T is significantly higher than that of wt GFP-MEF2C. Figure 5C, which is a merge of the curve shown in Figure 5B and the curve obtained with the GFP-MEF2C_R3T protein, indicates that the mobility of MEF2C_K4R in differentiating muscle cell nuclei is intermediate between the two MEF2C proteins which display the highest (MEF2C_WT) and the lowest (MEF2C_R3T) DNA-binding affinity. This is reflected by the calculated t1/2 for the three proteins in differentiating muscle cells, i.e. 0.37 s (±0.02 s SE) for MEF2C_R3T, 0.46 s (±0.03 s SE) for MEF2C_K4R and 0.64 s (±0.05 s SE) for MEF2C_WT (Figure 5D, DM 24 h). Collectively, and consistently with the results obtained with EMSA experiments, these data indicate that Lys4 acetylation is not a requisite for MEF2 association to DNA, but increases it at the early stage of muscle cell differentiation.
Effect of Lys4 acetylation on MEF2C activity and p300 coactivation
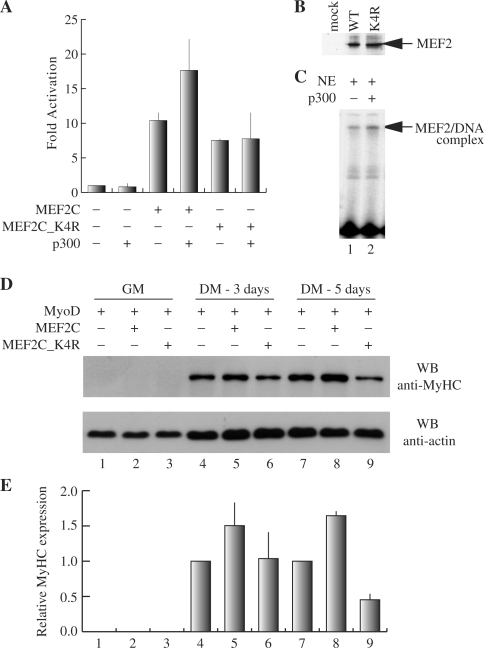
Reporter assays were performed by co-transfecting COS1 cells with wt or mutant MEF2C and a plasmid construct containing the Luciferase reporter gene under the control of three MEF2 sites from the Desmin gene regulatory region. It has been previously shown that p300 physically interacts with MEF2 proteins and enhances their transcriptional activity (43). To investigate the role played by the acetylation of Lys4 in mediating the p300 co-activation of MEF2C-dependent transcription, p300 was co-transfected in the cells. Figure 6A shows that the K4R mutation causes a modest, although reproducible, reduction (35%) of MEF2-dependent transcriptional activity.
Figure 6.
Effect of Lys4 acetylation on MEF2 activity and p300 co-activation. (A) Luciferase reporter activity of COS1 cells co-transfected with pGL3(desMEF2)3, pRSVβ-gal and the empty plasmid expression vector pcDNAI/Amp or the vectors encoding HA-p300, MEF2C (WT) and MEF2C_K4R (K4R) alone or in the indicated combinations. The obtained luciferase activity values were normalized to β-galactosidase activity and are shown as fold the value obtained with the empty pcDNAI/Amp expression vector (MEF2C-, p300−). The results shown are the average ± SD of at least three independent transfection experiments. (B) Western blot analysis of MEF2 proteins present in the cell lysates used in the experiments shown in panel A. (C) An oligonucleotide encompassing the MEF2 site of the MCK enhancer was used as probe in EMSA assays performed with nuclear extracts (NE) of C2C7 cells (lane1) and the same extracts incubated with p300 (lane 2). The specific MEF2–DNA complex is indicated by an arrow. (D) Myogenic conversion assay of C3H10T1/2 fibroblasts co-transfected with MyoD and either an empty vector, MEF2C_WT or MEF2C_K4R. After 48 h in GM, cells were allowed to differentiate in DM for 3 or 5 days before lysis and analysis of myosin heavy chain expression by western blot with the specific antibody. Actin levels were also determined as a loading control. The results of densitometric quantification of the MyHC bands obtained in the western blot are reported in panel E. (E) The relative density of each MyHC band is normalized to the density of the corresponding actin band and is expressed relative to the amount of MyHC protein in cells transfected with MyoD alone. The results of two independent experiments were averaged with the SD indicated.
Figure 6B is a western blot analysis of the extracts of transfected cells, showing that the mutant MEF2C protein is expressed at a level comparable with the wt protein. We also found that the stability of the mutant protein is comparable to that of wt MEF2C (data not shown). The reduced transcriptional activity of the K4R mutant was observed in myogenic C2C7 and L6 cell lines, as well as in C3H10T1/2 and NIH 3T3 cells (data not shown). In addition, the same effect was observed when reporter transcription was controlled by the MCK enhancer or the Myogenin proximal control region (data not shown). These data collectively indicate that the effect of Lys4 modification is not dependent on the cell and promoter context. Interestingly, Figure 6A clearly shows that the p300-dependent enhancement of MEF2 transcriptional activity is clearly reduced when MEF2C_K4R is used, suggesting that Lys4 acetylation substantially contributes to the co-activation mechanism. The observations that Lys4 acetylation results in an enhancement of MEF2 DNA-binding properties and that p300 promotes the acetylation of MEF2 on Lys4 in vitro and in cell culture, prompted us to investigate if p300 can influence the DNA-binding activity of MEF2C. To test this hypothesis we performed EMSA where a 32P-radiolabelled oligonucleotide representing the MEF2 site from the enhancer of the MCK gene was incubated with the nuclear extracts of C2C7 cells alone or added of the P300 enzyme purified from baculovirus. As shown in Figure 6C, the addition of p300 to the C2C7 nuclear extracts cells results in an enhancement of the endogenous MEF2-dependent DNA-binding activity. This effect is dependent on the enzymatic activity of p300, since the p300-specific enzymatic inhibitor LysCoA, abolishes it (data not shown). It has been extensively reported that MEF2 proteins cannot induce myogenic conversion in transfected fibroblasts, but, when co-expressed with the myogenic basic helix-loop-helix (bHLH) proteins, they enhance myogenic conversion (44). In order to investigate the contribution of Lys4 acetylation to the MEF2-dependent enhancement of myogenic conversion, we transfected C3H10T1/2 fibroblasts with MyoD and the expression vectors for either wt MEF2C or MEF2C_K4R. To evaluate the extent of myogenic conversion, we measured the expression level of the sarcomeric protein MyHC by western blotting followed by densitometric analysis of the signals associated to MyHC (Figure 6D and E). When wt MEF2C was co-expressed with MyoD, induction of myogenesis, assessed after 3 and 5 days in differentiation medium, was synergistic: compare lanes 4 and 7 (MyoD alone) to lanes 5 and 8 (MyoD + MEF2C). On the contrary, when C3H10T1/2 fibroblasts were cotransfected with MyoD and the MEF2C_K4R mutant no synergistic effect was observed, but rather a decrease of the MyHC signal after 5 days in differentiation medium: compare lanes 4 and 7 (MyoD alone) to lanes 6 and 9 (MyoD + MEF2C_K4R). The data presented suggest that the acetylation of MEF2 proteins on Lys4, specifically observed in differentiated muscle cells, contributes substantially to the increase of the DNA-binding affinity of MEF2C during muscle differentiation and thus to the activation of transcription of muscle-specific genes. In addition, since p300 acetylates Lys4 in MEF2C, this might be part of the mechanism at the basis of p300 co-activation of MEF2-dependent transcription.
DISCUSSION
MEF2 proteins play a pivotal role in skeletal and cardiac myogenesis, as well as in several other important cellular processes, such as neuronal survival and T-lymphocyte apoptosis (45). To fulfil such a variety of functions, the activity of MEF2 proteins must be subjected to a fine regulation that allows them to respond promptly to various signalling pathways. Several mechanisms contribute to the transcriptional regulation of MEF2 during myogenesis. They include the interaction with a complex network of transcriptional co-activators and co-repressors as well as post-translational modifications (10,15,46–50). In the present report, we present evidence for a regulation of the DNA-binding properties of MEF2C that occurs at the transition from myoblasts to differentiating myotubes and is based on a newly identified post-translational modification, i.e. Lys4 acetylation. Lys4 acetylation greatly enhances MEF2 binding to DNA in vitro and to chromatin in vivo, thus indicating that it might contribute to the activation of the MEF2-dependent transcriptional program at the onset of muscle cell differentiation.
We show that ‘the DNA-binding properties of MEF2 proteins change substantially at the transition from proliferating to differentiated C2C7 muscle cells’, a well-established and reproducible model of myogenesis (25). Although many mechanisms of regulation of MEF2 have been well described, most of them modulate the transactivation potential of MEF2 without apparent modification of its DNA-binding activity. Evidence of a regulatory mechanism for the DNA-binding activity of MEF2 was suggested in embryonic and fetal muscle cells, where a DNA-binding activity at the MEF2 site of the MCK enhancer was observed in nuclear extracts of fetal but not embryonic muscle cells, in spite of the presence of MEF2 proteins in both extracts. The treatment of the embryonic myoblast nuclear extracts with a phosphatase inhibitor restored the MEF2-dependent DNA-binding activity (33). MEF2A, C and D have similar DNA-binding properties in vitro and induce comparable transcriptional activation of reporter genes in transfected cultured cells (8,51–53). Although the interaction of MEF2 with co-repressors, like the class II histone deacetylases and Cabin I, involves the MADS box, several lines of evidence indicate that this takes place without interfering with the binding of MEF2 to the cognate DNA sequence (54–57). Nevertheless, it is conceivable that the interaction of MEF2 proteins with class II HDACs, inhibits the association of MEF2 with co-factors having HAT activity, which render chromatin more accessible to transcription factors. For example, it has been recently reported that the binding of MEF2A to the Glut4 promoter in skeletal muscle in vivo is activated by exercise. CaMK signalling is involved in this regulatory mechanism, likely through a disruption of MEF2/classII HDACs complexes (58). In the past, a casein kinase II (CKII) phosphorylation site was identified, that is conserved in the MADS box of all known MEF2 proteins (59). Phosphorylation of this site dramatically enhances the DNA-binding activity of MEF2C. This site appears to be phosphorylated constitutively in vivo, with no evidence for regulation. Moreover, p300-dependent acetylation of MEF2C was observed in differentiated C2C12 cells. Surprisingly, the acetylation of lysine residues outside the MADS box enhances the binding to the MEF2 site (15). It was suggested that in cardiomyocytes the novel protein Ki-1/57 physically interacts with MEF2C and this interaction inhibits the MEF2C–DNA-binding activity (60). In order to explore the DNA-binding properties of MEF2 proteins during muscle differentiation, we performed a time-course ChIP experiment that allowed us to establish that MEF2 proteins bind the Myogenin promoter in chromatin only upon induction of differentiation of C2C7 myogenic cells (Figure 1A and C). Similarly, it has been reported that MEF2 proteins are selectively recruited to an AT-rich DNA sequence of the MyoD distal enhancer only in differentiated C2C12 muscle cells (61).
In this context, FRAP represents an important complementary tool to ChIP to study the dynamic properties of protein–DNA interactions in living cells (32). By this technique we could demonstrate that the mobility of ectopically expressed GFP-MEF2C is highly and reproducibly reduced in differentiating C2C7 cells: the protein half-life of GFP-MEF2C was estimated to be 0.64 s in cells induced to differentiate for 24 h and 0.34 s in proliferating myoblasts, indicating that MEF2 proteins bind more stably to DNA upon induction of differentiation (Figure 1D and E). The decrease of GFP-MEF2C mobility in differentiating cells is mainly due to the interaction with DNA, since a similar effect was not observed with the MEF2C_R3T mutant that is unable to bind DNA.
The increased affinity of MEF2 for DNA at the onset of muscle cell differentiation correlates with the observation that, as shown by mass spectrometry analysis, ‘MEF2C is acetylated on Lys4 only in differentiated muscle cells’ (Figure 2A). Several recent reports demonstrated that MEF2 proteins are a target for acetylation. Zhao et al. (19) showed that Lys424 in MEF2D is subjected alternatively to sumoylation and acetylation that result respectively in the repression or activation of MEF2. Analogous results were reported for the corresponding lysine of MEF2A (50). Ma et al. (15) described several MEF2C acetylated residues only in differentiated muscle cells, which are located in the C-terminal region of the protein (Lys116, Lys 119, Lys 234, Lys 239, Lys 252 and Lys 264) and thus are unlikely to modulate MEF2 binding to DNA. Our mass spectrometry analysis of MEF2C, ectopically expressed in C2C7 cells, did not reveal peptides with masses expected from the acetylation events reported in the literature (15,19,50). On the contrary, we did observe the peptides corresponding to the unmodified residues. These discrepancies are possibly due to the fact that the acetylated peptides do not efficiently ionize or, alternatively, that the percentage of acetylated residues is too low to be revealed in our experimental conditions. In addition, acetylation of some of the residues, interfering with trypsin digestion, may result in the generation of peptides too large to be analysed. The acetylation events on MEF2 proteins summarized above take place in the C-terminus, thus they unlikely modulate the MEF2 DNA-binding properties. On the contrary, ‘Lys4 acetylation strongly enhances the DNA-binding activity of MEF2’, as we observed in EMSA experiments performed with the in vitro translated protein bearing the K4Q mutation, which mimics permanent acetylation, and in FRAP experiments with the GFP-MEF2C_K4R mutant, which does not allow Lys4 acetylation. Moreover, we show that MEF2C ectopically expressed in COS1 cells binds DNA less efficiently when Lys4 is substituted with arginine. The contribution of Lys4 acetylation to the high-affinity binding of MEF2C to its chromatin sites can be inferred by the behaviour of the non-acetylatable MEF2C_K4R mutant in FRAP experiments, which shows a higher mobility than wt MEF2C in differentiating muscle cells (protein half-life of 0.46 and 0.64 s for the MEF2C_K4R mutant and the wt protein, respectively). Lys4 acetylation does not represent a prerequisite for MEF2C binding to chromatin: the K4R mutant still exhibits a mild but significant reduction of its mobility in differentiating muscle cells (the half-life of 0.31 s in myoblasts is prolonged to 0.46 s in cells stimulated to differentiate). Coherently with these observations, Lys4 is located in the MADS box, the MEF2 domain directly involved in DNA binding, which is conserved in all MEF2 proteins and it was shown to play a role in determining the DNA-bending properties of MEF2 (62). The crystal and solution structures of the MEF2A–DNA complex have been solved, revealing that Lys4 interacts with the phosphodiester backbone to push the Arg3 side chain into the DNA minor groove (40,41). Lys4 acetylation neutralizes the side-chain positive charge involved in the interaction with DNA, however it enhances the DNA-binding affinity of MEF2C. The NMR analysis of the complex indicates that Lys4 establishes hydrophobic contacts with DNA, therefore it can be hypothesized that these contacts are stabilized by the acetylation event or, alternatively, that acetylated Lys4 mediates the interaction of MEF2C with additional co-factors resulting in a higher affinity DNA binding. Collectively, these data indicate that Lys4 acetylation is required for achieving the high DNA-binding affinity that is necessary for a prompt firing of the MEF2-dependent transcriptional programme in differentiating muscle cells. In order to evaluate the functional role of Lys4 acetylation of MEF2C, we performed transactivation and myogenic conversion assays.
Indeed we show that the ‘MEF2C_K4R mutant exhibits a weak but reproducible reduction (30%) of its transcriptional activity in comparison with the wt MEF2C protein in transactivation assays’ (Figure 6A) more importantly, the co-activation of MEF2 mediated by p300 depends on the presence of Lys4, since the activity of the mutant protein K4R is not enhanced by p300 in transactivation assays.
We do not know which enzymes are responsible for the acetylation of Lys4. It has been reported that p300 acetylates the C-terminus of MEF2C in vivo (15). The observations that: (i) p300 promotes the acetylation of Lys4 in MEF2C in vitro and in vivo; (ii) p300 directly enhances the DNA-binding affinity of MEF2C in EMSA; (iii) p300 co-activation of MEF2C transcriptional activity requires an intact Lys4 residue, together with the role played by this co-activator in muscle cell differentiation (63,64), collectively indicate p300 as a likely candidate for directly acetylating MEF2 proteins. In keeping with this hypothesis, we observed that p300 associates to chromatin on the same Myogenin gene DNA regulatory region that is bound by MEF2 (data not shown). This physical proximity could allow p300-dependent acetylation of MEF2C and thus stabilization of MEF2–DNA interactions.
The DNA-binding domain of MEF2 also mediates the interaction of MEF2 proteins with the myogenic bHLH proteins and thus their cooperation in promoting skeletal muscle differentiation (44). Thus, we investigated if Lys4 acetylation is involved in regulating the MEF2-MyoD synergy by performing myogenic conversion assays. As shown in Figure 6D and E we observed that lack of acetylation on Lys4 abolishes the functional interaction between MyoD and MEF2C, since the co-expression of MyoD and the mutant protein MEF2C_K4R does not result in an enhancement of myogenic conversion. On the contrary, we observed a decrease of MyHC expression, suggesting that the MEF2C_K4R mutant could behave as a dominant negative protein. The effect of MEF2C_K4R on MyHC expression is coherent with recent reports showing that MEF2 proteins are required for the correct expression of thick filament proteins and for muscle sarcomere integrity (6,7). We propose a model where MEF2 proteins are maintained functionally silent in myoblasts, by deacetylation of Lys4 and other lysine residues. Deacetylated MEF2 binds DNA with lower affinity, even though is still able to recognize its cognate DNA sites and thus to locate in the appropriate chromatin domains. A change of the equilibrium towards the MEF2 acetylated form, more stably bound to chromatin, induced directly or indirectly by p300, would cause the prompt and efficient firing of MEF2 transcriptional activity, in coincidence with the induction of muscle cell differentiation. It has been recently shown that the class I histone deacetylase HDAC3 physically interacts with MEF2 proteins and deacetylates them, moreover HDAC3 associates to the same DNA sequence on chromatin that are contacted by MEF2 (65). Upon induction of skeletal muscle differentiation, the MEF2/HDAC3 association is strongly reduced by a regulatory mechanism that still needs to be clarified. In addition to HDAC3, it has been widely described a mechanism of negative regulation of MEF2 activity by the class II HDACs 4/5/7/9. These deacetylases have not been shown to directly decetylate MEF2 proteins but they could contribute indirectly to maintain the deacetylated status of MEF2 proteins, by recruiting class I HDACs or by regulating the activity of p300. Class II HDACs associate with MEF2 proteins in C2C7 myoblasts; during skeletal muscle differentiation they are phosphorylated and translocate to the cytoplasm. Class I and class II deacetylases interact with the DNA-binding domain of MEF2, as well as p300, suggesting a mutual exclusive association. We suggest that the regulation of MEF2 acetylation, and thus of its DNA-binding activity, concerns HATs as well as HDAC3 and HDAC4/5/9 localization and activity and that Lys 4 is a nodal target for HATs and HDACs during myogenesis.
ACKNOWLEDGEMENTS
We are grateful to Roberto Mantovani for baculovirus expressed p300, Phil Cole for the p300 inhibitor LysCoA, Margaret Buckingham for the C2C7 muscle cell line, Stefano Schiaffino for the pGL3(desMEF2)3 reporter vector. We are indebted to Rossella Tupler, Margaret Buckingham and Graziella Messina for critically reading the manuscript and to Francesca Fanelli and Laurence Vandel for helpful discussions. We are also indebted to Andrea Tombesi (Centro Grandi Strumenti, University of Modena and Reggio Emilia) for technical assistance with the confocal microscope and to Alessandra Agresti for precious advices regarding the FRAP experiments. We thank Renata Battini for assistance and for numerous discussions and Alessio Polacchini for the purification and acetylation of the recombinant bacterial protein. This work was funded by Telethon (GP0210Y01); MURST-COFIN’04 and MURST COFIN'06 to S.F. Funding to pay the Open Access publication charges for this article was provided by MURST COFIN'06.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 2.Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 3.Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev. Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 4.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 5.Ornatsky OI, Andreucci JJ, McDermott JC. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 6.Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell. Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitbart RE, Liang CS, Smoot LB, Laheru DA, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- 9.Buchberger A, Ragge K, Arnold HH. The myogenin gene is activated during myocyte differentiation by pre-existing, not newly synthesized transcription factor MEF-2. J. Biol. Chem. 1994;269:17289–17296. [PubMed] [Google Scholar]
- 10.Lazaro JB, Bailey PJ, Lassar AB. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 2002;16:1792–1805. doi: 10.1101/gad.U-9988R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berdeaux R, Goebel N, Banaszynski L, Takemori H, Wandless T, Shelton GD, Montminy M. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat. Med. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 13.Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl Acad. Sci. USA. 2005;102:8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell. Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, Gocke CB, Yu H. Phosphorylation-facilitated sumoylation of MEF2C negatively regulates its transcriptional activity. BMC Biochem. 2006;7:5. doi: 10.1186/1471-2091-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguere V, et al. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- 18.Riquelme C, Barthel KK, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J. Cell Mol. Med. 2006;10:132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell. Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin JF, Schwarz JJ, Olson EN. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl Acad. Sci. USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borghi S, Molinari S, Razzini G, Parise F, Battini R, Ferrari S. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J. Cell Sci. 2001;114:4477–4483. doi: 10.1242/jcs.114.24.4477. [DOI] [PubMed] [Google Scholar]
- 22.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 23.Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126:2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- 24.Pinset C, Montarras D, Chenevert J, Minty A, Barton P, Laurent C, Gros F. Control of myogenesis in the mouse myogenic C2 cell line by medium composition and by insulin: characterization of permissive and inducible C2 myoblasts. Differentiation. 1988;38:28–34. doi: 10.1111/j.1432-0436.1988.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 25.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 26.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 27.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 28.Herbomel P, Bourachot B, Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984;39:653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- 29.Molinari S, Relaix F, Lemonnier M, Kirschbaum B, Schafer B, Buckingham M. A novel complex regulates cardiac actin gene expression through interaction of Emb, a class VI POU domain protein, MEF2D, and the histone transacetylase p300. Mol. Cell. Biol. 2004;24:2944–2957. doi: 10.1128/MCB.24.7.2944-2957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontoglio M, Faust DM, Doyen A, Yaniv M, Weiss MC. Hepatocyte nuclear factor 1alpha gene inactivation impairs chromatin remodeling and demethylation of the phenylalanine hydroxylase gene. Mol. Cell. Biol. 1997;17:4948–4956. doi: 10.1128/mcb.17.9.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phair RD, Gorski SA, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- 32.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, et al. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari S, Molinari S, Melchionna R, Cusella-De Angelis MG, Battini R, De Angelis L, Kelly R, Cossu G. Absence of MEF2 binding to the A/T-rich element in the muscle creatine kinase (MCK) enhancer correlates with lack of early expression of the MCK gene in embryonic mammalian muscle. Cell Growth Differ. 1997;8:23–34. [PubMed] [Google Scholar]
- 34.Catala F, Wanner R, Barton P, Cohen A, Wright W, Buckingham M. A skeletal muscle-specific enhancer regulated by factors binding to E and CArG boxes is present in the promoter of the mouse myosin light-chain 1A gene. Mol. Cell. Biol. 1995;15:4585–4596. doi: 10.1128/mcb.15.8.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Chait BT. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 2000;72:2482–2489. doi: 10.1021/ac991363o. [DOI] [PubMed] [Google Scholar]
- 39.Schaaf MJ, Willetts L, Hayes BP, Maschera B, Stylianou E, Farrow SN. The relationship between intranuclear mobility of the NF-kappaB subunit p65 and its DNA binding affinity. J. Biol. Chem. 2006;281:22409–22420. doi: 10.1074/jbc.M511086200. [DOI] [PubMed] [Google Scholar]
- 40.Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 41.Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 2000;19:2615–2628. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 45.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 46.Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 47.Cox DM, Du M, Marback M, Yang EC, Chan J, Siu KW, McDermott JC. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J. Biol. Chem. 2003;278:15297–15303. doi: 10.1074/jbc.M211312200. [DOI] [PubMed] [Google Scholar]
- 48.De Luca A, Severino A, De Paolis P, Cottone G, De Luca L, De Falco M, Porcellini A, Volpe M, Condorelli G. p300/cAMP-response-element-binding-protein (‘CREB’)-binding protein (CBP) modulates co-operation between myocyte enhancer factor 2A (MEF2A) and thyroid hormone receptor-retinoid X receptor. Biochem. J. 2003;369:477–484. doi: 10.1042/BJ20020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 2001;276:17007–17013. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 50.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 51.Martin JF, Miano JM, Hustad CM, Copeland NG, Jenkins NA, Olson EN. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D, Lipton SA, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol. Cell. Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 54.Chan JK, Sun L, Yang XJ, Zhu G, Wu Z. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 2003;278:23515–23521. doi: 10.1074/jbc.M301922200. [DOI] [PubMed] [Google Scholar]
- 55.Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 56.Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 57.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 58.Smith JA, Collins M, Grobler LA, Magee CJ, Ojuka EO. Exercise and CaMK activation both increase the binding of MEF2A to the Glut4 promoter in skeletal muscle in vivo. Am. J. Physiol. Endocrinol. Metab. 2007;292:E413–E420. doi: 10.1152/ajpendo.00142.2006. [DOI] [PubMed] [Google Scholar]
- 59.Molkentin JD, Li L, Olson EN. Phosphorylation of the MADS-Box transcription factor MEF2C enhances its DNA binding activity. J. Biol. Chem. 1996;271:17199–17204. doi: 10.1074/jbc.271.29.17199. [DOI] [PubMed] [Google Scholar]
- 60.Kobarg CB, Kobarg J, Crosara-Alberto DP, Theizen TH, Franchini KG. MEF2C DNA-binding activity is inhibited through its interaction with the regulatory protein Ki-1/57. FEBS Lett. 2005;579:2615–2622. doi: 10.1016/j.febslet.2005.03.078. [DOI] [PubMed] [Google Scholar]
- 61.L’Honore A, Rana V, Arsic N, Franckhauser C, Lamb NJ, Fernandez A. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol. Biol. Cell. 2007;18:1992–2001. doi: 10.1091/mbc.E06-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West AG, Shore P, Sharrocks AD. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell. Biol. 1997;17:2876–2887. doi: 10.1128/mcb.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem. 2000;275:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- 64.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 65.Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol. Cell. Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]