Abstract
The nucleoid-associated protein HU plays an important role in bacterial nucleoid organization and is involved in numerous processes including transposition, recombination and DNA repair. We show here that HU binds specifically DNA containing mismatched region longer than 3 bp as well as DNA bulges. HU binds single-stranded DNA (ssDNA) in a binding mode that is reminiscent but different from earlier reported specific HU interactions with double-helical DNA lesions. An HU dimer requires 24 nt of ssDNA for initial binding, and 12 nt of ssDNA for each additional dimer binding. In the presence of equimolar amounts of HU dimer and DNA, the ssDNA molecule forms an U-loop (hairpin-like) around the protein, providing contacts with both sides of the HU body. This mode differs from the binding of the single-strand-binding protein (SSB) to ssDNA: in sharp contrast to SSB, HU binds ssDNA non-cooperatively and does not destabilize double-helical DNA. Furthermore HU has a strong preference for poly(dG), while binding to poly(dA) is the weakest. HU binding to ssDNA is probably important for its capacity to cover and protect bacterial DNA both intact and carrying lesions.
INTRODUCTION
The bacterial chromosome is highly condensed and associated with several ‘architectural’ proteins. In Escherichia coli, one of the most abundant of such proteins is HU, which was the first one proposed to be a histone-like protein as indicated by its name: ‘H’ for histone-like and ‘U’ for U93 [and not ‘heat unstable’ as sometimes found in publications. At the time of its discovery, we have shown (and verified many times) that HU is in fact a very heat stable protein], an RNase− strain used at that time to study the bacterial nucleoid (1–3). HU is associated in vivo with the E. coli nucleoid (4,5) and was shown to be a very conserved protein in the prokaryotic world (6,7). The degree of conservation of the HU sequence is of the same order as that of histone H2B (8). HU is also present in chloroplasts (9), in an eukaryotic virus (10) and has an homolog HM in yeast mitochondria (11). Bacteria also contain an ‘HU-like protein’, IHF (Integration Host Factor), that belongs to the same family of DNA-binding proteins, but has different specific functions (12,13). The quasi totality (98%) of prokaryotes with sequenced genomes encode for at least one HU, while IHF is only found in proteobacteria (Oberto,J., personal communication). In enteric bacteria such as E. coli, HU, which is encoded by two genes, hupA and hupB, exists in solution as a homodimer, HUα2 or HUβ2, or as a HUαβ heterodimer (14,15). In vivo, the heterodimer is predominant (90%) after the transition to stationary phase (15), and this was the form we used in the present study. For brevity, we refer in this article to the HUαβ heterodimer as simply ‘HU’.
HU-dependent assembly of higher order nucleoprotein complexes is required, among other functions, for site-specific recombination reactions (16,17), the initiation of replication at oriC (18–26), and phage Mu transposition (27–29). The histone-like character of HU is linked to its capacity to introduce, like the histones, negative supercoiling into a relaxed DNA molecule in presence of topoisomerase I and to condense DNA (2,3,30). HU has as well a high homology in its amino acids sequence with the histone H2B (1,31).
This role of HU in the nucleoid organization was confirmed by the existence of a cross-talk between HU and topoisomerase I detected in vivo in E. coli cells (32). In vitro, the HU binding to negatively supercoiled DNA was shown to be three times stronger than to linear relaxed DNA [(4,33) and J.R.Y. and Dudnick,Y., unpublished data). This preferential binding of HU to supercoiled DNA is in agreement with the recent HU–DNA co-crystal structure, where the dsDNA fragment is bent so that the overall DNA trace corresponds the path of a negatively supercoiled DNA rather than that of a relaxed double helix (34).
HU-binding targets are not limited to generic dsDNA. This protein avidly binds distorted DNA structures such as forks, three/four-way junctions, nicks and overhangs (35,36). We have also shown that HU also binds dsRNA (33,37). The binding parameters for short DNA, RNA duplexes, as well as DNA–RNA hybrids are very similar, with one dimer packed every 9–11 bp (33). The HU binding to dsDNA (B-form), dsRNA (A-form) and DNA–RNA hybrids is considered non-specific because it is salt-dependent and characterized by a low affinity (32,41). The apparent dissociation constants for negatively supercoiled DNA, relaxed dsDNA and total bacterial RNA, are, respectively, of 450, 1300 and 2500 nM, per binding site. In contrast, binding to distorted DNA structures is strong even under high salt conditions (150 mM NaCl), with dissociation constants ranging from 1 to 10 nM (35,36). This global DNA, RNA and DNA–RNA recognition suggests that HU is able to interact with both the A and B conformations of nucleic acids either with regular double-strand structures or with specific, multi-branches structures. This broad specificity of HU is difficult to rationalize in terms of a simple unified mechanism. The X-ray structure of the HU–DNA complex (34,39) clarifies some issues, but it does not explain, for instance, how the protein can interact with the A form of dsRNA.In order to clarify the limits of the specificity and the putative-binding modes of the HU protein, we studied here its interaction with dsDNA containing mismatches and/or bulges, and with ssDNA. Since HU is involved in DNA repair, it could be expected to recognize mismatches and bulges that represent frequent DNA lesions. HU binding to ssDNA has been previously observed (40–42), however it has never been further characterized. We show here that HU binds dsDNA mismatches of four or more bases and bulges with high affinity and that these non-canonical DNA structures belong to the group of its specific targets. In contrast, histone-like U93 (HU) interacts with ssDNA with an affinity close to that for dsDNA. Both biochemical data and FRET measurements indicate that ssDNA makes an U-loop (hairpin-like) and wraps around the protein, which probably provides contacts with both sides of HU body as well as its β-arms. With high HU:DNA ratios, an HU dimer requires 24 nt of ssDNA for the initial binding, and 12 nt of ssDNA for each additional HU dimer. The HU–ssDNA interactions show a sequence preference and qualitatively differ from the action pattern of the single-stranded DNA-binding protein (SSB). This work confirms that HU behaves as an ubiquitous and versatile nucleic acid-binding protein due to its broad structural flexibility.
MATERIALS AND METHODS
DNA sequences
The series of the ssDNA oligonucleotides with the lengths from 20 to 48 nt was obtained by 3′-end truncation of the following basic sequence ‘D-48’: AGTCTAGAGT GCAGTTGAGT CCTTGCTACG ACGGATCCCT TAGGTCAG. The oligonucleotides in this series exhibited a smoothly descending regular dependence of gel mobility upon the chain length, as expected for ssDNA without secondary structures. Sequence D-48 represents an elongated sequence X: AGTCTAGACT GCAGT TGAGT CCTTGCTAGG ACGGATCCCT that was used previously (35,36), with two nucleotides replaced (underlined). We first tried to produce the test series of shorter fragments from sequence X, but found that the X-31 sequence AGTCTAGACT GCAGTTGAGT CCTTGCTAGGACGGATCCT exhibits an anomalous gel mobility, that was higher than both X-33 and X-29, probably due to a hairpin formation by the boldfaced bases. For this reason, sequence X was changed to D, and the latter was checked for the possibility of hairpin base pairing by a special computer program. In addition, sequence X′ complementary to X was used in a similar series of experiments. An independent series of oligomers produced from this sequence was also free from gel mobility anomalies and it showed results identical to those described in the text.
The dsDNA was constructed from oligonucleotide D-40 and the complementary oligonucleotide. The nicked DNA fragment was constructed from oligonucleotide D-40 and oliogonucleotides AGGGATCCGTCGTAGCAA GG and ACTCAACTGCACTCTAGACT. DNA containing mismatched region was constructed from oligonucleotide X and: partially complementary sequences AGGGATCCGTCCTA TTTGATGTAGCT CTGCAGTCTAGACT for mismatch 12, (regions complementary to X are underlined); Similarly, for shorter mismatches:
AGGGATCCGTCCTAG TTGATGTAGC
ACTGCAGTCTAGACT (10)
AGGGATCCGTCCTAGC TGATGTAG
AACTGCAGTCTAGACT (8)
AGGGATCCGTCCTAGCA GATGTA
CAACTGCAGTCTAGACT (6)
AGGGATCCGTCCTAGCAA ATGT
TCAACTGCAGTCTAGACT (4)
AGGGATCCGTCCTAGCAAA TG
TTCAACTGCAGTCTAGACT (2)
AGGGATCCGTCCTAGCAAAT A
TTCAACTGCAGTCTAGACT (1).
The dsDNAs were obtained by annealing the appropriate oligonucleotides with one of them 5′-labeled and the complementary non-labeled taken in 5× molar excess. Annealing reactions were carried out by incubating the oligonucleotides (300 nM) for 3 min at 80°C in 20 mM Tris–HCl (pH 8.0), 200 mM NaCl, and then allowing them to cool slowly.
Gel mobility shift assay
HU protein was purified from E. coli strain as described earlier (43). Varying amounts of HU protein were incubated with [5′-32P]-labeled DNAs (1 nM) for 15 min in 10 µl of the binding buffer, 20 mM Tris–HCl (pH 8.0), 0.05 mg/ml bovine serum albumin, 7% glycerol, 0.1 mM EDTA and indicated concentration of NaCl. Samples were loaded onto 8% polyacrylamide gels (29:1) buffered with 27 mM Tris–borate, 0.1 mM EDTA for 10 mM NaCl samples or with 55 mM Tris–borate, 0.2 mM EDTA for 150 mM NaCl samples, or with 95 mM Tris–borate, 0.2 mM EDTA for 200 mM NaCl samples for DNA mismatches and electrophoresed.
Agarose gels (0.8%) buffered with 27 mM Tris–borate/0.1 mM EDTA were used for separation of HU and SSB complexes with ssDNA of phage phiX (200 ng, NewEngland Biolabs). After electrophoresis, DNA was stained in the gel by soaking in the running buffer supplemented with 0.2 mg/l of ethidium bromide and photographed under UV light.
Quantification of dissociation constants
Based on different gels analysis and concentrations, the dissociation constants were calculated as described in (33). Assuming that HU binding to DNA is not sequence specific, and that its concentration is far from saturation the corresponding binding constant per binding site is given as Kdsite =[proteinfree] × [DNAfree]/[DNAbound], where [proteinfree] = [proteintotal] − [proteinbound] is the concentration of free HU, [DNAbound] = [proteinbound] is the concentration of HU–DNA complexes, and [DNAfree] is the concentration of the protein-binding sites. The latter value can be estimated as the free DNA concentration multiplied by the number of HU-binding modes on a given DNA fragment estimated as n = (N − L + 1), where N is the DNA chain length and L is the length of the DNA fragment covered by one HU molecule in the bound state. If f (free) and b (bound) represent the relative radioactivity of the corresponding gel bands, we can estimate [DNAfree] as [DNAfree] = [DNAtotal] × [nf/(f + b)] and obtain a convenient expression for the dissociation constant per binding site Kdsite = [proteintotal] × (nf/b) − [DNAtotal] × [nf/(f + b)]. The corresponding dissociation constant per DNA molecule is given as Kdmol = Kdsite/n For non-sequence-specific binding, the value of Kdmol should decrease with the chain length whereas Kdsite should be approximately constant. If a DNA molecule contains one specific binding site, with the non-specific interactions being negligible, the corresponding binding constant is Kdspecific = Kdmol = [proteintotal] × (f/b) − [DNAtotal] × [f/(f + b)]. The radioactivity of the bands corresponding to free and bound DNA was determined by a PhosphorImager analysis of gels. The best fit over several protein concentrations was taken as Kd.
Binding of the second HU dimer might be facilitated by the protein–protein interactions that can be measured by the factor of co-operativity, ω. The ω factor is determined as Kd1/Kd2, where Kd1 and Kd2 are the dissociation constants of the first and second complexes, respectively. For binding of HU to DNA, the ω has been determined as described previously (44,33).
Fluorescence studies
The same sequences D-28 and D-36 as in the gel mobility shift assays were used. The double-labeled oligonucleotides were purchased from Eurogentec containing the donor, fluorescein, at the 5′ DNA end, and the acceptor, tetramethylrhodamine, at the 3′ DNA. All measurements were taken on a Spex Fluorolog DM1B instrument, using bandwidth of 1.8 nm and 0.2 cm × 1 cm quartz cuvettes. For emission spectra, the excitation wavelength was set at the wavelength where absorption of the donor was maximal (495 nm). For excitation spectra, the emission wavelength was 580 nm. All experiments were performed in 10 mM cacodylate buffer pH 7, 25°C and 1 µM of oligonucleotide and increasing amounts of HU protein. The other experimental conditions are as specified in the figures. The spectra were corrected for the dilution factor.
Conformational calculations
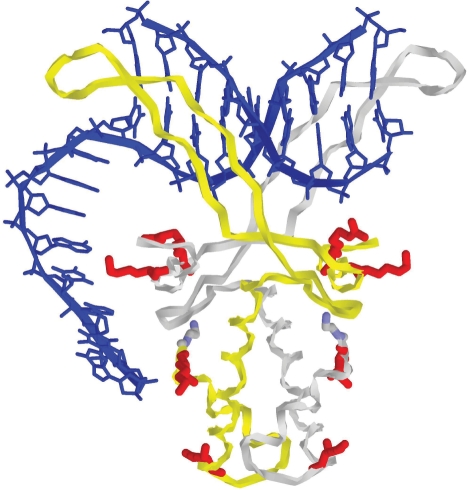
The conformation of the HU complex with 3′-overhand shown in Figure 5 was obtained by homology modeling based upon the X-ray structure of HU–DNA complex (34). The protein coordinates were taken from the PDB entry 1p51. The double-helical DNA fragment was first built in a canonical B-DNA conformation with the base-pair sequence corresponding to the PDB entry. The abasic site originally present in this DNA segment was removed. This double helix was docked to the HU-binding site by energy minimization with all relevant atom positions restrained to X-ray coordinates. The 3′-overhang was built by continuing the double-helical DNA with an AT-alternating sequence of appropriate length in the initial conformation corresponding to a single strand of a canonical B-DNA. The whole structure was next energy minimized with X-ray restraints as well as a few additional restraints corresponding to putative contacts of the ssDNA segment with the charged residues on the surface of HU core, as discussed in our previous studies (36).
Figure 5.
Complex of HU dimer with 3′-overhang. A hypothetical conformation obtained by homology modeling based on the X-ray structure of HU–DNA complex (34,45). The coordinates of the central part, including the protein and the double-helical DNA, are close to those in the original PDB entry 1p51. The double-helical DNA fragment (in blue) was first built in a fiber canonical B-DNA conformation with the base-pair sequence corresponding to the PDB entry. The abasic site originally present in this DNA segment was removed and the structure was energy minimized with all relevant atom positions restrained to X-ray coordinates. The 3′-overhang was built by continuing the double-helical DNA with an AT-alternating sequence of appropriate length in the initial conformation corresponding to a single strand of a canonical B-DNA. The whole structure was next energy minimized with X-ray restraints as well as with a few additional restraints corresponding to putative contacts of the ssDNA segment with the charged residues (in red) on the surface of HU core, discussed in our previous studies (35,36).
RESULTS
HU binds dsDNA mismatches
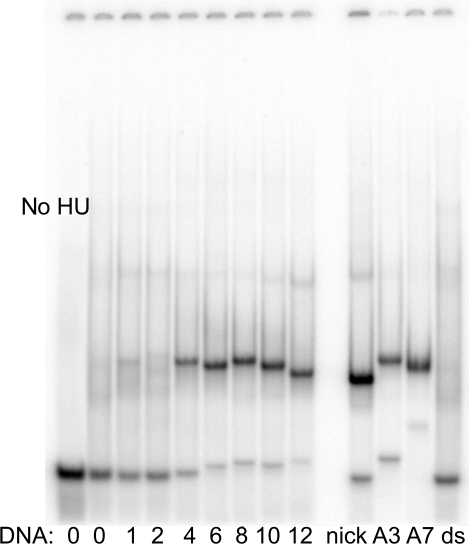
The band-shift analysis of HU binding to dsDNA with internal mismatches of 1 to 7 consecutive base pairs is shown in Figure 1. Clearly HU binds DNA containing more than 3-bp long mismatches, under stringent salt conditions (150 mM salt), indicating a structure-specific interaction. In contrast, under these high salt conditions, there is no binding of HU to dsDNA (‘ds’ in Figure 1). This non-specific binding of HU to plain ds DNA is only observed in low salt conditions (10 mM NaCl, see Figure 2). When mismatches are superior to 4 bp, the apparent binding constant, Kd (measured as described in the Materials and Methods section), depends only slightly on the length of the mismatch: 21, 6, 6, 6 and 3 nM for mismatches of 4, 6, 8, 10 and 12 bp, respectively. This interaction is structure specific and the observed Kd are in fact similar of those of other HU specific targets: nick (8 nM), DNA overhang (18 nM) and DNA ss fork (4 nM). In sharp contrast, under these stringent conditions, no binding was observed with mismatches of 1 and 2 bp, probably because they are not flexible enough for structure-specific HU binding. This flexibility issue will be discussed subsequently. In the case of 3, 4 and longer mismatches (=<>=), the binding is probably linked to a DNA bending at its flexible hinges, which will allow HU to interact with DNA simultaneously by its arms and body.
Figure 1.
HU binding to DNA mismatches and bulges in stringent conditions (high salt). HU protein (50 nM) was mixed with a dsDNA containing in its middle a mismatched region of varying size (from 1 to 12) as indicated (left) or with dsDNA containing an insert of three or seven adenines (lanes A3 and A7). DsDNA (ds) or the duplex containing a nick (nick) were used as control. All the DNAs were 40-nt long and originated from sequence ‘X’. Samples were incubated in 200 mM NaCl and run in 95 mM Tris–borate. Lane no HU is the 40-mer dsDNA in the absence of HU.
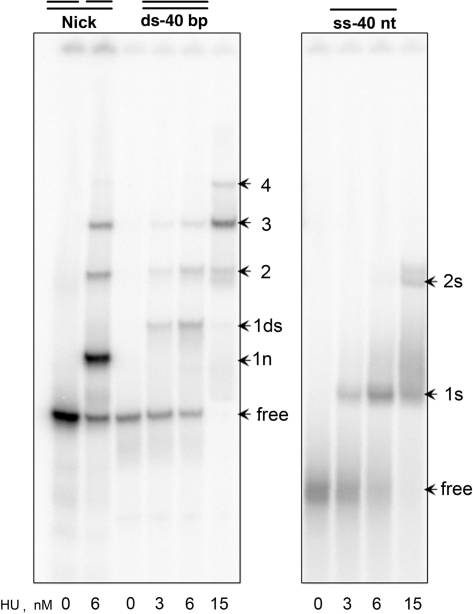
Figure 2.
HU binding to ssDNA, dsDNA and nick in low salt conditions. HU protein, at the concentration indicated in nM on the bottom, was mixed with end-labeled DNA, 40-bp long dsDNA, the same DNA with a nick in the middle and the 40 nt ssDNA, at 10 mM NaCl and analyzed by polyacrylamide gel buffered with 18 mM Tris–borate. All DNAs are originated of the sequence ‘X’. Arrows ‘1n’ and ‘1ds’ mark the bands corresponding to HU dimer–nick (1:1) and HU–dsDNA (1:1) complexes, respectively; arrows ‘2’, ‘3’ and ‘4’ mark the bands corresponding to two, three and four HU dimers bound to nick or dsDNA, while arrows ‘1s’ and ‘2s’ mark the bands corresponding to one or two HU dimers bound to ssDNA, respectively. Position of the free DNA is marked as ‘free’.
To study HU binding to DNA bulges, we used a 40-mer dsDNA fragments containing on one strand either 3 or 7 adenines, (called A3 and A7, respectively in Figure 1). The A3 and A7 bulges form in solution a V-like structure with very different angles between the double-strand branches. Figure 1 shows that HU binds both structures specifically, with a higher affinity for the more flexible A7 bulge. The corresponding dissociation constants are 10 and 1 nM, respectively. These results suggest that HU uses the DNA flexibility rather than pre-formed ligand shapes for binding DNA bulges.
To further characterize the mode of binding of HU to these structures containing single-stranded islands, we next investigated how HU binds to plain ssDNA.
HU binds ssDNA
The band-shift data shown in Figure 2 compare HU binding to ssDNA with representative DNA substrates studied previously. This study is performed, in contrast to the one in Figure 1, in low salt conditions (10 mM salt). We observed that a 40 bp dsDNA binds up to four HU dimers at high HU concentration (15 nM). With nicked DNA, HU forms a structure-specific complex easily distinguished by its high intensity (see band ‘1n’), which is the only one observed in high salt conditions (Figure 1). As seen in Figure 2, the gel mobility of this specific complex is strongly increased compared to the first band observed for the dsDNA complex (‘1ds’). In contrast, the multimeric HU complexes with nicked DNA do not differ from those with intact DNA (bands 2, 3 and 4). The data for ssDNA evidence that, under these low salt conditions, HU binds a 40 nt ssDNA with an affinity similar to that for dsDNA. The values of the apparent dissociation constants are: Kdmol = 3, 7 and 4 nM, and the Kdsite = 3, 250 and 70 nM for nick, dsDNA and ssDNA, respectively. For nick DNA and dsDNA, these data are in good agreement with values previously reported (41,44).
There are evident differences between ssDNA and dsDNA binding. At most two HU dimers are able to bind the 40 nt ssDNA, contrary to the four complexes observed with dsDNA. Moreover, the HU binding to dsDNA is cooperative (33,44) whereas its binding to ssDNA is not. The co-operativity factor is ω ∼ 0.4, indicating that the second HU binds ssDNA less avidly than the first one (no co-operativity). For comparison, this factor is ω ∼ 30 for dsDNA (33)
The size of HU-binding site for ssDNA
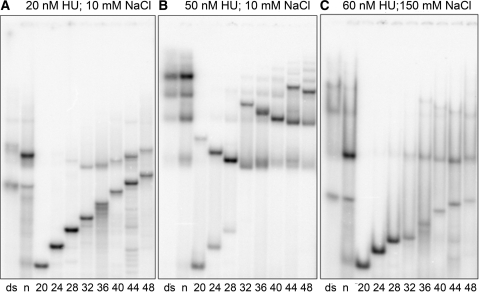
According to the results in Figure 3 and data not shown, a HU dimer requires ∼24 nt of ssDNA for initial binding. This minimum ssDNA length, required for a successful HU binding, should be distinguished from the portion of DNA that is occupied by the bound protein (binding site). For instance, the HU-binding site for dsDNA is only 9 bp (41). To obtain more accurate estimates, we studied the interaction of HU with ssDNA varying in size from 20 to 48 nt. Figure 3 is representative of these experiments. As expected, the gel mobility of free ssDNA gradually decreases with increased DNA length. This observation is important since it confirms that the ssDNA molecules used in our experiments do not form secondary structures that could affect HU–ssDNA interactions. The HU-binding affinity to ssDNA increases with the DNA length and becomes significant starting from 24 nt. Binding of a second HU dimer is only detected when the ssDNA fragment exceeds 32 nt and with a high HU concentration.
Figure 3.
HU binding to ssDNA of various lengths. Binding of labeled DNA to HU protein was analyzed by polyacrylamide gel electrophoresis. DNA samples were: dsDNA (ds), nick (n), both 36-bp long; and ssDNA of sequence ‘D’ with the length varying from 20 to 48 nt (indicated at the bottom in nt). HU concentration was 20 nM (A), 50 nM (B) and 60 nM (C). Depending on the salt conditions, the gel was buffered with 18 mM Tris–borate for 10 mM NaCl (panels A and B), or with 55 mM Tris–borate for 150 mM NaCl (C).
Figure 3A also shows an unexpected result. Usually, complexes with short ds or ssDNA migrate in the gel faster than complexes with longer DNA, which is natural and usual, but the behavior of the HU–ssDNA complex does not follow the same rule (Figure 3). Instead, complexes with the shortest DNA exhibit very low gel mobility while mobility increases with the DNA length, reaching a clear maximum at 32 nt. Only for HU complexes with longer ssDNA the behavior of the gel mobility corresponds to the usual decreasing trend. These results suggest that the shape of the HU–ssDNA complexes significantly changes with the length of the bound DNA, namely, the complex of HU with a DNA longer than 32 nt, is more compact than that observed with shorter DNA (24 or 28 nt).
Similar effects are observed under saturating protein concentrations as shown in Figure 3B. These data clarify the ssDNA length required for HU binding. The ssDNA of 24–28 nt binds only one HU dimer, even under high concentration of HU. A complex with two HU dimers is detectable for ssDNA of 32 nt and longer, and three HU dimers can be accommodated by ssDNA of 44–48-nt long. Therefore, although the binding of the first HU to ssDNA requires at least 24 nt, 12 nt are apparently sufficient to accommodate every additional HU dimer on ssDNA. This observation was critical for the understanding of the interaction of HU with ssDNA and was the reasons to initiate FRET analysis. The dissociation constants for HU–ssDNA binding are Kdmol = 755, 114, 80, 54, 40, 41 nM and Kdsite = 3800, 1030, 1050, 930, 850, 1050 nM for 28, 32, 36, 40, 44 and 48-nt long ssDNA, respectively. The median Kdsite value is ∼1000 nM suggesting that ssDNA binding under low salt conditions is three times weaker than that of dsDNA.
No co-operativity in HU binding to ssDNA could be detected in contrast to the positive co-operativity observed for HU binding to dsDNA under the same conditions [(33,44) and Figure 3], as with lower HU concentrations (Figure 3A). According to Figure 3B, the chain length dependence of the gel-mobility profile for HU multimers bound to ssDNA exhibit the same convex profile as that for the HU dimer. The descending branch of this profile, inversed with respect to the common behavior, spans ∼12 nt for the HU dimer (20–32 nt) as well as for two HU dimers (32–44 nt) reaching the maximum mobility at 44 nt. The maximum mobility of the ssDNA complex with three HU dimers, should be expected with a 56-nt long ssDNA (data not shown).
Binding of HU to ssDNA is salt dependent
It was shown earlier that HU binding to dsDNA is drastically reduced at 200 mM NaCl, while it continues to bind, under these high salt conditions, to a variety of DNA structural distortions, such as nick or overhang, even under high salt conditions (35,36). This illustrates the specific binding of HU to DNA lesions under high salt conditions compared to its non-specific binding to dsDNA only observed under low salt conditions. This observation prompted us to check how HU binds ssDNA in high salt. In contrast to low salt binding, for which gels are buffered with 22 mM Tris–borate, such experiments are commonly carried out in gels buffered with 90 mM Tris–Borate, Unfortunately, in 90 mM gels, dsDNA––HU complexes do not form sharp bands and migrate as smears so that DNA binding is detected mainly by a decreased intensity of the free DNA bands (44). To avoid these difficulties, we carried out the high-salt ssDNA–HU-binding assays with an intermediate Tris–borate concentration of 55 mM. Under these conditions, the bands of dsDNA–HU complexes of different stoichiometry remain detectable (Figure 3C). The smears are caused by the dissociation of the complexes during gel migration. As expected, the 1:1 HU–nick complex forms a sharp band with a strongly increased relative intensity. On the contrary, the HU binding to ssDNA is strongly reduced with respect to the low salt conditions (Figure 3B). As in the low salt conditions, ssDNA forms 1:1 and 1:2 complexes with HU, depending upon the DNA length, but the characteristic DNA chain lengths are shifted upward with respect to the low salt conditions. Notably, the 1:1 and 1:2 complexes are formed starting from 28 and 36 nt, respectively. The chain-length dependence of their gel mobility remains convex, with the mobility maxima observed at a longer ssDNA (44 nt for the 1:1 complex). The Kd suggest that HU binding is less salt dependent for ssDNA than for dsDNA. Otherwise the salt effect looks quite similar for ssDNA and dsDNA, suggesting a non-specific character of the ssDNA–HU binding.
HU does not bind two ssDNA molecules simultaneously
Our interpretation of the results shown in Figures 2 and 3 implies that HU always binds only a single DNA chain. However, the X-ray structure of a HU–dsDNA complex has demonstrated that the HU arms contact the minor groove of the double helix (34), whereas ssDNA has no minor groove. Therefore, we decided to check another hypothesis, namely, that HU could grap two ssDNA chains simultaneously, perhaps enforcing a partial hybridization. To this end, we studied HU binding with a mixture of labeled ssDNA of two different sizes, respectively, 32 and 48 nt. If HU were able to bind both ssDNA simultaneously, heterocomplexes with an intermediate mobility should appear. The experiment (data not shown) clearly indicates that such heterocomplexes are undetectable even under saturating concentrations of HU. All the results obtained in different conditions prove that HU binds one ssDNA molecule.
HU binding to ssDNA does not resemble SSB–DNA interactions
When bound to long ssDNA, SSB unfolds the ‘random’ base pairing and opens the DNA molecule all along its totality. DNA structures that arise from such partial base pairing contain single-stranded forks, >=<, i.e. motifs that the HU protein binds with high affinity (35,36). We expected, therefore, that HU would not unfold long ssDNA, in contrast to SSB. To check this ssDNA of phage phi (5454 nt) bound to either HU or the SSB protein was loaded onto an agarose gel and, after electrophoresis, stained with ethidium bromide. The SSB binding reduces the staining of the bands in agreement with DNA unfolding. In contrast, the HU binding did not decrease ethidium bromide staining at least if HU: ssDNA ratio does not exceed 1:15 nt (data not shown). Thus, HU did not change the ss/dsDNA equilibrium. This protein property should also affect the melting of dsDNA. Noteworthy, SSB decreases the DNA melting temperature by 10–20° (45), whereas HU shows a significant opposite effect which is particularly strong for A/T-rich DNA regions (31). This result is in agreement with our findings that HU has a relatively low affinity for poly(dT) and poly(dA) single-strands (data not shown; see Discussion section).
HU binding induces FRET between labeled ssDNA ends
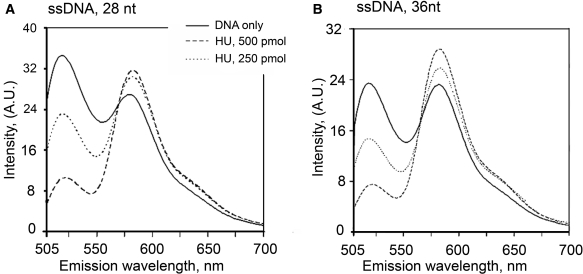
Very long apparent DNA-binding sites and abnormalities in gel mobility of protein–DNA complexes often indicate that protein binding induces bends in DNA (46). Based on the relatively long ssDNA length required for its initial binding and the high flexibility of ssDNA, we hypothesized that HU could induce an U-loop when bound, similarly to IHF in the X-ray structure of the IHF–dsDNA complex (46). The fluorescence excitation energy transfer (FRET) provides valuable information about the structure of biologically relevant molecules, including nucleic acids–proteins interactions, because of its distance and orientation dependence (38,47). In this study, we attached the donor (fluoresceine) to the 5′ end of the 28 and 36 nt oligonucleotides and the acceptor (tetramethylrhodamine) to the 3′ end. These lengths were chosen to be shorter and longer than 32 nt, which makes the most compact complex with HU, according to the gel mobility observed in Figure 3. We expect to observe a transfer of excitation energy from the donor to the acceptor only when the protein folds the oligonucleotide in such a way that the ends of the oligonucleotides are close to each other. In the unfolded form, little transfer is expected [the distance between the two dyes is well above the Föster critical distance (ca 4.5 nm)]. Upon addition of increasing concentrations of HU, a FRET is clearly observed (Figure 4), the two ends of the oligonucleotide are brought close to each other. The FRET signal reaches a plateau for a molar ratio of proteins per oligonucleotide higher than 2–3. Interestingly, the stimulated emission of the acceptor is quenched for the 28 nt, maybe because of the proximity of the protein. As expected, for other DNA structures, dsDNA, nick, 3′ and 5′ DNA overhangs, we did not detect changes in FRET upon HU binding (data not shown). Altogether, these results are in agreement with an U-shaped ssDNA structure bound to HU.
Figure 4.
Fluorescence resonance energy transfer between 5′ fluorescein and 3′- tetramethyl rhodamine (FAM) for 28-nt-long (Panel A) and 36-nt-long (Panel B) ssDNA is induced by HU binding. To 1 µM of oligonucleotides (solid lines) was added HU in (dimer:DNA) ratio 1:2 (dashed lines) and 1:1 (dotted line). On the right, the fluorescence signal of the donor (circles) and of the acceptor [IA/(ID+IA), squares] is shown for different ratios of HU and ssDNA.
DISCUSSION
Previous studies have shown that HU binds specifically many DNA repair intermediates such as dsDNA with nicks or ss forks (=<) (36,44,48). These different structures have one common trait, namely, they all contain a stretch of dsDNA interrupted by a flexible region or a hinge. If this is a common feature for HU binding, HU should then also specifically bind to DNA containing mismatches and bulges since they are intrinsically flexible structures. Here we show that this is indeed the case. Long mismatches of 4–12 bp and the bulge of 7 nt are better binders than the corresponding shorter analogs, strongly indicating that the hinge flexibility is a main condition for efficient interaction with HU. In longer DNA mismatches, HU can recognize the ss fork (=<) structure as shown earlier (36). Therefore, this interaction is based on the HU recognition of the 3′ DNA overhang. Mismatches of 1 or 2 bp apparently cannot be bent by HU. Our study presents examples of HU substrates where the DNA flexibility is locally increased without introducing ss breaks.
The observed requirement of a ‘flexible region’ in dsDNA for strong and specific HU binding prompted us to investigate HU binding to ssDNA. The binding of HU to ssDNA has been reported but never studied thoroughly (4,40–42). Here we demonstrate that the protein binds ssDNA fragments starting from a certain threshold chain length. This minimal length is estimated to ca 24 nt, slightly varying with the base sequence. Similar experiments with the complementary ssDNA strand showed no detectable binding for 20-nt long ssDNA even with the highest HU concentrations, and only weak binding for the corresponding 23-mer ssDNA (data not shown). Interactions with shorter ssDNA are much weaker or non-existent. Together with the FRET data, the relatively long threshold length suggests that, in the ssDNA–HU complex, the DNA chain wraps around the HU dimer and forms an U-shaped complex. To evaluate the putative shape and parameters of such complexes, we took advantage of the recently solved HU–dsDNA co-crystal structure (34).
In this X-ray structure (34), HU was co-crystallized with a deformed DNA duplex, containing several single nucleotide mismatches and bulges. By computer homology modeling, we estimated the trace of ssDNA on the protein surface in the corresponding conformation of a dsDNA overhang. Figure 5 shows the computed conformation of a short 3'-DNA overhang bound to HU. This structure corresponds to the results obtained by biochemical methods (35,36) with minimal deviations from the X-ray data (34). The two long protruding β-hairpin arms contact the double-helical part of the overhang similarly to the original structure. On both sides of the protein core, there are several positively charged amino acid residues (colored in red in Figure 5). The biochemical studies (35,36) suggest that strong structure-specific binding is achieved when HU can interact with DNA by both its arms and body. The DNA contacts with the surface residues shown in Figure 5 are probably involved in these interactions (33,35,36,48–50), which explains the dominating role of the DNA flexibility that we observed experimentally in our work. It is clear that these HU–DNA contacts are possible only if the DNA fragment contains a flexible hinge as in bulges, mismatches and earlier studied non-canonical DNA structures (36).
Assuming that the HU binding to ssDNA occurs by the same electrostatic contacts, the structure shown in Figure 5 can be used to model the possible ssDNA traces in the bound state as well as the length of ssDNA fragments necessary to join different interaction sites. Accordingly, ∼6 bp are needed for DNA to run from the extremity of the HU arms to the closest positively charged region of the HU body. This agrees with the minimal length of the ss segment required for specific HU binding to overhangs (35,36). The double-helical part of the overhang that interacts with the HU arms involves 9 bp steps, which equals to the minimal estimated HU-binding site on dsDNA (36). Based on these measurements, it should take 9 nt (HU arms) + 6 nt (HU side) = 15 nt to form a Γ-shaped complex in the case of ssDNA. Since we do not detect stable complexes for such short ssDNA, we think that simultaneous contacts of DNA with the HU arms and both its sides are probably necessary for binding. Therefore, ssDNA in order to bind to HU must be at least 9 nt + 2 × 6 nt = 21 nt. The latter agrees with our results, we found that ssDNA must be around 24 nt to form a complex with HU. Finally, the FRET observed for both 28 and 36 nt, indicate that the DNA ends run in parallel as it should be in an U-shaped structure.
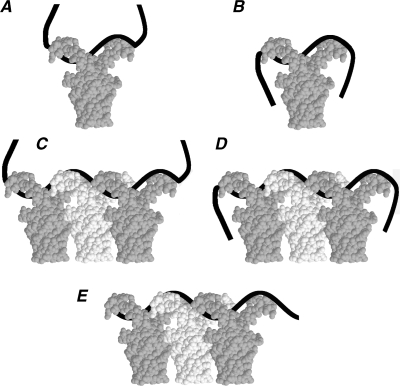
Based on the above considerations, the unusual behavior of HU–ssDNA complexes in band-shift experiments can be explained as follows. By increasing the ssDNA length, the gel mobility does not change or slightly grows with charge if the size of the minimal plane projection of the complex remains constant. If this projection increases, the gel mobility strongly decreases regardless of the charge. The complex of the HU dimer with 24 nt ssDNA is schematized in Figure 6A and B. The tails of the ssDNA can be already absorbed by the opposite sides of the HU body, but this binding mode is in dynamic equilibrium with conformations where one or both DNA ends are free (Figure 6A), therefore the gel mobility is reduced as the length of the ssDNA in the complexes increases (Figure 3A). With increased DNA length, the U-shaped binding mode becomes predominant; the average effective size of the complex is reduced and its gel mobility increased. The ssDNA length of 32 nt apparently corresponds to the most compact configuration of the U-shaped complex and it is thus characterized by the highest mobility (Figure 6B). With longer ssDNA, its ends start to protrude beyond the minimal cross-section and the gel mobility of the complexes progressively decreases (Figure 3A). For a 32-nt long ssDNA, one can estimate that the side contacts are 11-nt long, while the arm contacts are 9-nt long. A similar effect was earlier observed, but not explained, for HU bound to 3′-DNA overhangs (36). In this case, the gel-mobility reached a maximum with a 3′-overhang of 10 nt in good agreement with the present data.
Figure 6.
The putative model of HU:ssDNA binding in complexes of different stoichiometry. The ssDNA is shown as a black band. The HU dimers are shown in white and gray for clarity. Plates A–B and plates C–E display sketches of complexes of a ssDNA strand with one and three HU, respectively. HU forms two types of complexes with ssDNA. Complexes of the first type have low affinity and low gel mobility (plates A and C). Complexes of the second type are more compact and have higher affinity and gel mobility (plates B and D). The conformation of the ssDNA part, which is not covered by HU protein, explains the difference between the high- and low-affinity complexes. Plate E represents the complex formed by ssDNA absorbed on HU oligomers so that the DNA extremities are hidden by the protein.
Similar factors determine the gel mobility of multimeric HU complexes formed with higher protein concentrations (Figure 3B). The possible overall shape of such complexes is shown in Figure 6C and D. Their gel mobility varies with the DNA length in the same way as for the binding of a single HU dimer. Based upon the results shown in Figure 3B, we estimate that each additional HU on ssDNA needs 12 nt. The most compact U-shaped complex is formed when at least 10 nt are allowed to interact with both HU sides, corresponding to a series of gel mobility maxima at 32, 44, 56 nt and so forth, as observed in Figure 3B. The observed FRET signal can be mainly attributed to the U-shaped complexes of stoichiometry 1:1 and 1:2. In both cases, the distance between the DNA ends is small enough to give FRET.
HU binding to dsDNA is cooperative (44), whereas HU binding to ssDNA is not, as shown here. This difference may have a simple structural explanation. Straight dsDNA cannot make contacts with HU sides, leaving some protein surface unused. This surface contributes to the binding of a second HU which results in a positive co-operativity. In contrast, the U-shaped ssDNA–HU complex employs both protein sides. Binding of another HU must destroy some contacts of the initial complex, which results in the apparent absence of co-operativity or even in a negative co-operativity.
We suppose that the numerous weak bands seen in Figure 3B are due to the complexes sketched in Figure 6E. Because the ssDNA is bound by the arms of HU dimers, it is almost hidden and does not interact with the gel matrix. The gel mobility of such complexes should only slightly grow with the ssDNA length due to the increased charge, which is seen in Figure 3B. This binding is significantly reduced in high salt, which can be attributed to the electrostatic screening of protein–protein and/or protein–DNA interactions. A hypothesis that HU multimers can form a 1D scaffold that plays a structuring role in bacterial chromatin in vivo has been discussed in the literature (51,52). Recent studies of a HU mutant indicate that protein–protein contacts and multimer HU complexes can be specifically involved in DNA binding (53,54). It has also been shown that, under nearly physiological HU concentrations, dsDNA–HU complexes look like rigid nucleoprotein filaments in which HU appears to arrange helically around DNA (55).
HU binding to ssDNA has the following sequence preference: poly(dG) > ‘generic’ > (dC) >(dT) >> (dA) (data not shown). HU exhibits a negative co-operativity for poly(dT) binding, so that we never detected binding of three HU dimers to poly(dT) of 36, 5′ and 72 nt. A strong HU preference to short GC-rich ssDNA was earlier demonstrated with generic oligodeoxyribonucleotide microchips (56) in conditions in which any secondary structure formation can be excluded. Noteworthy, the G-tracts exhibit an anomalously high rates of base-pair opening in dsDNA (57), that the GpG stack is the weakest among dinucleotides (58).
Our interest in ssDNA originated from its presence in several structures binding specifically HU. It cannot, however, be excluded that HU–ssDNA interactions play a distinct role in vivo. Another protein, SSB, whose specific function consists in binding ssDNA, is present in bacteria, but in a much smaller number of copies [300 for SSB versus 30 000 for HU, (4,31,59)]. Binding of HU to ssDNA is qualitatively different from the one of SSB. The SSB–ssDNA binding is highly cooperative (59), while the HU–ssDNA binding is not. The binding is sequence dependent in both cases, but the sequence preferences are different. Notably, the SSB protein has a strong preferences to poly(dT): (dT) > (dC) >> generic DNA > (dA) (59). In contrast, HU binds with the following rank: generic DNA > (dC) > (dT) >> (dA). SSB and HU have opposite effects on the ds/ssDNA equilibrium. SSB facilitates DNA melting while HU inhibits the melting of AT-rich regions (31,45). Due to the high co-operativity and a relatively small number of copies, SSB is mainly engaged in replication forks and transcription bubbles where long stretches of ssDNA are formed temporarily (59–61). In contrast, since HU binding to ssDNA is non-cooperative, HU can bind and protect various smaller DNA lesions scattered in the inactive part of the bacterial genome.
The capacity to bind ssDNA can also be necessary for the function of the HU protein in DNA repair mechanisms. The specific binding of HU to short dsDNA mismatches points to its role in the repair of some particular DNA lesions. DNA alkylating agents as well as UV light produce bulky interstrand DNA lesions that structurally resemble short mismatches. Accordingly, the HU deletion mutants exhibit increased sensitivity to irradiation as well as to H2O2 treatment compared to the wild-type E. coli [(16) and J.Y.R. and Claret,L., personal communication). The suggested participation of the HU protein in repairing DNA mismatches should also cause an enhanced mutation rate in E. coli, in which HU is absent. Truly it was shown that spontaneous mutation rapidly accumulate into the double hupAB mutants (62). The ‘real’ HU mutants (i.e. without compensatory mutations) are morphologically different from the wild-type as they form very tiny colonies, but this phenotype is reverted very rapidly to give normal size colonies in the revertants (62,63). The observed extremely high reversion rate could be due to an increased mutation rate in bacteria lacking HU protein. Extensive base-pair mismatching should also appear within converged DNA regions during homologous recombination. Protective degradation of such DNA proceeds via recognition of mismatched zones by mutS protein that initiates the degradation cascade (64). The specific HU binding to extended mismatches can inhibit the homologous recombination by facilitating the recruitment of mutS and other proteins directly involved in degradation of DNA.
ACKNOWLEDGEMENTS
We are indebted to Olivier Pellegrini for purifying the HU protein. We thank Patricia Carandi for her help during the preparation of the manuscript, Ciaran Condon, Brigitte Hartmann, Jacques Oberto and Murat Saparbaev for valuable discussions. This work was supported by the CNRS (UPR 9073) and fellowships from ARC (Association de la Recherche sur le Cancer) et la Fondation de la Recherche Medicale. Funding to pay the Open Access publication charges for this article was provided by Inserm (U565).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rouviere-Yaniv J, Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl Acad. Sci. USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouviere-Yaniv J, Yaniv M, Germond JE. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 3.Drlica K, Rouviere-Yaniv J. Histone like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouviere-Yaniv J. Localization of the HU protein on the Escherichia coli nucleoid. Cold Spring Harb. Symp. Quant. Biol. 1978;42:439–447. doi: 10.1101/sqb.1978.042.01.047. [DOI] [PubMed] [Google Scholar]
- 5.Wery M, Woldringh CL, Rouviere-Yaniv J. HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie. 2001;83:193–200. doi: 10.1016/s0300-9084(01)01254-8. [DOI] [PubMed] [Google Scholar]
- 6.Haselkorn R, Rouviere-Yaniv J. Cyanobacterial DNA-binding protein related to Escherichia coli HU. Proc. Natl Acad. Sci. USA. 1976;73:1917–1920. doi: 10.1073/pnas.73.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oberto J, Rouviere-Yaniv J. Serratia marcescens contains a heterodimeric HU protein like Escherichia coli and Salmonella typhimurium. J.Bacteriol. 1996;178:293–297. doi: 10.1128/jb.178.1.293-297.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aitken A, Rouviere-Yaniv J. Amino and carboxy terminal sequences of the DNA-binding protein HU from the Cyanobacterium Synechocystis PCC 6701 (ATCC 27170) Biochem. Biophys. Res. Commun. 1979;91:461–467. doi: 10.1016/0006-291x(79)91544-4. [DOI] [PubMed] [Google Scholar]
- 9.Briat JF, Letoffee S, Mache R, Rouviere-Yaniv J. Similarity between the bacterial histone-like HU and a protein from spinach chloroplasts. FEBS Lett. 1984;172:75–79. [Google Scholar]
- 10.Neilan JG, Lu Z, Kutish GF, Sussman MD, Roberts PC, Yozawa T, Rock DL. An African swine fever virus gene with similarity to bacterial DNA binding proteins, bacterial integration host factors, and the Bacillus phage SPO1 transcription factor, TF1. Nucleic Acids Res. 1993;21:1496. doi: 10.1093/nar/21.6.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caron F, Jacq C, Rouviere-Yaniv J. Characterization of a histone-like protein extracted from yeast mitochondria. Proc. Natl Acad. Sci. USA. 1979;76:4265–4269. doi: 10.1073/pnas.76.9.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberto J, Drlica K, Rouviere-Yaniv J. Histones, HMG, HU, IHF: Meme combat. Biochimie. 1994;76:901–908. doi: 10.1016/0300-9084(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 13.Nash HA. In: Regulation of Gene Expression in Escherichia coli.. Lin ECC, Lynch AS, editors. Chapter 8. R.G. Landes Company; 1996. pp. 149–179. [Google Scholar]
- 14.Rouviere-Yaniv J, Kjeldgaard NO. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979;106:297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- 15.Claret L, Rouviere-Yaniv J. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J. Mol. Biol. 1997;273:93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- 16.Boubrik F, Rouviere-Yaniv J. Increased sensitivity to gamma irradiation in bacteria lacking protein HU. Proc. Natl Acad. Sci. USA. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Waters R. Escherichia coli strains lacking protein HU are UV sensitive due to a role for HU in homologous recombination. J. Bacteriol. 1998;180:3750–3756. doi: 10.1128/jb.180.15.3750-3756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornberg A. Enzyme studies of replication of the Escherichia coli chromosome. Adv. Exp. Med. Biol. 1984;179:3–16. doi: 10.1007/978-1-4684-8730-5_1. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg A. Enzyme systems initiating replication at the origin of the Escherichia coli chromosome. J. Cell Sci. Suppl. 1987;7:1–13. doi: 10.1242/jcs.1987.supplement_7.1. [DOI] [PubMed] [Google Scholar]
- 20.Hwang DS, Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 21.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 22.Polaczek P, Kwan K, Campbell JL. Unwinding of the Escherichia coli origin of replication (oriC) can occur in the absence of initiation proteins but is stabilized by DnaA and histone-like proteins IHF or HU. Plasmid. 1998;39:77–83. doi: 10.1006/plas.1997.1328. [DOI] [PubMed] [Google Scholar]
- 23.Polaczek P, Kwan K, Liberies DA, Campbell JL. Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol. Microbiol. 1997;26:261–275. doi: 10.1046/j.1365-2958.1997.5701931.x. [DOI] [PubMed] [Google Scholar]
- 24.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 2002;46:113–124. doi: 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 25.Morales P, Rouviere-Yaniv J, Dreyfus M. The histone-like protein HU does not obstruct movement of T7 RNA polymerase in Escherichia coli cells but stimulates its activity. J. Bacteriol. 2002;184:1565–1570. doi: 10.1128/JB.184.6.1565-1570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe A, Vinella D, D’Ari R. The Escherichia coli histone-like protein HU affects DNA initiation, chromosome partitioning via MukB, and cell division via MinCDE. J. Bacteriol. 1997;179:3494–3499. doi: 10.1128/jb.179.11.3494-3499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavoie BD, Shaw GS, Millner A, Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 28.Lavoie BD, Chaconas G. A second high affinity HU binding site in the phage Mu transpososome. J. Biol. Chem. 1994;269:15571–15576. [PubMed] [Google Scholar]
- 29.Kobryn K, Lavoie BD, Chaconas G. Supercoiling-dependent site-specific binding of HU to naked Mu DNA. J. Mol. Biol. 1999;289:777–784. doi: 10.1006/jmbi.1999.2805. [DOI] [PubMed] [Google Scholar]
- 30.Rouviere-Yaniv J, Bonnefoy E, Huisman O, Almeida A. In: The Bacterial Chromosome.. Drlica K, Riley M, editors. Washington, DC 20005: Chapter 19. American Society for Microbiology; 1990. pp. 247–257. [Google Scholar]
- 31.Rouviere-Yaniv J, Gros F, Haselkorn R, Reiss C. In: The Organisation and Expression of the Eukaryotic Genome.. Bradbury EM, Javaherian K, editors. New York: Academic Press; 1977. pp. 211–231. [Google Scholar]
- 32.Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. Cross-talk between topoisomerase I and HU in Escherichia coli. J. Mol. Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 33.Balandina A, Kamashev D, Rouviere-Yaniv J. The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J. Biol. Chem. 2002;277:27622–27628. doi: 10.1074/jbc.M201978200. [DOI] [PubMed] [Google Scholar]
- 34.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamashev D, Balandina A, Rouviere-Yaniv J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999;18:5434–5444. doi: 10.1093/emboj/18.19.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamashev D, Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balandina A, Claret L, Hengge-Aronis R, Rouviere-Yaniv J. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 2001;39:1069–1079. doi: 10.1046/j.1365-2958.2001.02305.x. [DOI] [PubMed] [Google Scholar]
- 38.Berney C, Danuser G. FRET or no FRET: a quantitative comparison. Biophys. J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Berthold V, Geider K. Interaction of DNA with DNA-binding proteins. The characterization of protein HD from Escherichia coli and its nucleic acid complexes. Eur. J. Biochem. 1976;71:443–449. doi: 10.1111/j.1432-1033.1976.tb11132.x. [DOI] [PubMed] [Google Scholar]
- 41.Bonnefoy E, Rouviere-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991;10:687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holck A, Kleppe K. Affinity of protein HU for different nucleic acids. FEBS Lett. 1985;185:121–124. doi: 10.1016/0014-5793(85)80753-5. [DOI] [PubMed] [Google Scholar]
- 43.Pellegrini O, Oberto J, Pinson V, Wery M, Rouviere-Yaniv J. Overproduction and improved strategies to purify the three native forms of nuclease-free HU protein. Biochimie. 2000;82:693–704. doi: 10.1016/s0300-9084(00)01151-2. [DOI] [PubMed] [Google Scholar]
- 44.Pinson V, Takahashi M, Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 45.Genschel J, Litz L, Thole H, Roemling U, Urbanke C. Isolation, sequencing and overproduction of the single-stranded DNA binding protein from Pseudomonas aeruginosa PAO. Gene. 1996;182:137–143. doi: 10.1016/s0378-1119(96)00535-5. [DOI] [PubMed] [Google Scholar]
- 46.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 47.Jares-Erijman EA, Jovin TM. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 48.Castaing B, Zelwer C, Laval J, Boiteux S. HU protein of Escherichia coli binds specifically to DNA that contains single-strand breaks or gaps. J. Biol. Chem. 1995;270:10291–10296. doi: 10.1074/jbc.270.17.10291. [DOI] [PubMed] [Google Scholar]
- 49.Bonnefoy E, Takahashi M, Yaniv JR. DNA-binding parameters of the HU protein of Escherichia coli to cruciform DNA. J. Mol. Biol. 1994;242:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 50.Pontiggia A, Negri A, Beltrame M, Bianchi ME. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 51.White SW, Wilson KS, Appelt K, Tanaka I. The high-resolution structure of DNA-binding protein HU from Bacillus stearothermophilus. Acta Crystallogr. D Biol. Crystallogr. 1999;55:801–809. doi: 10.1107/s0907444999000578. [DOI] [PubMed] [Google Scholar]
- 52.Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. J. Mol. Biol. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 53.Guo F, Adhya S. Spiral structure of Escherichia coli HUalphabeta provides foundation for DNA supercoiling. Proc. Natl Acad. Sci. USA. 2007;104:4309–4314. doi: 10.1073/pnas.0611686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription. J. Biol. Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- 55.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krylov AS, Zasedateleva OA, Prokopenko DV, Rouviere-Yaniv J, Mirzabekov AD. Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips. Nucleic Acids Res. 2001;29:2654–2660. doi: 10.1093/nar/29.12.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dornberger U, Leijon M, Fritzsche H. High base pair opening rates in tracts of GC base pairs. J. Biol. Chem. 1999;274:6957–6962. doi: 10.1074/jbc.274.11.6957. [DOI] [PubMed] [Google Scholar]
- 58.Sponer J, Gabb HA, Leszczynski J, Hobza P. Base-base and deoxyribose-base stacking interactions in B-DNA and Z-DNA: a quantum-chemical study. Biophys. J. 1997;73:76–87. doi: 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 60.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 61.Wold S, Crooke E, Skarstad K. The Escherichia coli FIS protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 1996;24:3527–3532. doi: 10.1093/nar/24.18.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huisman O, Faelen M, Girard D, Jaffe A, Toussaint A, Rouviere-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J. Mol. Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 64.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]