Abstract
Different RNA species are rigorously discriminated and exported by distinct export factors, but this discrimination mechanism remains largely unknown. We previously showed, by RNA microinjection experiments, that intronless mRNAs are discriminated from U snRNAs based on their difference in RNA length. However, it was unclear how they are discriminated in the natural situation in which their nascent transcripts emerge progressively during transcription. We hypothesized that transcription from the corresponding promoters is important for this discrimination. Here we show that contrary to our hypothesis, the discrimination process was not significantly influenced by whether transcription occurred from an mRNA- versus a U snRNA-type promoter. Rather, the features of transcribed RNAs determined the RNA identity, consistent with our previous results of RNA microinjection. Moreover, we found that the poly (A) tail can function as an identity element for mRNA export. The presence of a poly (A) tail of an appropriate length committed otherwise short Pol II transcripts to the mRNA export pathway in a dominant manner, indicating that the poly (A) tail either contributes to increasing the RNA length or functions as a platform to recruit mRNA export factors. Our results reveal a novel function of the poly (A) tail in mRNA export.
INTRODUCTION
The nuclear envelope separates eukaryotic cells into two major compartments, the nucleus and the cytoplasm. This separation requires transport between the two compartments through the nuclear pore complexes, the channels embedded in the nuclear envelope (1). The vast majority of RNA species, following their transcription and processing in the nucleus, are exported to the cytoplasm. Different RNA species, such as tRNAs, U snRNAs, mRNAs and rRNAs, utilize distinct export pathways, i.e. distinct sets of export factors (2,3). Accumulating evidence shows that the pathway of RNA export can influence the fate of a given RNA in the cytoplasm (4–8), indicating the biological importance of the choice of RNA export pathway. This means that the cellular export machinery must be able to discriminate distinct RNA species, and therefore each RNA species should have identifying features that specify its export pathway.
The mechanism of discrimination between U snRNAs and mRNAs has been a focus of our research interest because these two classes of RNA have similarities. Both classes of RNA are transcribed by RNA polymerase II and therefore both initially acquire a 7-methyl guanosine cap (m7G-cap) structure. This common m7G-cap structure is essential for U snRNA export and stimulatory for mRNA export (9–14). Importantly, neither class of RNA has clearly conserved RNA sequences or structures that may function as its distinguishing features, with the only exceptions being the poly (A) tail for mRNAs and the Sm-binding site for U snRNAs. Neither of these two elements, however, is essential for RNA export per se (9,10). Despite these similarities, these two classes of RNA utilize distinct export factors.
Major spliceosomal U snRNAs such as U1, U2, U4 and U5 are initially exported from the nucleus in Metazoa (15,16). U snRNA export is mediated by the leucine-rich nuclear export signal receptor CRM1, a member of the importinβ family (17–19). CRM1 binds indirectly to U snRNAs through two adaptor proteins. One is the heterodimeric Cap Binding Complex (CBC), which binds specifically to the essential export signal of U snRNAs, the m7G-cap structure (11,20,21). The other adaptor is PHAX, which bridges the interaction between CRM1 and the CBC/U snRNA complex (22). PHAX has a leucine-rich NES to which CRM1 binds cooperatively with RanGTP. In this way these five proteins and a U snRNA assemble into the export complex.
On the other hand, mRNA export is mediated by distinct export factors. The principal export receptor for mRNAs is TAP-p15 heterodimer (23–25). TAP is one of the few non-importinβ family transport receptors known to date. Although TAP itself can bind RNA, its recruitment to mRNAs requires adaptor proteins. RNA-binding adaptor proteins, including Aly/REF and shuttling SR proteins, are first recruited to mRNA, and these adaptor proteins in turn recruit mRNA export receptor TAP-p15 heterodimer onto the RNA through protein–protein interactions (26–32). Interestingly, SR proteins only in their dephosphorylated state can recruit TAP-p15 (36,37). The m7G-cap structure of mRNAs is bound by CBC, and the CBC binding can stimulate the recruitment of adaptor proteins (13,14). However, the contribution of the cap structure and CBC to mRNA export is lower than in U snRNA export.
In attempts to search for the distinguishing features between mRNAs and U snRNAs, we previously found that the presence of introns in mRNA precursors is one such feature. When a pre-mRNA intron was artificially inserted into U1 snRNA, the spliced U1 snRNA was exported via the mRNA export pathway instead of the U snRNA pathway, indicating that introns can function as a distinguishing feature of mRNAs, i.e. as an identity element for mRNA export (7). It is very likely that this identity element functions through splicing-dependent deposition of protein complexes such as the exon junction complex (EJC) and/or the transcription/export (TREX) complex, both of which contain mRNA-specific RNA-binding protein Aly/REF (13,31,33–35). Aly/REF as well as some shuttling SR proteins in their dephosphorylated state functions in recruiting the principal mRNA export receptor, TAP/p15 heterodimer, onto the RNA, thus inducing mRNA export (26–28,31,32,36,37).
We subsequently found that RNA length plays an important role in the choice of RNA export pathway (7,12). When U snRNA was elongated by inserting an unstructured RNA fragment longer than 200–300 nt, the elongated U snRNA behaved like an mRNA in export. Conversely, if an intronless mRNA was shortened (100 nt or shorter) by deletion, the shortened mRNA behaved like a U snRNA in export. Taken together, these findings indicate that the presence of a stretch of unstructured RNA of a certain length can function as a splicing-independent mRNA identity element (12).
The above two identity elements were identified by RNA microinjection experiments using Xenopus oocytes in which in vitro transcribed RNAs were microinjected into the nucleus, and therefore in which RNA export was not dependent on in vivo transcription. This experimental system was advantageous for focusing on RNA export per se, but on the other hand, it was not useful for examining the influence of transcription on the identity of the transcripts in nuclear export. More specifically, it was not clear how these mRNA identity elements, especially ‘RNA length’, are recognized in the natural situation in which the nascent transcripts emerge progressively as transcription proceeds. Therefore, we hypothesized that transcription from the two corresponding promoters, namely the mRNA- or U snRNA-type promoter, may be important for the discrimination between the nascent transcripts of mRNAs and U snRNAs in vivo.
The mRNA-type promoters and the U snRNA-type promoters have distinct cis-elements for transcription (38). Although RNA polymerase II is involved in transcribing both types, the transcription from the U snRNA-type promoters requires an additional set of transcription factors termed the snRNA activating protein complex (SNAPc) on top of the basic Pol II transcription factors (38). It is well documented that different promoters can load different protein factors, especially RNA processing factors, onto the nascent transcripts (reviewed in 39–41, also see 42–44). Some splicing factors and cleavage/polyadenylation factors are loaded through transcription from the mRNA-type promoters (39–41,45,46), whereas a protein complex for 3'-end cleavage of U snRNAs, termed ‘Integrator’, is loaded through transcription from the U snRNA-type promoters (47). Therefore, it was plausible to hypothesize that specific export factors for mRNAs and U snRNAs are loaded through transcription from the corresponding promoters, leading to commitment to the corresponding export pathways. In yeast, the recruitment of adaptor proteins is coupled to transcription. Aly/REF (Yra1p in yeast) together with DExD-box RNA helicase UAP56 (Sub2p in yeast) interacts with the transcription elongation complex (THO complex), forming a larger protein complex termed TREX (transcription/export) complex (48,49). Owing to this complex formation, Aly/REF and UAP56 are recruited onto the nascent mRNA transcript during transcription elongation (48,49). Although the recruitment of the TREX complex is known to be splicing-dependent in vertebrates (35), it is still possible that transcription may be coupled to RNA export in the case of intronless vertebrate genes.
In this study, we focused on the effect of transcription on the process of discrimination between intronless mRNAs and UsnRNAs. Contrary to our initial expectation, whether transcription occurred from the mRNA- or U snRNA-type promoter did not influence the discrimination process. Instead, the features of the transcribed RNAs, rather than the ways they were transcribed, were important for determining the identities of the RNAs for nuclear export, as was previously shown by us in RNA microinjection experiments (7,12). Moreover, during the course of this study, we serendipitously found that the presence of the poly (A) tail in the transcript can function as an identity element for mRNA nuclear export. If a poly (A) tail of an appropriate length was present on the transcript, the transcript was committed to the mRNA export pathway in a dominant manner, even when the transcript without the poly (A) tail was too short to behave like an mRNA, indicating that the poly (A) tail either contributes to increasing the RNA length or functions as a platform to specifically recruit mRNA export factors. Our results reveal a novel role of the poly (A) tail in mRNA nuclear export.
MATERIALS AND METHODS
DNA constructs
U1 gene derivatives: The Xenopus U1 gene was PCR-amplified from Xenopus genomic DNA and the amplified fragment was cloned in the EcoRI-HindIII sites of the vector pUC118 (Takara). The Sm site of the cloned U1 gene was mutagenized by PCR to an XhoI site (9). Various cDNA fragments were inserted into the XhoI site as described previously (12). A DNA fragment containing A70 sequence with XhoI and SalI sites was made by annealing A70-containing oligo DNA with T70-containing oligo DNA. One or two copies of this DNA fragment were inserted into the XhoI site of the U1 gene. cDNA expression constructs without U1 sequence: Various fragments were PCR-amplified from β-globin cDNA, DHFR cDNA or the U5ΔSm gene as described previously (12), except that EcoRV and XbaI sites were added to the upstream and downstream DNA ends, respectively. The 100-nt DHFR fragment was PCR-amplified by the two primers (5′-GGAAGATCTGCATCATGGTTCGACCATTGA-3′, 5′-AACTCGTTCCTGAGCGGAGGCGGATCCGCG-3′). The fragment was digested by BglII and BamHI and ligated to multimerize when necessary. The multimerized fragments were PCR-amplified again to add EcoRV and XbaI sites. The U1 promoter and the U1 terminator were PCR-amplified from the U1 gene with the following two primer sets; 5′-CCGGAATTCAAGCTTTTGTACAAGGATTCA-3′ and 5′-ACGATATCGAGTTGAACAAGAAATTTTCAA-3′, or 5′-GCTCTAGAATTTTGTTTGTTTAAAGATAGA-3′ and 5′-CATAAGCTTGATCTCTATCTTTAAACAAAC-3′, respectively. The CMV promoter and the BGH poly (A) signal were PCR-amplified from the pcDNA3 plasmid (Invitrogen) with the following two primer sets; 5′-CCGGAATTCTGTACGGGCCAGATATACGCG-3′ and 5′-TCAGATATCTCTAGTTAGCCAGAGAGCTCT-3′, or 5′-TGCTCTAGACTAATAAAATGAGGAAATTGC-3′ and 5′-AAACTGCAGTTCCGCCTCAGAAGCCATAGA-3′, respectively. Various cDNA fragments were cloned between one of the two promoter fragments and one of the two terminator fragments, using EcoRI-PstI or EcoRI-HindIII sites of pUC118.
Assay of RNA export coupled to in vivo transcription
Microinjection into Xenopus oocytes was performed as previously described (12), except that 17 ng/oocyte of various plasmids was microinjected instead of RNA, and nuclear and cytoplasmic RNA was recovered at 3–4 h after microinjection and visualized by northern blotting with specific probes. CTE and PHAXΔNES were prepared as described (7,22).
Northern blotting
RNA samples were fractionated by 8% PAGE (7M Urea) and transferred to Hybond N+ membranes (Amersham). Hybridization with oligo DNA probes, 32P-labeled at the 5′ termini, was performed in PerfectHybPlus (Sigma). For detecting only polyadenylated transcripts, 5′-TTTTTTTTTTTTGCGATGCAATTTCCT-3′ was used as a probe at 53°C. For detecting other transcripts, the following probes were used at 40°C; U1ΔSm: 5′-ACTACCAGCTCGAGTGCAGTCG-3′, other U1 derivatives: 5′-CTCGAGTGCAGTCGAGTTTCCCGCA-3′, DHFR100: 5′-TCTAGAAACTCGTTCCTGAGCGGAGGC-3′, β-globin cDNA-derived transcripts: 5′-AGTGAACACAGTTGTGTCAGAAGCAAATGTAAGCAAGCTTGTATTC-3′, Xenopus endogenous U6 RNA: 5′-CAGGGGCCATGCTAATCTTCTCT-3′, Xenopus endogenous 5.8S RNA: 5′-CGAAGTGTCGATGATCAATGTGTCC-3′. Analysis and quantitation of the northern signal was performed with BAS-2500 (FujiFilm) and Image Gauge version 3.45 (FujiFilm).
RNA immunoprecipitation
Nuclear extract was prepared at 3 h after microinjection and immunoprecipitation was performed as described previously (12). The precipitated RNA was treated with RQ1 DNase (Promega) and analyzed by quantitative PCR, using SuperScript III Platinum SYBR Green One-Step qRT-PCR (Invitrogen) and a 7500 RealTime PCR system (Applied Biosystems). The following PCR primers were used. Non-polyadenylated β-globin transcripts: 5′-GAATACAAGCTTGCTTACATTTGC-3′ and 5′-TGCTAGTGAACACAGTTGTG-3′. Polyadenylated β-globin transcripts: 5′-GAATACAAGCTTGCTTACATTTGC-3′ and 5′-TTTTTTTTTTTTTTTTTTTTTGCGATGCAA-3′. U1-derived transcripts: 5′-ATACTTACCTGGCAGGGGAG-3′ and 5′-TCCCCCACTACCAGCTCGAG-3′.
RESULTS
RNA length is an important determinant of RNA identity in nuclear export, even in an RNA export assay system that is coupled to in vivo transcription
To examine whether transcription in vivo influences the discrimination between intronless mRNAs and U snRNAs, we developed an RNA export assay system that is coupled to in vivo transcription. To this end, DNA fragments derived from U snRNA and/or mRNA sequences were cloned downstream of either the U1 snRNA promoter (as a representative of U snRNA-type promoters) or the cytomegalovirus (CMV) promoter (as a representative of mRNA-type promoters) to initiate transcription in vivo. We also inserted either the U1 snRNA terminator or the poly (A) signal from the bovine growth hormone (BGH) gene downstream of the DNA fragments to terminate transcription. The resultant plasmids were microinjected into the nucleus of Xenopus oocytes, and export of RNA transcripts produced in vivo from the injected plasmids was examined by northern blotting analysis with the nuclear and cytoplasmic RNA fractions. The export pathways of the transcripts were analyzed using pathway-specific export inhibitors.
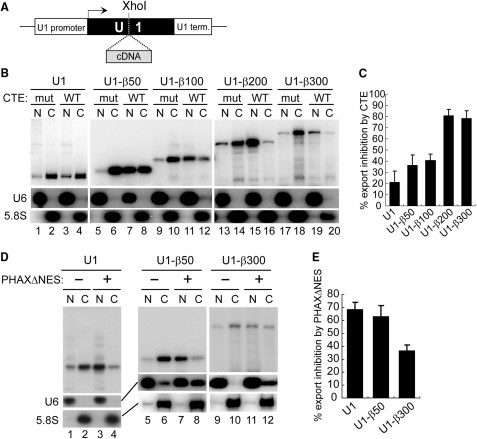
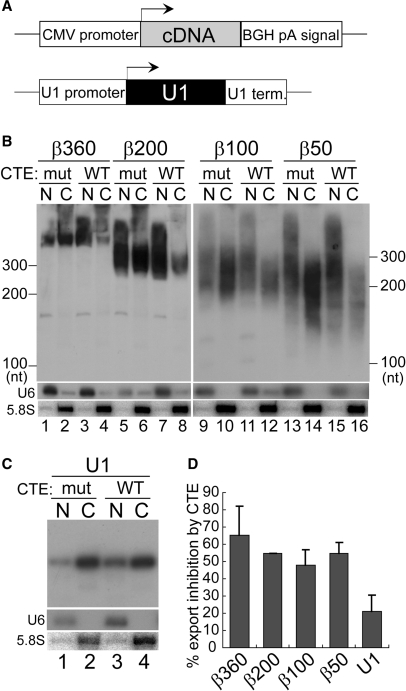
We first cloned the Xenopus laevis U1 snRNA gene including its promoter and terminator, and introduced an XhoI restriction site in place of the Sm-binding site near the middle of the U1 sequence (Figure 1A). The U1 RNA transcript with this modification, called U1ΔSm RNA, is exported efficiently but is defective in re-import into the nucleus (9). When this mutant U1 plasmid was microinjected into the nucleus of Xenopus laevis oocytes, the U1 transcript produced in vivo from the plasmid was quite uniform, forming a discrete single band as judged by northern blotting analysis (Figure 1B, lanes 1, 2; Figure 1D, lanes 1, 2). This is because the U1 terminator, unlike the poly (A) signal, directs cleavage of the transcript at a discreet site without adding additional nucleotides (50). Examination of the nucleocytoplasmic distribution of the U1 transcript revealed that the transcript was mainly cytoplasmic (e.g. Figure 1D, lanes 1,2), indicating that the U1 transcript was exported efficiently. To determine the export pathway of the U1 transcript, two pathway-specific RNA export inhibitors were employed. The first was CTE (constitutive transport element) from type D retroviruses, which specifically binds to TAP-p15 and thereby inhibits mRNA export when a saturating amount is injected (23,51). The second was a dominant-negative mutant of a U snRNA export factor, PHAX that lacks the NES sequence (PHAXΔNES) and thus blocks U snRNA-CRM1 complex formation (22). The nucleocytoplasmic distribution of the U1 transcript was only slightly affected by the co-injected wild-type CTE, and not at all by a non-functional CTE mutant (Figure 1B, compare lanes 1, 2 with 3, 4, see Figure 1C for quantitation). In contrast, when the PHAXΔNES protein was co-injected in place of CTE, export of the U1 transcript was strongly inhibited (Figure 1D, compare lanes 1, 2 with 3, 4, and Figure 1E for quantitation). The distributions of the endogenous U6 and 5.8S RNAs, which are nuclear and cytoplasmic markers, respectively, verified the efficacy of the nuclear-cytoplasmic fractionation (Figure 1B and D; also see subsequent figures). This result confirmed that the U1 transcript produced from the injected plasmid in the nucleus of Xenopus oocytes was clearly exported by the canonical export pathway, the U snRNA export pathway.
Figure 1.
Effect of RNA length on RNA export in the transcription-coupled system (I). (A) Diagram of the DNA constructs. Human β-globin cDNA fragments of various lengths were inserted into the XhoI site of the U1ΔSm gene. (B) The plasmids harboring the U1ΔSm genes with or without the insertion of cDNA fragments of various lengths (50–300 nt) were microinjected into the nucleus of Xenopus oocytes, with 17 ng/oocyte of either wild-type (WT) or mutant (mut) CTE, and the nucleocytoplasmic distribution of the transcripts was analyzed by northern blotting of nuclear (N) and cytoplasmic (C) RNA fractions after 4 h. The distribution of endogenous U6 snRNA (U6) and 5.8S rRNA (5.8S) was also analyzed by northern blotting to verify the efficacy of the nucleocytoplasmic fractionation. (C) Quantitation of RNA export inhibition by CTE WT from the experiments as in B. Average inhibition and standard deviation were calculated (with CTE mut taken as standard) from three independent experiments. (D) Plasmids harboring the U1ΔSm genes with or without the insertion of the cDNA fragments of 50 or 300 nt were microinjected into the nucleus, with 4.1 ng/oocyte of the PHAXΔNES protein (+) or buffer (−), and export of the transcripts was examined as in B, except that RNA was isolated 3 h after injection. (E) Quantitation of RNA export inhibition by PHAXΔNES from the experiments as in D. Average inhibition and standard deviation were calculated (with buffer taken as standard) from three independent experiments.
We previously showed, in microinjection experiments with in vitro transcribed RNAs, that if U1 RNA was progressively elongated by the insertion of cDNA fragments of increasing length, the elongated U1 RNA was progressively converted to utilize the mRNA export pathway (12). If the insert was as short as 50 nt, the elongated U1 RNAs still behaved like a U snRNA during nuclear export, but if the insert was as long as 200–300 nt, the elongated U1 RNAs behaved overall like an mRNA (12). To test if this was also the case with our current RNA export assay system that is coupled to in vivo transcription, the U1ΔSm gene was progressively elongated by the insertion of human β-globin cDNA fragments of various lengths (50–300 nt) into the XhoI site (Figure 1A), and the resultant plasmids carrying these elongated U1 genes were microinjected into the nucleus of Xenopus oocytes for the RNA export assay (Figure 1B–E).
The U1-derived transcripts were overall exported efficiently (Figure 1B–D). The export of relatively short U1-derived transcripts, in which a β-globin cDNA sequence of 50 or 100 nt was inserted, was only weakly affected by CTE, but was strongly inhibited by PHAXΔNES (Figure 1B, lanes 5–12, Figure 1D, lanes 5–8, and 1C and E for quantitation), indicating that these relatively short transcripts utilized mostly the U snRNA pathway. In contrast, the export of relatively long U1-derived transcripts, in which a β-globin cDNA sequence of 200 or 300 nt was inserted, was strongly inhibited by CTE, but relatively weakly inhibited by PHAXΔNES (Figure 1B, lanes 13–20, Figure 1D, lanes 9–12, and 1C and E for quantitation), indicating that these relatively long transcripts utilized mostly the mRNA pathway. When the β-globin cDNA fragments were inserted in the antisense orientation or when cDNA fragments derived from the DHFR gene were employed, similar results were obtained (see Supplementary data). These results were in good agreement with the results obtained from our previous RNA microinjection experiments (7,12), and indicated that RNA length is an important determinant of RNA identity in nuclear export, even in an RNA export assay system that is coupled to in vivo transcription.
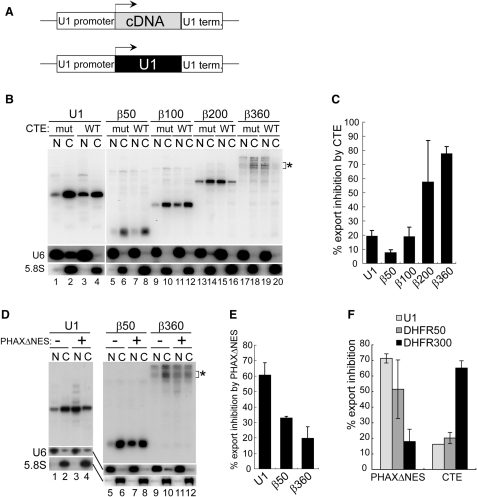
To confirm the importance of RNA length, we next constructed plasmids in which cDNA sequences of various lengths (50–360 nt), without any U1 RNA sequence, were sandwiched between the U1 promoter and the U1 terminator (Figure 2A). The resultant plasmids were microinjected into the nucleus for similar RNA export assays as in Figure 1. Consistent with the results in Figure 1, the export of relatively short transcripts, in which a β-globin cDNA sequence of 50 or 100 nt was used, was only weakly affected by CTE, but relatively strongly inhibited by PHAXΔNES (Figure 2B, lanes 5–12, Figure 2D, lanes 5–8, and 2C and E for quantitation), indicating the overall utilization of the U snRNA pathway. In contrast, the export of relatively long transcripts, in which a β-globin cDNA sequence of 200 or 360 nt was used, was strongly inhibited by CTE, but relatively weakly inhibited by PHAXΔNES (Figure 2B, lanes 13–20, Figure 2D, lanes 9–12, and 2C and E for quantitation), indicating the overall utilization of the mRNA pathway. Since the β360 construct produced multiple bands of transcripts for unknown reason, the bands near the expected size were quantified (Figure 2B and D, asterisks). When the DHFR cDNA fragments were employed or when the β-globin cDNA fragments were inserted in the antisense orientation, similar results were obtained (Figure 2F and Supplementary data).
Figure 2.
Effect of RNA length on RNA export in the transcription-coupled system (II). (A) Diagram of the DNA constructs. Human β-globin cDNA fragments of various lengths were cloned between the promoter and terminator of the U1 gene. The U1ΔSm gene was also used as a control. (B) Export of the transcripts produced from the microinjected plasmids harboring the cDNA fragments of various lengths (50–360 nt) between the promoter and terminator of the U1 gene was analyzed as in Figure 1B. The U1ΔSm gene was also used as a control. The bands indicated by the asterisk correspond to the 360-nt transcript whose 3′-end was properly formed. (C) Quantitation of RNA export inhibition by CTE WT from the experiments like that shown in B. Since the β360 construct produced multiple bands for unknown reason, the bands near the expected size were quantified (B, asterisk). (D) Export of the transcripts produced from the microinjected plasmids harboring the cDNA fragments of 50 or 360 nt between the promoter and terminator of the U1 gene was analyzed as in Figure 1D. The U1ΔSm gene was also used as a control. The bands indicated by the asterisk correspond to the 360-nt transcript whose 3′-end was properly formed. (E) Quantitation of RNA export inhibition by PHAXΔNES from the experiments like that shown in D. Since the β360 construct produced multiple bands, the bands near the expected size were quantified (D, asterisk). (F) Similar experiments to those in C and E except that cDNA fragments from the DHFR gene instead of the β-globin gene were used.
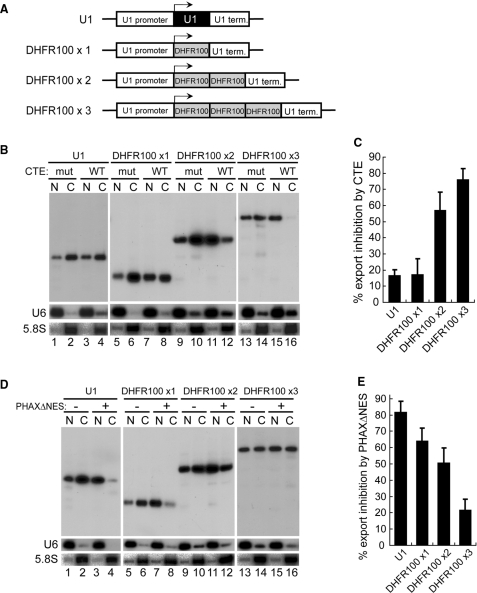
To further confirm that RNA length, independent of RNA sequence, is important, 1–3 copies of a 100-nt DNA fragment from the DHFR gene were cloned between the promoter and terminator of the U1 gene (Figure 3A). These plasmids were subjected to similar RNA export assays. RNA export progressively shifted from U snRNA export to mRNA export as the copy number increased, as judged by the requirement for TAP and PHAX proteins (Figure 3B, lanes 5–16, Figure 3D, lanes 5–16 and 3C, 3E for quantitation). These results strongly suggested that RNA length per se, independent of RNA sequence, is the determinant of the choice of RNA export pathway.
Figure 3.
Effect of RNA length on RNA export in the transcription-coupled system (III). (A) Diagram of the DNA constructs. One to three copies of a 100-nt DNA fragment from the DHFR gene were cloned between the promoter and terminator of the U1 gene. The U1ΔSm gene was also used as a control. (B) Export of the transcripts produced from the microinjected plasmids harboring 1–3 copies of a 100-nt DNA fragment from the DHFR gene between the promoter and terminator of the U1 gene was analyzed as in Figure 1B. The U1ΔSm gene was also used as a control. (C) Quantitation of RNA export inhibition by CTE WT from the experiments like that shown in B. (D) Export of the transcripts produced from the microinjected plasmids as in B was analyzed as in Figure 1D. The U1ΔSm gene was also used as a control. (E) Quantitation of RNA export inhibition by PHAXΔNES from the experiments like that shown in D.
Promoter sequence per se does not have a clear influence on the choice of export pathway of the transcript
It should be pointed out that the effect of RNA length on the RNA export pathway choice was dominant over the putative effect, if any, of the U snRNA-type transcription regulatory sequences (promoter and terminator). If the transcript was long enough, it behaved like an mRNA even though it was produced from the U snRNA-type transcription unit (Figures 1–3). This means that transcription from the U1 promoter or termination by the U1 terminator cannot induce U snRNA export in a dominant manner.
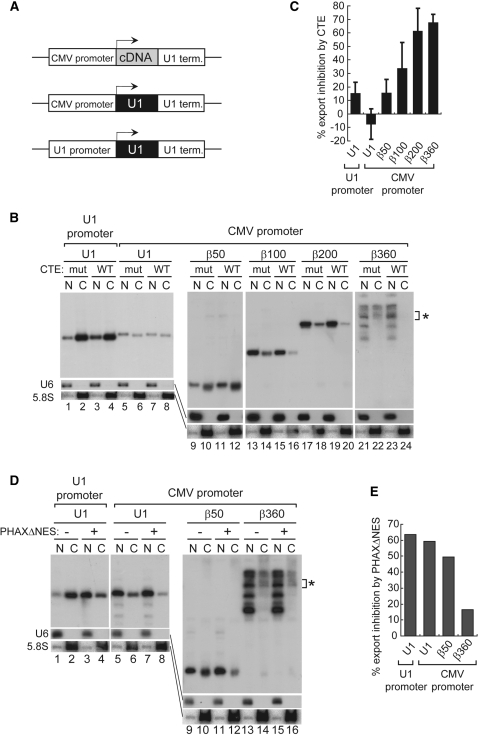
Moreover, when the U1 RNA sequence was inserted between the CMV promoter (an mRNA-type promoter) and the U1 terminator (Figure 4A), and the resultant plasmid was microinjected into the nucleus, the U1 transcript initiated from the CMV promoter and terminated by the U1 terminator was clearly detected by northern blotting (Figure 4B, lanes 5–8), although transcription from mRNA-type promoters tends to read through the U snRNA-type terminators (43,44). The U1 transcript produced from the plasmid behaved like a U snRNA, as judged by the effects of CTE and PHAXΔNES (Figure 4B and D, lanes 5–8, and C and E for quantitation), indicating that the CMV promoter could not convert the U1 transcript to utilize the mRNA export pathway. This was also the case with the plasmid in which the β-globin cDNA sequence of 50 nt was inserted between the CMV promoter and the U1 terminator. The CMV promoter could not convert the β50 transcript to utilize the mRNA export pathway (Figure 4B and D, lanes 9–12, and C and E for quantitation), confirming that the CMV promoter could not induce mRNA export in a dominant manner. It should be pointed out that the effect of RNA length on the choice of RNA export pathway was also clearly seen when the CMV promoter and the U1 terminator were employed (Figure 4B, lanes 9–24 and D, lanes 9–16, and C and E for quantitation), further confirming the importance of RNA length in the export pathway choice. Since the β360 construct produced multiple bands of transcripts for unknown reason, the bands near the expected size were quantified (Figure 4B and D, asterisks). These results taken together strongly suggest that the promoter sequence per se has no significant influence on the choice of export pathway of the transcript. They also suggest that the terminators of the U snRNA genes have no clear influence on the choice of export pathway of the transcript.
Figure 4.
Effect of transcription from the CMV promoter on RNA export. (A) Diagram of the DNA constructs. Human β-globin cDNA fragments of various lengths or the U1ΔSm sequence were cloned between the CMV promoter and U1 terminator. The U1ΔSm gene was also used as a control. (B) Export of the transcripts produced from the microinjected plasmids harboring the cDNA fragments of various lengths (50–360 nt) or the U1ΔSm sequence between the CMV promoter and the U1 terminator was analyzed as in Figure 1B. The U1ΔSm gene was also used as a control. The bands indicated by the asterisk correspond to the 360 nt transcript whose 3′-end was properly formed. (C) Quantitation of RNA export inhibition by CTE WT from experiments like those shown in B. Since the β360 construct produced multiple bands for unknown reason, the bands near the expected size were quantified (B, asterisk). (D) Export of the transcripts produced from the microinjected plasmids harboring the cDNA fragments of 50 or 360 nt or the U1ΔSm sequence between the CMV promoter and the U1 terminator was analyzed as in Figure 1D. The U1ΔSm gene was also used as a control. The bands shown by the asterisk correspond to the 360 nt transcript whose 3′-end was properly formed. (E) Quantitation of RNA export inhibition by PHAXΔNES from the experiment shown in D. Since the β360 construct produced multiple bands, the bands near the expected size were quantified (D, asterisk).
Presence of poly (A) tail of appropriate length can induce mRNA export in a dominant manner
Surprising results were obtained from experiments in which transcription of β-globin cDNA sequences of various lengths was initiated from the CMV promoter and terminated by the BGH poly (A) signal (Figure 5A). Since transcripts that were not terminated at the BGH poly (A) signal were present at significant levels (data not shown), only properly polyadenylated transcripts were detected by northern blotting using a specific probe (see ‘Materials and Methods’ section for details). When the polyadenylated transcripts were specifically visualized, the bands of the transcripts were smeared due to the variation of the poly (A) tail length. Interestingly, the transcripts with a very long poly (A) tail (>250 nt) appeared to be retained in the nucleus (Figure 5B). This is most evident in lanes 13 and 14 of Figure 5B, in the area above the 300 nt marker. In contrast, the transcripts with ‘proper’ lengths of poly (A) tail were exported efficiently (Figure 5B, lanes 1, 2, 5, 6, 9, 10, 13, 14). More importantly, CTE strongly inhibited the export of polyadenylated RNAs overall, regardless of the lengths of the cDNA sequences, while the export of the control U1 transcript was hardly affected by CTE (Figure 5B, C and D for quantitation), suggesting that these polyadenylated transcripts utilized mostly the mRNA export pathway. Even the transcript with the shortest cDNA sequence (β50) behaved overall like an mRNA. This unexpected phenomenon appeared to be due to the presence of either the CMV promoter or the poly (A) signal, and given the results in Figure 4 showing that the CMV promoter had no significant influence, it was likely that the poly (A) signal was responsible.
Figure 5.
Effect of transcription from an mRNA transcription unit on RNA export. (A) Diagram of the DNA constructs. Human β-globin cDNA fragments of various lengths were cloned between the CMV promoter and the poly (A) signal from the bovine growth hormone (BGH) gene. The U1ΔSm gene was also used as a control. (B) Export of the transcripts produced from the microinjected plasmids harboring the cDNA fragments of various lengths (50–360 nt) between the CMV promoter and the BGH poly (A) signal was analyzed as in Figure 1B. Only the polyadenylated transcripts were visualized (see ‘Materials and Methods’ section). (C) The transcript from the plasmid harboring the U1ΔSm gene was analyzed as a control as in Figure 1B. (D) Quantitation of B and C.
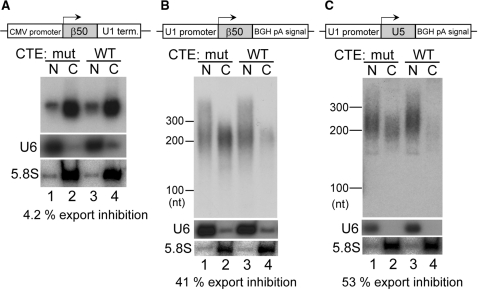
To confirm that the poly (A) signal rather than the CMV promoter was responsible for this phenomenon, the behavior of the transcripts from two additional constructs was examined. The first construct had the CMV promoter and the U1 terminator (Figure 6A, same as the β50 construct in Figure 4), and the second the U1 promoter and the poly (A) signal (Figure 6B). Since transcription from the U1 promoter tends to ignore the poly (A) signal (42), only properly polyadenylated transcripts from the second construct were detected by northern blotting using a specific probe (see ‘Materials and Methods’ section for details). The transcript from the first construct behaved like a U snRNA (4.2% export inhibition by CTE, Figure 6A, also see Figure 3C), whereas the transcript from the second construct behaved like an mRNA (41% export inhibition by CTE, Figure 6B). These results clearly indicated that it was the poly (A) signal that was responsible. This conclusion was further confirmed using the third construct (Figure 6C). If the transcription of U5 snRNA was terminated by the poly (A) signal, the U5 transcript behaved like an mRNA (53% export inhibition by CTE, Figure 6C), indicating that even a U snRNA could be committed to the mRNA export pathway. These results indicated that either the poly (A) signal-mediated 3′-end processing or the presence of a poly (A) tail induced mRNA export in the transcripts that should otherwise behave like a U snRNA.
Figure 6.
Effect of 3′-end processing signals on RNA export. Export of the transcripts produced from the plasmids harboring the β-globin 50-nt cDNA fragment between the CMV promoter and the U1 terminator (panel A) or between the U1 promoter and the BGH poly (A) signal (panel B), or from the plasmid harboring the U5 snRNA sequence between the U1 promoter and the BGH poly (A) signal (panel C) was analyzed as in Figure 5.
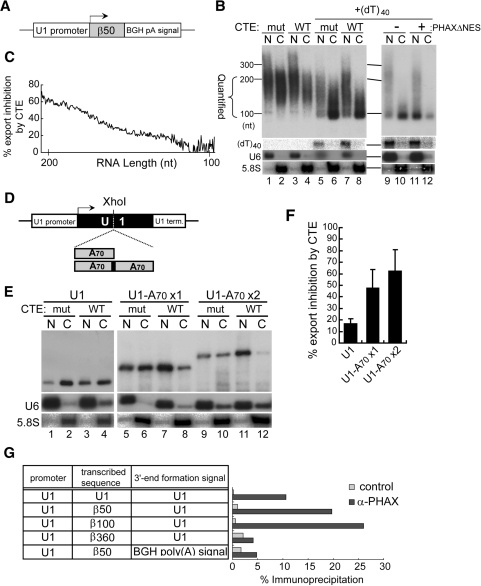
To distinguish between the above two possibilities, the inhibitory effect of CTE on the export of the polyadenylated β50 transcript was calculated and plotted as a function of RNA length (Figure 7B, lanes 1–4 and 7C). The inhibitory effect of CTE, i.e. the degree of commitment to mRNA export, was clearly positively correlated with the RNA length, and hence the poly (A) tail length (Figure 7C). The longer the poly (A) tail within the measured range, the more strongly the RNA was committed to mRNA export, strongly suggesting that the presence of the poly (A) tail rather than the cleavage/polyadenylation per se induced mRNA export. Again the transcripts with very long poly (A) tails above the tested range were retained in the nucleus (above the 300 nt marker, Figure 7B, lanes 1, 2).
Figure 7.
Effect of poly (A) tail length on RNA export. (A) Diagram of the DNA construct as in Figure 6B is shown. (B) Effect of CTE and PHAXΔNES on export of the transcript from the plasmid was analyzed. For lanes 5–12, 0.8 pmol/oocyte of (dT)40 was coinjected with the plasmid. RNA length markers are shown on the left. The range of RNA length quantified in C is marked. (C) The RNA export inhibition by CTE observed in B, lanes 1–4, was calculated and plotted as a function of RNA length in the marked range in B. (D) One or two copies of a DNA fragment containing 70-nt poly (A) sequence were inserted into the XhoI site of the U1ΔSm gene. (E) Effect of CTE on export of the transcripts from the plasmids was analyzed. (F) Quantitation of RNA export inhibition by CTE from the experiments like that shown in E. (G) Various DNA constructs indicated in the table were microinjected into the nucleus of Xenopus oocytes. The nuclear fraction was prepared after 3 h, and immunoprecipitation was performed with either anti-PHAX antibody (α-PHAX) or anti-mouse IgG antibody (control). The co-precipitated RNA was recovered and analyzed by real-time RT-PCR. The efficiency of immunoprecipitation by each antibody is shown on the right.
To further confirm that the presence of a poly (A) tail of an appropriate length was responsible for inducing mRNA export, an attempt was made to artificially shorten the poly (A) tail of the transcript by microinjecting (dT)40 single-stranded DNA oligonucleotide into the nucleus, since the (dT)40 should hybridize to poly (A) tails and the endogenous RNaseH activity should then degrade the poly (A) tails. As anticipated, the poly (A) tail length of the transcript was significantly shortened (Figure 7B, lanes 5–12). A large fraction of the transcript was seen around the 100 nt marker (Figure 7B, lanes 5–12), indicating that this fraction had very short poly (A) tail. This poly (A) shortening appeared to have occurred in the nucleus, since the injected (dT)40 stayed in the nucleus (Figure 7B) and since the vast majority of RNase H activity is nuclear (52). Importantly, CTE hardly inhibited the export of this fraction of the transcript (lanes 5–8) while PHAXΔNES strongly inhibited its export (lanes 9–12), indicating that this fraction of the transcript utilized the U snRNA pathway. These results strongly suggest that the presence of a poly (A) tail of appropriate length, rather than the cleavage/polyadenylation per se, is important for inducing mRNA export.
To examine whether the presence of poly (A) sequence functions in inducing mRNA export independent of its position, one or two copies of a DNA fragment containing 70-nt poly (A) sequence were inserted into the XhoI site of the U1ΔSm gene. These plasmids were subjected to similar RNA export assays. RNA export progressively shifted from U snRNA export to mRNA export as the poly (A) length increased, as judged by the requirement for TAP protein (Figure 7E, lanes 5–12 and 7F for quantitation). These results clearly indicated that the presence of poly (A) sequence of appropriate length can induce mRNA export independent of its position and without the polyadenylation signal.
To confirm that the changes in RNA export pathway were reflected by the changes in export RNP composition, immunoprecipitation with an anti-PHAX antibody (7,12) was carried out to analyze the association of PHAX with the transcripts (Figure 7G). When the transcripts were produced from the U1 promoter and the U1 terminator, relatively short transcripts (produced from the first three constructs, Figure 7G), which behaved overall like a U snRNA, were efficiently bound by PHAX, whereas the β360 transcript (produced from the fourth construct), which behaved like an mRNA, was poorly bound by PHAX. Moreover, the polyadenylated β50 transcript (produced from the last construct), which behaved overall like an mRNA, was also poorly bound by PHAX. We suspected that the association of Aly/REF with these transcripts would be the opposite to that of PHAX, since their binding to RNA is mutually exclusive (7,12). However, the Aly/REF antibodies could not immunoprecipitate any transcripts above the background level in the current DNA microinjection system, although the same antibodies worked well in the RNA microinjection system (7,12). The reason for this is not clear, but it may have been because the amount of transcripts produced from the microinjected plasmids was much smaller than the amount of RNA used in the RNA microinjection experiments. In any case, these results with the anti-PHAX antibody were in good agreement with the changes of RNA export pathway in response to changes of the RNA length and the presence of the poly (A) tail.
DISCUSSION
Importance of RNA length in determining mRNA identity for nuclear export
In this report, we first showed unequivocally that RNA length is an important determinant of the choice of RNA export pathway, even in a transcription-coupled RNA export assay system. Regardless of whether the transcription was initiated from an mRNA-type promoter or a U snRNA-type promoter, relatively short transcripts preferentially recruited a U snRNA export factor, PHAX, and therefore they were committed overall to the U snRNA export pathway, whereas relatively long transcripts did not associate with PHAX and they were committed overall to the mRNA export pathway (Figures 1–4). Neither the CMV nor the U1 promoter could override this RNA length effect (Figures 1–4), suggesting that transcription does not affect the recruitment of export factors onto the RNA in our assay system. This is consistent with the fact that splicing rather than transcription has a role in loading export factors onto RNA in vertebrates (13,31,33–35). Accordingly, our initial hypothesis that transcription from the corresponding promoter may influence the RNA identity was disproved.
It was surprising to find that RNA length had a similar effect on the choice of RNA export pathways in both the transcription-coupled system and the RNA microinjection system, since the transcript would gradually emerge from the plasmids in the transcription-coupled system, whereas an RNA with a defined length is introduced into the nucleus in the RNA microinjection system. These results indicate that the features of the transcribed RNAs, rather than the ways they are transcribed, are important for determining the RNA identity for nuclear export.
However, these results do not necessarily mean that the RNA identity is determined only after the transcription is completed. If the RNA contains an intron, it may be spliced before the transcription termination (45,46). In that case, the RNA may immediately qualify as an mRNA, even before the transcription termination. Also, in the case of intronless mRNAs, as soon as a relatively unstructured RNA region of few hundreds nucleotides has been generated during transcription, the RNA may immediately qualify as an mRNA. Since the binding of an mRNA export factor, Aly/REF, and a U snRNA export factor, PHAX, to a given RNA is mutually exclusive (7,12), it will be of interest to examine whether PHAX is first recruited to the cap structure of mRNAs at the very early stage of transcription but is released from the RNA as soon as the RNA qualifies as an mRNA.
Role of the poly (A) tail in mRNA nuclear export
Transcription termination by the U1 snRNA gene terminator had no significant effect on the choice of RNA export pathway (Figures 1 and 2). In contrast, transcription termination by the poly (A) signal had a significant dominant effect to induce mRNA export of the transcript. What is responsible for inducing mRNA export is not the cleavage/polyadenylation per se but the presence of a poly (A) tail of an appropriate length (Figures 5–7). When the short β50 transcript was polyadenylated in vivo, the transcript lost affinity for PHAX and utilized mostly the mRNA export pathway (Figure 7B, C and G). Moreover, the longer the poly (A) tail within a certain range, the more the transcript behaved like an mRNA (Figure 7B and C). Furthermore, the poly (A) sequence could induce mRNA export independent of its position (Figure 7D–F). This may mean that the poly (A) sequence simply contributes to increasing the RNA length. In fact, the polyadenylated β50 transcript (total length, ∼200 nt) behaved very similarly to the β200 transcript without a poly (A) tail (total length, ∼200 nt) in nuclear export (60% export inhibition by CTE, Figure 2). Hence it is not possible to say that the effect of poly (A) sequence is stronger than the RNA length effect.
Although the poly (A) sequence can induce mRNA export, when the poly (A) tail was very long (>250 nt), the transcript was not transported to the cytoplasm but rather retained in the nucleus (Figures 5–7). Similar observations were made in previous studies using yeast mutant strains in which hyper-adenylated mRNAs are retained at the transcription gene loci via the action of the nuclear exosome. These observations indicate the existence of a checkpoint mechanism that controls the quality of mRNAs by blocking or allowing their nuclear export (53–55). Our result suggests that such a mechanism is evolutionarily conserved in vertebrates.
Alternatively, the poly (A) tail may not simply contribute to increase the RNA length, since the poly (A) tail is thought to be covered by poly (A) binding protein(s) (56,57), and therefore mRNA export factors may not be able to directly bind to the poly (A) tail. It is possible that a poly (A) tail of an appropriate length may function as a platform to recruit mRNA export factors. In this latter scenario, poly (A) binding protein(s) should function in recruiting mRNA export factors. The importance of the poly (A) tail and its binding proteins (Pab1p and Nab2p) has been already suggested in yeast (reviewed in 56,57). Although the nuclear functions of the yeast poly (A) binding proteins appear to be related to poly (A) tail length control and therefore to the above-mentioned exosome-mediated quality control mechanism, rather than to mRNA export per se (58–62), some studies suggested more direct role of Nab2p in mRNA export (63–65). The vertebrate homolog of yeast Nab2p, however, has not been identified to our knowledge. In any case, our finding clearly demonstrates that the poly (A) tail is directly linked to the mRNA export pathway in vertebrates, and it remains to be investigated whether the poly (A) tail simply contributes to increasing the RNA length or plays more active role in mRNA export.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Makoto Kitabatake and Mr Ichiro Taniguchi for suggestions and criticisms of the manuscript. This work was supported by Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (JST), Tokyo 102-0075, Japan and grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding to pay the Open Access publication charges for this article was provided by CREST, JST, Tokyo, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Nuclear RNA export. J. Cell. Sci. 2003;116:587–597. doi: 10.1242/jcs.00268. [DOI] [PubMed] [Google Scholar]
- 3.Komeili A, O'Shea EK. New perspectives on nuclear transport. Ann. Rev. Genet. 2001;35:341–364. doi: 10.1146/annurev.genet.35.102401.090720. [DOI] [PubMed] [Google Scholar]
- 4.Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML. Tap and NXT promote translation of unspliced mRNA. Genes Development. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuersten S, Segal SP, Verheyden J, LaMartina SM, Goodwin EB. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell. 2004;14:599–610. doi: 10.1016/j.molcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Development. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno M, Segref A, Kuersten S, Mattaj IW. Identity elements used in export of mRNAs. Mol. Cell. 2002;9:659–671. doi: 10.1016/s1097-2765(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 8.Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl Acad. Sci. USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 10.Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell. Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 12.Masuyama K, Taniguchi I, Kataoka N, Ohno M. RNA length defines RNA export pathway. Genes Development. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 14.Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 15.Mattaj IW. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Berlin. pp100–114: Springer-Verlag; 1988. U snRNP assembly and transport. [Google Scholar]
- 16.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 17.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 19.Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 20.Kataoka N, Ohno M, Moda I, Shimura Y. Identification of the factors that interact with NCBP, an 80 kDa nuclear cap binding protein. Nucleic Acids Res. 1995;23:3638–3641. doi: 10.1093/nar/23.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno M, Kataoka N, Shimura Y. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 1990;18:6989–6995. doi: 10.1093/nar/18.23.6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- 23.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 24.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl Acad. Sci. USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA (New York, NY) 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- 32.Masuyama K, Taniguchi I, Kataoka N, Ohno M. SR proteins preferentially associate with mRNAs in the nucleus and facilitate their export to the cytoplasm. Genes Cells. 2004;9:959–965. doi: 10.1111/j.1365-2443.2004.00774.x. [DOI] [PubMed] [Google Scholar]
- 33.Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Development. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl Acad. Sci. USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 38.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 39.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell. Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell. Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Development. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 42.Dahlberg JE, Schenborn ET. The human U1 snRNA promoter and enhancer do not direct synthesis of messenger RNA. Nucleic Acids Res. 1988;16:5827–5840. doi: 10.1093/nar/16.13.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Vegvar HE, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- 45.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 46.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 47.Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 48.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 50.Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 52.Cazenave C, Frank P, Toulme JJ, Busen W. Characterization and subcellular localization of ribonuclease H activities from Xenopus laevis oocytes. J. Biol. Chem. 1994;269:25185–25192. [PubMed] [Google Scholar]
- 53.Jensen TH, Dower K, Libri D, Rosbash M. Early formation of mRNP: license for export or quality control? Mol. Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 54.Saguez C, Olesen JR, Jensen TH. Formation of export-competent mRNP: escaping nuclear destruction. Curr. Opin. Cell. Biol. 2005;17:287–293. doi: 10.1016/j.ceb.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Sommer P, Nehrbass U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr. Opin. Cell. Biol. 2005;17:294–301. doi: 10.1016/j.ceb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochimica et Biophysica Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 58.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA (New York, NY) 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dower K, Kuperwasser N, Merrikh H, Rosbash M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA (New York, NY) 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunn EF, Hammell CM, Hodge CA, Cole CN. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes Development. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thakurta AG, Ho Yoon J, Dhar R. Schizosaccharomyces pombe spPABP, a homologue of Saccharomyces cerevisiae Pab1p, is a non-essential, shuttling protein that facilitates mRNA export. Yeast (Chichester, England) 2002;19:803–810. doi: 10.1002/yea.876. [DOI] [PubMed] [Google Scholar]
- 62.Chekanova JA, Belostotsky DA. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA (New York, NY) 2003;9:1476–1490. doi: 10.1261/rna.5128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suntharalingam M, Alcazar-Roman AR, Wente SR. Nuclear export of the yeast mRNA-binding protein Nab2 is linked to a direct interaction with Gfd1 and to Gle1 function. J. Biol. Chem. 2004;279:35384–35391. doi: 10.1074/jbc.M402044200. [DOI] [PubMed] [Google Scholar]
- 65.Green DM, Johnson CP, Hagan H, Corbett AH. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl Acad. Sci. USA. 2003;100:1010–1015. doi: 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.