Abstract
Family B DNA polymerases from archaea such as Pyrococcus furiosus, which live at temperatures ∼100°C, specifically recognize uracil in DNA templates and stall replication in response to this base. Here it is demonstrated that interaction with uracil is not restricted to hyperthermophilic archaea and that the polymerase from mesophilic Methanosarcina acetivorans shows identical behaviour. The family B DNA polymerases replicate the genomes of archaea, one of the three fundamental domains of life. This publication further shows that the DNA replicating polymerases from the other two domains, bacteria (polymerase III) and eukaryotes (polymerases δ and ε for nuclear DNA and polymerase γ for mitochondrial) are also unable to recognize uracil. Uracil occurs in DNA as a result of deamination of cytosine, either in G:C base-pairs or, more rapidly, in single stranded regions produced, for example, during replication. The resulting G:U mis-pairs/single stranded uracils are promutagenic and, unless repaired, give rise to G:C to A:T transitions in 50% of the progeny. The confinement of uracil recognition to polymerases of the archaeal domain is discussed in terms of the DNA repair pathways necessary for the elimination of uracil.
INTRODUCTION
Family B DNA polymerases from the archaea, for example the enzyme from Pyrococcus furiosus (Pfu-Pol) commonly used in the PCR, strongly bind to template-strand uracil and stall polymerization in response to this base (1). The ability to recognize uracil arises from a specialized binding pocket in the amino-terminal domain of the polymerase (2), which interacts tightly with uracil in single-stranded DNA (3). Deamination of cytosine converts G:C base-pairs to pro-mutagenic G:U mismatches, replication of which results in 50% of the progeny containing a G:C→A:T transition mutation (4). Repair of G:U mis-pairs is usually initiated by uracil–DNA–glycosylases (UDGases), enzymes that remove uracil from DNA by glycosidic bond hydrolysis. In double stranded DNA, UDGase action initiates a base excision repair pathway that, ultimately, restores the G:C base-pair (5–7). Polymerase mediated uracil-induced stalling of replication is probably the first step of an additional DNA repair pathway that prevents copying of the G:U mismatch and permanent fixation of a transition mutation; replication might therefore offer the final opportunity for mutation avoidance. The pathways that follow stalling probably involve recombinational daughter-strand ‘gap’ processes, often used to repair and restart stalled replication forks (8,9). Thus far uracil recognition has only been observed for hyperthermophilic archaeal DNA polymerases and may be an adaptation to high temperatures, expected to promote cytosine deamination (10), at which many archaea live. In this article a mesophilic archaeal Family-B DNA polymerase has been investigated to elucidate whether uracil recognition is strictly limited to hyperthermophiles. In addition archaea constitute one of the three fundamental domains of life (11,12), the other two being bacteria and eukarya. This article also investigates whether the polymerases responsible for the replication of DNA in these two domains are able to recognize uracil.
MATERIALS AND METHODS
Polymerase purification
The purification of the family B DNA polymerase from P. furiosus (13), Saccharomyces cerevisiae DNA polymerases ε (14) and δ (15) and the family B DNA polymerase from Methanosarcina acetivorans (16) have been described. The Escherichia coli PolIII holoenzyme used in this publication was PolIII*, which contains the α, ε, θ, χ, ψ, δ, δ’, γ and τ sub-units but lacks the β clamp (17), prepared as described (18). The two sub-unit human mitochondrial DNA polymerase γ was purified from human embryonic kidney cells expressing the SV40 large-T antigen (HEK293T cells) (19) containing two plasmids that encode the catalytic and accessory sub-units of the polymerase (20). The plasmids, pcDNA3.1(-)/Myc-His A (geneticin antibiotic marker for the catalytic sub-unit, hygromycin for the accessory) (Invitrogen) place the coding sequences in-frame with a Myc-His tag (19). Growth and harvesting of the HEK293T overproducing cells (growth media containing 2 mg/ml geneticin and 100 μl/ml hygromycin) has been described (19). Cells were suspended in ice cold (all subsequent purification steps were performed at 4°C) 20 mM sodium phosphate pH 7.4, 10 mM NaCl, 20 mM imidazole, 250 mM sucrose, complete protease inhibitor (Roche), lysed using a French press and clarified by centrifugation at 1000 g for 10 min. DNase I (10 μg/ml final concentration and Triton X-100 (1% v/v final concentration) were added to the lysate and, after incubation for 30 min, the solution was centrifuged at 1000 g for 30 min. The crude cell extract was applied to a 1 ml His-Trap column (Amersham-Pharmacia), which was washed with 20 mM sodium phosphate, pH 7.4, 10 mM NaCl, 20 mM imidazole, 0.1% (v/v) Triton-X100. The polymerase was eluted using a gradient to 500 mM imidazole (total gradient volume 90 ml); appropriate fractions (monitored by SDS-PAGE) were pooled and dialysed against 10 mM Tris, pH 7.5, 10 mM NaCl, 0.1% (v/v) Triton X-100. The volume was reduced to one-fifth of the original (VivaSpin column, 10 kDa cut-off; Sartorius, Epsom, UK) and the concentrate applied to a 1 ml Hep-Trap column (Amersham-Pharmacia). Following washing the polymerase was eluted using a gradient to 2 M NaCl (gradient volume 60 ml); appropriate fractions (monitored by SDS-PAGE) were pooled and dialysed against 100 mM Tris, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 5 mM dithiothreitol, 30% (v/v) glycerol. Bovine serum albumin was added (0.5 mg/ml final concentration) and the samples rapidly frozen using liquid nitrogen and stored at −80°C. Saccharomyces cerevisiae PCNA, RPA and RFC were purified as described (21,22).
Protein-sequence alignment
Sequences were aligned using protein blast (http://www.ncbi.nlm.nih.gov/BLAST/).
Primer-template extension assays
A 16-mer primer (5′-GCAGTCCTAGACGCAG-3′) and 32-mer template (5′-CATCCG(T/U)GG(T/U)GCTATCCTGCGTCTAGGACTGC-3′) were used; annealing produces a duplex region of 16 base pairs with a single stranded template-strand extension of 16 bases. A single uracil is located at one of the two positions indicated (thymine for controls), which places uracil either seven or ten bases ahead of the primer-template junction. Extension assays (20 μl final volume) contained 2.5 nM primer-template (primer labelled at the 5′-terminus with 32P), 500 μM of each dNTP, polymerase specific buffer (archaeal and E. coli polymerases; 20 mM Tris–HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 5 mM MgCl2, 0.1% Triton X-100, 0.1 mg/ml bovine serum albumin: eukaryotic polymerases; 40 mM Tris–HCl (pH 7.8), 75 mM NaCl, 8 mM magnesium acetate, 1 mM dithiothreitol, 0.2 mg/ml bovine serum albumin) to which was added polymerase (1 μM final concentration) to initiate the reaction. Polymerizations were carried out at 30°C for mesophilic DNA polymerases, 72°C for thermophilic DNA polymerases for times of between 0.5 and 30 min. Reactions were stopped by addition of a 2-fold excess of EDTA (over Mg2+) and 40% formamide and products detected using denaturing polyacrylamide (15%) gel electrophoresis followed by phosphorimaging. Some reactions with yeast Pol δ and ε were carried out in the presence of additional replisome components. Here the primer-template was pre-incubated with 1 μM PCNA, 5 μM RFC 1 μM RPA, 1 μM UDGase inhibitor (UGI), 0.5 μM ATP, 5 μM dNTPs and 8 mM MgCl2 (other buffer components as above) for 30 s. Reaction was initiated by adding polymerase at levels that varied between 0.04 and 1 μM.
Binding of Mac-Pol to uracil-containing DNA
The affinity of Mac-Pol for uracil in DNA was determined using fluorescence anisotropy with hexachlorofluorescein (hex)-labelled oligodeoxynucleotides (3,23). Assays were carried out in 1 ml volumes containing 6 nM Hex-GCCCGCGGGAUATCGGCCCTTA in 10 mM Hepes (pH 7.5), 100 mM NaCl and 1 mM EDTA and Mac-Pol (0–160 nM) was added until saturation was observed. Data fitting to give KD has been described (3,23).
RESULTS
Amino acid sequence alignment
The N-terminal domain of the family B DNA polymerase from P. furiosus (Pfu-Pol) contains a pocket responsible for specific binding of uracil (2), important amino acids of which are shown in Figure 1. Y7 and P36/Y37 constitute a lid and base respectively and amino acids 90–97 and 111–116 form the two sides of the pocket. R119 and D123 are involved in a complex hydrogen bonding network, interlinking many of the pocket amino acids including Y37. The peptide backbone of Y37 makes hydrogen bonds to uracil and, hence, Y37 is a particularly important residue (2). These key uracil-recognizing amino acids are retained with Mac-Pol, a family B DNA polymerase from the mesophilic archaeon M. acetivorans (24), as shown in Figure 1. In general the amino acids shown in Figure 1 are very well conserved among all euryarchaeal family B DNA polymerases with no obvious distinction between thermophilic and mesophilic representatives (Supplementary Material). The eukaryotic replicative polymerases ε and δ are members of the family-B DNA polymerase family and show amino acid sequence homology with archaeal enzymes. Elements of the uracil-binding pocket are apparent for both yeast polymerases (and the relevant enzymes from a number of eukaryotes, Supplementary Information), as shown by alignment with Pfu-Pol in Figure 1. However, not all the key amino acids are retained and the homology is clearly less striking than between Pfu-Pol and Mac-Pol. There is no indication of a residue that could correspond to Y7, although a single aromatic amino acid, serving as a pocket lid, would be difficult to identify from sequence alignments as it might arise from a different region of the eukaryotic polymerase. The PY pocket base is perfectly conserved with Pol ε and reasonably retained, especially the Y, with Pol δ. Similarly segments corresponding to the pocket walls (amino acids 90–97 and 111–116 in Pfu-Pol) are apparent for both Pols δ and ε and particularly noticeable is the near complete conservation of the equivalents of R119 and D123, both amino acid identity and spacing being retained. With Pfu-Pol V93 is a key amino acid for uracil recognition, mutation to V93Q abolishing interaction (2). With eukaryotic polymerases this position is invariably occupied with an aliphatic hydrophobic amino acid.
Figure 1.
The amino acids that form the uracil-binding pocket of Pfu-Pol (2) are shown (numbers above refer to the positions of amino acids in the Pfu-Pol sequence). An alignment is shown for the corresponding amino acids in Mac-Pol and yeast Pols δ and ε. Homology is excellent between Pfu-Poland Mac-Pol and partial between Pfu-Pol and the two yeast enzymes. A more comprehensive table showing more species is given in the Supplementary Material.
Escherichia coli PolIII is a member of the Pol C family (17) and amino acid homology between the α sub-unit (the polypeptide that possess the polymerase activity) and Pol B family members is limited to key regions that contribute to the active sites (polymerase and proof-reading exonuclease) and are involved in dNTP recognition (25,26). The α sub-unit of PolIII does not contain a sequence with any similarity to the uracil-recognition region of archaeal family B polymerases and crystal structures of two representatives (27,28) show no evidence for the presence of a uracil-binding pocket. The polymerase sub-unit of mitochondrial Pol γ belongs to the A family (20), again polymerases with only limited homology to the B group (25,26) and no hint of sequence elements matching the uracil-binding region of Pfu-Pol. No crystal structures are yet available for Pol γ, but other family A members of known structure e.g. E. coli Klenow fragment (29) and Taq-Pol (30) show no semblance of a uracil-binding pocket. Thus amino acid alignments suggest that the mesophilic Mac-Pol should recognize uracil, eukaryotic Pols δ and ε may interact with the base and provide no evidence for this function with bacterial PolIII and mitochondrial Pol γ.
Primer-template extension assays
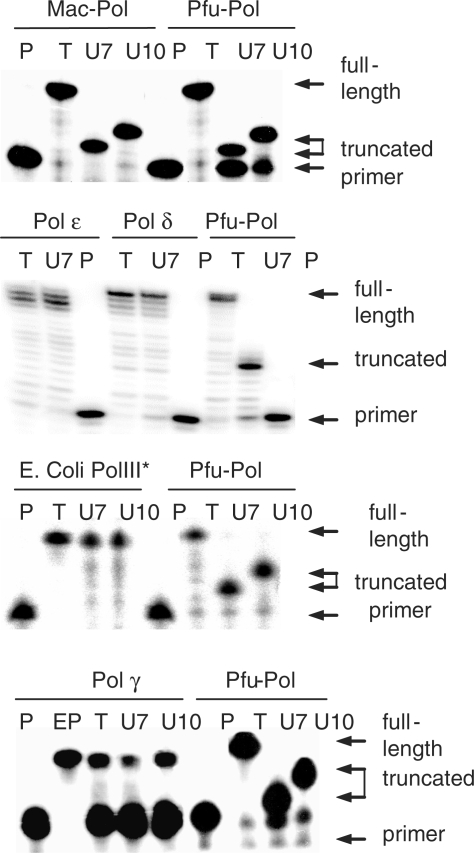
Hyperthermophilic archaeal DNA polymerases stall replication on encountering uracil, most easily assayed using template strands with a single uracil in defined positions (1–3). This assay has been extended to the study of a number of other replicating polymerases and Figure 2 shows that primer extension catalysed by Mac-Pol is halted by the presence of uracil. Figure 2 also shows data obtained with Pfu-Pol; it is apparent that the stalling position is the same for both the hyperthermophilic and the mesophilic polymerase i.e. four bases prior to encounter of uracil. In contrast, Figure 2 demonstrates that the replicating polymerases from yeast, S. cerevisiae DNA polymerases ε (Sce-Pol ε) and δ (Sce-Pol δ) are able to replicate past uracil, giving full length rather than truncated products. The results shown in Figure 2, for the two yeast polymerases, were obtained using an excess of PCNA, RFC and RPA, replisome components that increase polymerase processivity and accuracy (15,31,32). However, an additional experiment using Sce-Pol δ without added replisome components also resulted in read through of uracil, giving identical results to those shown in Figure 2 (data not shown). Thus PCNA, RFC and RPA are without influence in respect to uracil recognition. Yeast Pol δ unusually contains NY at positions 36/37, other eukaryotic Pol δs have PY or HY here (supplementary). However, the yeast enzyme is not an unusual outlier as identical results (not shown) were seen with human Pol δ. In Figure 2 the replicating DNA polymerase from E. coli (E. coli Pol III*) has been tested, as with the eukaryotic enzymes the presence of uracil did not result in cessation of replication and complete read through was observed. Finally Figure 2 evaluates human Pol γ, the two sub-unit enzyme responsible for mitochondrial DNA replication (20). This polymerase has relatively low activity, only extending a small fraction of the primer; nevertheless, it is apparent that stalling of replication in response to template strand uracil does not take place.
Figure 2.
Primer-template extension assays. In all cases the following primer-template was used: GCAGTCCTAGACGCAG CGTCAGGATCTGCGTCCTATCG(T/U)GG(T/U)GCCTAC. The templates contain a single uracil (thymine in controls) either 7 or 10 bases ahead of the primer-template junction. The polymerases under investigation are shown above each panel. Individual gel lanes are labelled: P (primer); polymerase not added, serves as marker for migration of unextended 16-mer primer. EP (extended primer, only used with Pol γ), chemically synthesized 32-mer corresponding to fully extended primer, serves as marker for full extension. T (thymine), control template lacking uracil. U7/U10; templates containing uracil either 7 or 10 bases ahead of primer-template junction. The archaeal polymerases Mac-Pol and Pfu-Pol fully extend the primer when the template lacks uracil. In contrast, the presence of uracil in the template strand leads to truncated products due to uracil-induced stalling of polymerization. All other polymerase (yeast Pols δ and ε, E. coli PolIII* and mitochondrial Pol γ) fully extend the primer regardless of whether uracil is present or not in the template.
Binding of mesophilic archaeal polymerase to uracil-containing DNA
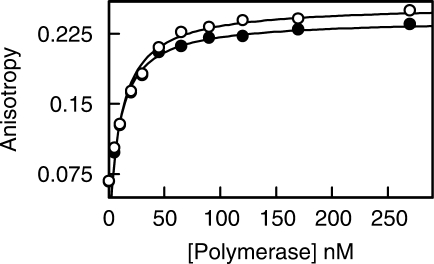
Primer-template extension assays are qualitative in nature, more accurate data can be obtained using fluorescence anisotropy to measure the affinity of the polymerase for uracil-containing DNA (3,24). Performing such an experiment with Mac-Pol (Figure 3) gave a KD of 9.7 nM for binding to a single-stranded 22-mer containing a centrally located uracil. As shown in Figure 3 an identical, within the error limits of the assay, KD of 8.5 nM was seen with Pfu-Pol (previously we also obtained a value of 8 nM (2,3) with Pfu-Pol). Thus the binding affinity of thermophilic and mesophilic archaeal polymerase for uracil appears to be the same.
Figure 3.
Binding titration for Pfu-Pol (black circles) and Mac-Pol (white circles). The polymerases were added to Hex-GCCCGCGGGAUATCGGCCCTTA (6 nM) and the fluorescence anisotropy measured (the same number of measurements were carried out for both polymerases; in some cases the white circles obscure the black). The titration was carried out three times to give a KD of 8.5 ± 1.3 nM for Pfu-Pol and 9.7 ± 1.6 nM for Mac-Pol.
DISCUSSION
The results presented in this publication confirm that family-B DNA polymerases from thermophilic archaea cease DNA polymerization in response to template strand uracil and this property is shared by the polymerase from the mesophilic archaeon M. acetivorans (25). In contrast DNA polymerases ε and δ from S. cerevisiae, E.coli DNA polymerase III and human mitochondrial polymerase γ all read-through uracil. Thus, at least as assessed with purified proteins, only the archaeal domain appears to possess a polymerase able to bind uracil tightly and subsequently stall replication. Several factors may be responsible for the differences between archaea and bacteria/eukaryotes.
Have appropriate polymerases been compared?
If uracil-induced stalling serves to protect from the consequences of copying G:U mismatches, it is important to compare polymerases responsible for genome replication, rather than playing, for example, a specialized role in DNA repair. PolIII has been unequivocally assigned as the replicating polymerase in E.coli (17) and with eukaryotes Pol ε appears to copy the leading strand and Pol δ the lagging (31,33) and, between them, these two polymerases are responsible for nuclear DNA replication. Similarly Pol γ has been established as the enzyme responsible for eukaryotic mitochondrial DNA replication (20). With the archaea the polymerase responsible for DNA replication awaits absolute confirmation; therefore it remains a possibility that the family-B polymerase is a specialized enzyme dedicated to uracil repair. However, the family-B enzymes are by far the most likely candidates for DNA replication, being the only polymerases present in all archaea (34–36) and clearly related to the eukaryotic replicative polymerases δ and ε. An unusual hetero-dimeric polymerase (classified as family D) has been identified in euryarchaea (37) and both the family B and family D polymerases are essential for euryarchaeal survival (38). Based on the biochemical properties of the family B and D polymerases from Pyrococcus abysii it was suggested that B copies the leading strand and D the lagging (39). Recently, the same group has proposed an alternative scenario; Pol D initially elongating RNA primers before a switch to Pol B directed synthesis (40), with Pol B elongating almost all the leading strand and an undefined fraction of the lagging. Crenarchaea lack Pol D, instead multiple forms of Pol B are found and are assumed to be solely responsible for DNA replication (34–36). In conclusion it appears that all the polymerases studied in this publication serve to replicate the genomes of the organisms from which they are derived.
Is uracil recognition a consequence of high temperature environments?
Prior to this publication read-ahead recognition of uracil has only been demonstrated with polymerases purified from archaea that occupy high temperature niches. The family-B polymerases from the hyperthermophiles P. furiosus, Pyrococcus woisei and Thermococcus literalis (1), Sulfolobus solfataricus (41) and Pyrococcus horikishii, Thermococcus gorgonarius, Sulfurisphaera ohwakuensis, Aeropyrum pernix and Pyrodictium occultum (unpublished observations) all stall replication in response to uracil. Here, for the first time, a family B polymerase from a mesophilic archaeon, M. acetivorans (optimum growth temperature 35–40°C) (24), has been shown to bind uracil-containing DNA as tightly as Pfu-Pol and to halt replication when this base is encountered. Thus recognition of uracil is not exclusively a property of enzymes isolated from hyperthermophiles and, although the bacterial and eukaryotic polymerases were from mesophiles (E. coli, yeast and humans), differences in uracil sensing as compared to archaea cannot simply be explained by dissimilarities in habitat temperature. Further information may be available by studying PolIII from hyperthermophilic bacteria such as Aquifex aeolicus (42), a polymerase very similar in organization to that from E. coli and anticipated to behave identically.
Are the archaea defective in standard uracil repair enzymes?
Uracil arises in DNA in two ways, polymerase catalysed de novo incorporation using dUMP (from dUTP) (44) and deamination of cytosine in G:C base-pairs (4). Both are detrimental to cells; A:U base-pairs, arising from de novo incorporation, may interfere with DNA binding proteins and lead to futile cycles of DNA repair (7) and G:U mis-pairs, the consequence of deamination, are mutagenic following replication (4–7). All organisms protect the genome from uracil using dUTPase (enzymes that hydrolyses dUTP to dUMP and PPi) and UDGase (enzymes that excise uracil from DNA to initiate base excision repair). Archaea possess both dUTPase (45) and UDGase (46,47) and their mutation rates appear similar to mesophilic bacteria and eukaryotes (48,49). Therefore, the archaea do not appear deficient in uracil-protecting enzymes or DNA repair pathways in general and, it is, therefore, unlikely that the archaea have a polymerase-based uracil repair pathway because they are lacking in the standard uracil defences.
Why do only the archaea possess a replicating polymerase that recognizes uracil?
Accurate removal of uracil, or indeed most damaged bases, from the genome relies on the double stranded nature of DNA; following excision of the aberrant base, the complementary strand is used as a template for re-synthesis of the correct sequence (5–7). Problems arise when DNA damage is encountered during replication and under such circumstances polymerization is often halted and appropriate repair initiated. Pathways include fork reversal followed by damage removal, template switching using the copy of the complementary strand to direct polymerization and recombination with the copying of a sister chromosome (8,9,50). A key step appears to be the cessation of DNA replication, which serves as the signal for initiation of the relevant mutli-step/multi-protein DNA repair system. Replication linked repair is, therefore, often considered in the context of DNA lesions that halt the replicating polymerase: examples include; single and double strand breaks, through which the polymerase cannot copy; DNA crosslinks, which cannot be unwound; and bulky damaged bases or abasic sites, to which the polymerase cannot easily match an incoming dNTP. Uracil presents a formidable problem for replication-coupled repair, as it is a near perfect analogue of thymine and mistaken for this base by most polymerases (7). In general polymerases, as with the bacterial and eukaryotic enzymes studied in this publication, do not stop on encountering uracil; rather they insert adenine opposite with high efficiency, resulting in irreversible fixing of a mutation, should the uracil have arisen by deamination of a G:C base-pair. Recognition of uracil by a specialized pocket in the archaeal DNA polymerases allows replication to be halted and presumably facilitates repair by the same pathways used with ‘intrinsically-stopping’ lesions such as strand breaks. A more pertinent question to the one posed as this sub-heading's title might be, therefore, ‘what happens when uracil is encountered during replication in bacteria and eukaryotes?’ One possibility, despite dUTPase and UDGase being present in all three domains of life, is that uracil defence is more active in bacteria/eukaryotes, such that uracil is encountered, during replication, at levels low enough to be tolerated without cessation of DNA polymerization. Such a scenario seems unlikely and may be especially problematic with mitochondria as, although the organelle contains UDGase activity, it is subject to a high mutagenic load due to reactive oxygen species arising from oxidative metabolism (20). An alternative is that UDGase itself might act a uracil sensor during replication. In eukaryotes, one particular UDGase (UNG) interacts with PCNA and RPA and is associated with replication foci, excising uracil arising from incorporation of dUMP during replication (51,52). Here, UNG appears to travel with the replication apparatus and following incorporation of dUMP ‘back tracks’ to initiate base excision repair of A:U base-pairs in the newly synthesized double-stranded DNA. UNG may also have a role in standard base excision repair of G:U mismatches (53,54) and it has been tentatively suggested that the enzyme may further act on G:U mismatches that have escaped repair prior to replication (6). Here, though, UNG would have to scan ahead of the polymerase to generate an abasic site that would then stall the approaching polymerase, allowing entry into recombination-based repair pathways. It is not clear how UNG is organized such that it can fulfil these quite different roles, in one case tracking backwards and acting post-replication, in the other reading forward and acting pre-replication. Interestingly the UDGase from archaea also interacts with PCNA (55). A final possibility is that bacteria and eukyarotes contain as yet unidentified uracil-binding proteins that travel with the replication apparatus and passively stall DNA synthesis in an analogous manner to the uracil-binding pocket of archaeal polymerases.
In summary, this publication indicates that uracil sensing by replicating polymerases is a property of archaea (both thermophilic and mesophilic), not shared with bacteria and eukaryotes. The repair processes used by archaea following uracil-induced stalling await elucidation, as do the mechanisms by which bacteria and eukaryotes deal with uracil encountered during replication.
ADDENDUM
Eukaryotes contain a third family-B DNA polymerase, Pol ζ (zeta), which shows homology to the archaeal family B DNA polymerases but is involved in the by pass of damaged bases rather than DNA replication (56). Primer-template extension assays clearly demonstrated that Pol ζ does not stall replication on encountering uracil and this result is included for completion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
J.W. is a PhD student supported by Cancer Research UK (CRUK). B.A.C. was supported by grants from UK BBSRC and the European Union. P.M.J.B. was supported, in part, by grant GM 32431 from the National Institutes of Health. E.J. was supported by the Swedish Research Council, the Swedish Cancer Society and Svenska Smärtafonden. P.M. was supported by the UK MRC and the Lister Institute of Preventive Medicine. I.K.O.C. was supported by National Science Foundation Grant MCB-023841. Funding to pay the Open Access publication charges for this article was provided by The University of Newcastle.
Conflict of interest statement. None declared.
REFERENCES
- 1.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects pro-mutagenic template-strand uracil. Proc. Natl Acad. Sci. USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 3.Shuttleworth G, Fogg MJ, Kurpiewski MR, Jen-Jacobson L, Connolly BA. Recognition of the pro-mutagenic base uracil by family B DNA polymerases from archaea. J. Mol. Biol. 2004;337:621–634. doi: 10.1016/j.jmb.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 6.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA – occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 7.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 8.Baynton K, Fuchs RPP. Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci. 2000;25:74–79. doi: 10.1016/s0968-0004(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 9.Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 11.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains archaea, bacteria and eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allers T, Mevarech M. Archaeal genetics – the third way. Nat. Rev. Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- 13.Evans SJ, Fogg MJ, Mamone A, Davis M, Pearl LH, Connolly BA. Improving dideoxynucleotide-triphosphate utilisation by the hyper-thermophilic DNA polymerase from Pyrococcus furiosus. Nucleic Acids Res. 2000;28:1059–1066. doi: 10.1093/nar/28.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chilkova O, Jonsson B-H, Johannson E. The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:14082–14086. doi: 10.1074/jbc.M211818200. [DOI] [PubMed] [Google Scholar]
- 15.Fortune JM, Stith CM, Kissling GE, Burgers PMJ, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase δ. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins JB, Murphy MC, White BA, Mackie RI, Ha T, Cann IKO. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 2004;279:6315–6326. doi: 10.1074/jbc.M304491200. [DOI] [PubMed] [Google Scholar]
- 17.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme – structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 18.Fay PJ, Johanson KO, McHenry CS, Bambara RA. Size classes of products synthesized processively by two subassemblies of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 1982;257:5692–5699. [PubMed] [Google Scholar]
- 19.Spelbrink JN, Toivonen JM, Hakkaart GAJ, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, et al. In vivo functional analysis of the human mitochondrial DNA polymerases POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 20.Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase γ in mitochondrial DNA replication and repair. Chem. Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- 21.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A – expression, complex formation and functional characterisation. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 22.Ayyagari R, Gomes XV, Gordenin DA, Burgers PMJ. Okazaki fragment maturation in yeast. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 23.Reid SL, Parry D, Liu H-H, Connolly BA. Binding and recognition of GATATC target sequences by the EcoRV restriction endonuclease: a study using fluorescent oligonucleotides and fluorescence polarisation. Biochemistry. 2001;40:2484–2484. doi: 10.1021/bi001956p. [DOI] [PubMed] [Google Scholar]
- 24.Sowers KR, Baron SF, Ferry JG. Methanosarcina-acetivorans Sp-Nov, an acetotrophic methane-producing bacterium isolated from marine-sediments. Appl. Environ. Microbiol. 1984;47:971–978. doi: 10.1128/aem.47.5.971-978.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 26.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamers MH, Georgescu RE, Lee S-G, O’Donnell M, Kuriyan J. Crystal structure of the catalytic α subunit of the E.coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 28.Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 29.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 30.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 31.Garg P, Burgers PMJ. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 32.Chilkova O, Stenlund P, Isoz I, Stith CM, Grabowski PM, Lundström E-B, Burgers PMJ, Johansson E. The leading and lagging strand DNA polymerases are loaded onto the primer-template via separate reactions but are equally processive in the presence of PCNA. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm741. Available online (doi: 10-1093/nar/gkm741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pursell ZF, Isoz I, Lundström E-B, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cann IKO, Ishino Y. Archaeal DNA replication: identifying pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelman Z, White MF. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Barry ER, Bell SD. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cann IKO, Komori H, Toh S, Kanai S, Ishino Y. A heterodimeric DNA polymerase: evidence that members of euryarchaeota possess a disctinct DNA polymerase. Proc. Natl Acad. Sci. USA. 1998;95:14250–14255. doi: 10.1073/pnas.95.24.14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berquist BR, DasSarma P, DasSarma S. Essential and non-essential DNA replication genes in the model halophilic archaeon Halobacterium sp. NRC-1. BMC Genet. 2007;8 doi: 10.1186/1471-2156-8-31. 31 (doi: 10.1186/1471-2156-8-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henneke G, Flament D, Hübscher U, Querellou J, Raffin JP. The hyperthermophilic euryarchaeota Pyrococcus abysii requires two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005;350:53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 40.Rouillon C, Henneke G, Flament D, Querellou J, Raffin JP. DNA polymerase switching on homotrimeric PCNA at the replication fork of the euryarchaea Pyrococcus abysii. J. Mol. Biol. 2007;369:343–355. doi: 10.1016/j.jmb.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 41.Grúz P, Shimizu M, Pisani FM, De Felice M, Kanke Y, Nohmi T. Proceesing of DNA lesions by archaeal DNA polymerases from Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:4024–4030. doi: 10.1093/nar/gkg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruck I, Yuzhakov A, Yurieva O, Jeruzalmi D, Skangalis M, Kuriyan J, O’Donnell M. Analysis of a multicomponent thermostable DNA polymerase III replicase from an extreme thermophile. J. Biol. Chem. 2002;277:17334–17348. doi: 10.1074/jbc.M110198200. [DOI] [PubMed] [Google Scholar]
- 43.Brynolf K, Eliasson R, Reichard P. Formation of Okazaki fragments in polyoma DNA-synthesis caused by misincorporation of uracil. Cell. 1978;13:573–580. doi: 10.1016/0092-8674(78)90330-6. [DOI] [PubMed] [Google Scholar]
- 44.Tye B-K, Chien J, Lehman IR, Duncan BK, Warner HR. Uracil-incorporation- source of pulse-labelled DNA fragments in replication of Escherichia coli chromosome. Proc. Natl Acad. Sci. USA. 1978;75:233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogrefe HH, Hansen CJ, Scott BR, Nelson KB. Archaeal dUTPase enhances PCR amplification with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl Acad. Sci. USA. 2001;99:596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H, Fitz-Gibbon S, Marcotte EM, Tai JH, Hyman EC, Miller J. Characterisation of a thermostable uracil DNA glycosylase specific for U/G and T/G mismatches from the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Bacteriol. 2000;182:1272–1279. doi: 10.1128/jb.182.5.1272-1279.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandigursky M, Franklin WA. Uracil-DNA glycosylase in the extreme thermophile archaeoglobus fulgidus. J. Biol. Chem. 2000;275:19146–19149. doi: 10.1074/jbc.M001995200. [DOI] [PubMed] [Google Scholar]
- 48.Grogan DW, Carver GT, Drake JW. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl Acad. Sci. USA. 2001;98:7928–7933. doi: 10.1073/pnas.141113098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grogan DW. Stability and repair of DNA in hyperthermophilic archaea. Curr. Issues Mol. Biol. 2004;6:137–144. [PubMed] [Google Scholar]
- 50.Longhese MP, Foiani M. Responses to replication of DNA damage. In: Siede W, Kow YW, Doetsch P, editors. DNA Damage Recognition. NY: Taylor and Francis Group; 2006. pp. 827–840. [Google Scholar]
- 51.Otterlei M, Warbrick E, Nagelhaus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakk O, et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Anderson S, Slupphaug G, Daly G, Krokan HE, Lindahl T, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 53.Akbari M, Otterlei M, Péna-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, et al. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 2004;32:5486–5498. doi: 10.1093/nar/gkh872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dionne I, Bell SD. Characterisation of an archaeal family 4 uracil DNA glycosylase and its interaction with PCNA and chromatin proteins. J. Biochem. 2005;387:859–863. doi: 10.1042/BJ20041661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolas NK, Durocher D. DNA repair: DNA polymerase ζ and Rev1 break in. Curr. Biol. 2006;16:R296–R299. doi: 10.1016/j.cub.2006.03.043. [DOI] [PubMed] [Google Scholar]