Abstract
Methylation of the cytosine is the most frequent epigenetic modification of DNA in mammalian cells. In humans, most of the methylated cytosines are found in CpG-rich sequences within tandem and interspersed repeats that make up to 45% of the human genome, being Alu repeats the most common family. Demethylation of Alu elements occurs in aging and cancer processes and has been associated with gene reactivation and genomic instability. By targeting the unmethylated SmaI site within the Alu sequence as a surrogate marker, we have quantified and identified unmethylated Alu elements on the genomic scale. Normal colon epithelial cells contain in average 25 486 ± 10 157 unmethylated Alu's per haploid genome, while in tumor cells this figure is 41 995 ± 17 187 (P = 0.004). There is an inverse relationship in Alu families with respect to their age and methylation status: the youngest elements exhibit the highest prevalence of the SmaI site (AluY: 42%; AluS: 18%, AluJ: 5%) but the lower rates of unmethylation (AluY: 1.65%; AluS: 3.1%, AluJ: 12%). Data are consistent with a stronger silencing pressure on the youngest repetitive elements, which are closer to genes. Further insights into the functional implications of atypical unmethylation states in Alu elements will surely contribute to decipher genomic organization and gene regulation in complex organisms.
INTRODUCTION
Progress in large-scale sequencing projects is critical to identify and decipher gene organization and regulation in many species including human. Nevertheless, cumulated evidences indicate that the complexity of living organisms is not just a direct outcome of the number of coding sequences and that the presence of multiple regulatory mechanisms accounts for a significant part of biological complexity (1,2). Among these mechanisms, repetitive elements may play a key role in gene regulation and genomic structure. Active transposable elements are involved in genome rearrangement and illegitimate recombination and can also influence gene expression by altering splicing or by acting as enhancers or promoters (3–7). Advances in the understanding of epigenetic mechanisms that regulate these repetitive elements may contribute to elucidate their specific participation in biological processes (8).
Silenced regions in mammals and other vertebrates are differentiated, although not exclusively, by the presence of DNA methylation (9). Methylation of the cytosine is an epigenetic modification of DNA that plays an important role in the control of gene expression and chromosome structure in mammalian cells (10–13). Most of the 5-methylcytosines are found in CpG-rich sequences within tandem and interspersed repeats (9,12) of which the previous estimates indicate that constitute up to 45% of the human genome (14). Among these repeats, Alu's, with more than one million copies per haploid genome, are considered the most successful family (15). Interestingly, Alu's are not randomly distributed within the human genome, as they tend to accumulate in gene-rich regions (14,16,17). Previous works have estimated that Alu elements harbor up to 33% of the total number of CpG sites in the genome (18) and have been reported to be highly methylated in most somatic tissues (18–20). Methylation represents the primary mechanism of transposon suppression and active transposons are demethylated in mammalian genomes (12). It has been proposed that regions of the genome containing repetitive elements might be masked by compartmentalization of the chromatin, resulting in a reduction of the effective size of the genome (21).
Noteworthy, even though a vast number of CpG dinucleotides are provided by the collection of repetitive sequences in the human genome, this dinucleotide is greatly under-represented throughout the genome, but it can be found at close to its expected frequency in small genomic regions (200 bp to a few kb), known as CpG islands (22). These areas are ‘protected’ from methylation and are located in the proximal promoter regions of 75% of human genes (12,13,22). Methylated CpG islands are strongly and hereditably repressed (12). Hence DNA methylation is usually considered as a sign of long-term inactivation (9,10,12).
Cancer cells are characterized by the accumulation of both genetic and epigenetic changes. Widespread genomic hypomethylation is an early alteration in carcinogenesis and has been associated with genomic disruption and genetic instability (23–27). Repeats unmasked by demethylation are likely to facilitate rearrangements due to mitotic recombination and unwanted transcription (28–30). Alternatively, aberrant de novo methylation of CpG islands is a hallmark of human cancers and is associated with epigenetic silencing of multiple tumor suppressor genes (31–37). Therefore, the screening for differentially methylated sequences in tumors appears as a key tool to further understand the molecular mechanisms underlying malignant transformation of cells. Although, the repertoire of methylation screening methodologies has expanded widely (37–39), and different approaches have been used to make bulk estimates of methylation in repetitive elements (40,41), there is still a lack of screening strategies that specifically allow a feasible identification of DNA methylation alterations in repetitive elements (21).
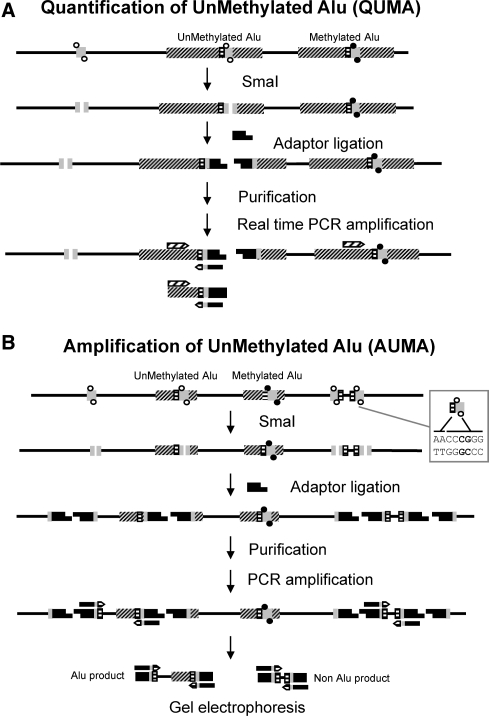
Here we report two variants of a novel methodology to quantify and identify unmethylated Alu sequences. The CpG site within the consensus Alu sequence AACCCGGG is used as a surrogate reporter of methylation. Unmethylated sites are cut with the methylation-sensitive restriction endonuclease SmaI (CCCGGG) and an adaptor is ligated to the DNA ends. Quantification of UnMethylated Alus (QUMA) is performed by real-time amplification of the digested and adaptor-ligated DNA using an Alu consensus primer that anneals upstream of the SmaI site and an adaptor primer extended with the TT dinucleotide in its 3′ end (Figure 1A). The product generated by this approach is completely inside the Alu element and hence it is not possible to make a unique identification. As an alternative approach, we have also performed restrained amplification of digested and adaptor-ligated DNA fragments that are flanked by two close SmaI sites. In this case, the same primer homologous to the adaptor with the additional TT nucleotides at the 3′ end to enrich for Alu sequences is used in absence of the Alu consensus primer (Figure 1B). This second approach is named Amplification of UnMethylated Alu's (AUMA) and results in a complex representation of unique DNA sequences flanked by two unmethylated SmaI sites. When resolved by high-resolution electrophoresis, the AUMA generated sequences appear as a fingerprint characteristic of each sample (Figure 2) and individual scoring and identification of each band can be performed. Because AUMA's stringency is based on a short sequence (AACCCGGG) that is found preferentially but not exclusively in Alu elements, other unmethylated sequences are also present in AUMA fingerprints.
Figure 1.
Schematic diagram of the QUMA and AUMA methods. DNA is depicted by a solid line, Alu elements are represented by dashed boxes. The QUMA and AUMA recognition sites (AACCCGGG) are represented by dashed/gray boxes. CpGs at SmaI sites are shown as full circles when methylated and as open circles when unmethylated. The methylation-sensitive restriction endonuclease SmaI can only digest unmethylated targets, leaving blunt ends to which adaptors can be ligated. (A) QUMA is performed by real-time PCR of an inner Alu fragment using a primer complementary to the Alu consensus sequence upstream of the SmaI site and the primer complementary to the adaptor to which two Alu homologous nucleotides (TT) have been added. (B) In AUMA, sequences flanked by two ligated adaptors are amplified by PCR using a single primer, the same adaptor primer plus the TT nucleotides. When only a few nucleotides are added to the primer, i.e. TT, as illustrated here, other non-Alu sequences may be amplified. This allows the amplification of a large number of sequences that typically range from 100 to 2000 bp.
Figure 2.
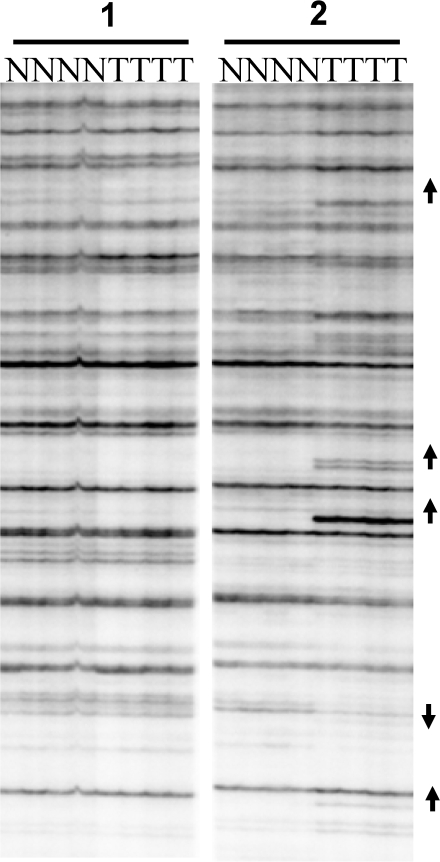
AUMA of normal (N)–tumor (T) pairs of two different patients performed using primer BAu-TT. A highly reproducible band patterning is observed among the four replicates. Representative bands showing gains (hypomethylations) and losses (hypermethylations) are marked with up and down arrowheads, respectively.
Application of QUMA and AUMA to a series of colorectal carcinomas and their paired normal mucosa has offered global estimates of unmethylation of Alu elements in normal and cancer cells and has revealed a large collection of unique sequences that undergo highly recurrent hypomethylation and hypermethylation in colorectal tumors.
MATERIALS AND METHODS
Tissues and cell lines
Fifty colorectal carcinomas and their paired non-adjacent areas of normal colonic mucosa were included in this analysis. Samples were collected simultaneously as fresh specimens and snap-frozen within 2 h of removal and then stored at −80°C. All samples were obtained from the Ciutat Sanitària i Universitària de Bellvitge (Barcelona, Spain). The study protocol was approved by the Ethics Committee. Human colon cancer cell lines (HT29, SW480, HCT116, LoVo, DLD-1, CaCo-2 and LS174T) were obtained from the American Type Culture Collection (ATCC; Manassas, VA). KM12C and KM12SM cells were generously provided by A. Fabra. DNA from tumor–normal pairs was obtained by conventional organic extraction and ethanol precipitation. DNA purity and quality was checked in a 0.8% agarose gel electrophoresis. RNA from cell lines was obtained by phenol–chloroform extraction and ethanol precipitation, following standard procedures.
Bioinformatic analysis
The distribution of SmaI sites, putative amplification hits, PCR homologies, CpG islands and repetitive elements was assessed using the human genome assembly 36.1 from NCBI. Data were obtained from the Repbase (http://www.girinst.org/repbase/index.html) and the Genome Browser Databases (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/). Only assembled chromosome fragments were considered. A Perl routine was used to score all positions containing the target sequences in all chromosomes (available from the authors upon request). Data were analyzed using Excel spreadsheets.
To calculate the proportion of unmethylated Alu elements at the genomic level, the number of AUMA hits identified in bioinformatic analysis were corrected according to the distribution of experimentally generated AUMA products performing Monte Carlo simulations. One thousand Monte Carlo simulations were performed using an Excel Add-in (available at www.wabash.edu/econometrics). In Monte Carlo simulations, it was assumed that 80–100% of SmaI sites at CpG islands are unmethylated and that 50–100% of SmaI sites in other genomic regions different from Alu's and CpG islands are unmethylated.
Quantification of QUMA
One microgram of DNA was digested with 20 U of the methylation sensitive restriction endonuclease SmaI (Roche Diagnostics GmbH, Mannheim, Germany) for 16 h at 30°C, leaving cleaved fragments with blunt ends (CCC/GGG). Adaptors were prepared incubating the oligonucleotides Blue (CCGAATTCGCAAAGCTCTGA) and the 5′ phosphorylated MCF oligonucleotide (TCAGAGCTTTGCGAAT) at 65°C for 2 min, and then cooling to room temperature for 30–60 min. One microgram of the digested DNA was ligated to 2 nmol of adaptor using T4 DNA ligase (New England Biolabs, Beverly, MA, USA). Subsequent digestion of the ligated products with the methylation insensitive restriction endonuclease XmaI (New England Biolabs) was performed to avoid amplifications from non-digested methylated Alu's. The products were purified using the GFX Kit (Amersham Biosciences, Buckinghamshire, UK) and eluted in 250 μl of sterile water.
Quantitative real-time PCR was performed using 1 ng (the equivalent of 333 genomes) of DNA in a LightCycler 480 real-time PCR system with Fast Start Master SYBR Green I kit (Roche). Mastermix was prepared to a final concentration of 3.5 mM MgCl2 and 1 μM of each primer. The downstream BAu-TT primer (constituted by the 3′ end of Blue primer, and the GGGTT sequence including the GGG 3′ side of the cut SmaI site and the Alu homologous TT dinucleotide, ATTCGCAAAGCTCTGAGGGTT) and the upstream primer was an Alu consensus sequence (CCGTCTCTACTAAAAATACA) (see Supplementary Data). Magnitudes were expressed as number of unmethylated Alus per haploid genome after DNA input normalization. The number of haploid genomes present in the test tube was determined in the same multiwell plate by quantification of Alu sequences irrespectively of the methylation state. A real-time PCR using Alu consensus primers upstream of the CCCGGG site was performed (see Supplementary Data) and the number of genomes was calculated against a standard curve constructed with a reference genomic DNA measured by UV spectrophotometry.
To determine the efficiency of the assay and to perform absolute quantification, an external Alu product generated by PCR from a DNA fragment containing an AluSx element was used as standard (Supplementary Methods). The number of copies of the external control were spectrophotometrically quantified and dilution curves were generated and treated as samples. Comparison of dilution curves before and after sample processing indicated that the mean recovery was 73%. DNA samples overdigested with the methylation insensitive XmaI endonuclease were spiked with different amounts of the external standard and processed. The sensitivity of the QUMA detection was 100 unmethylated Alu's per haploid genome (Supplementary Figure 1) using 1 ng of genomic DNA per PCR. A linear response was observed between 1000 and 100 000 unmethylated Alu's per haploid genome (Supplementary Data).
Amplification of AUMA
DNA digestion with SmaI enzyme and ligation to the linker was performed as described above for QUMA, except for the XmaI digestion that was skipped. The product was purified using the GFX Kit (Amersham Biosciences) and eluted in 250 μl of sterile water. Six different chimeric primers constituted by the 3′ end of the Blue primer sequence (ATTCGCAAAGCTCTGA), the cut SmaI site (GGG) and two, four or seven additional nucleotides homologous to the Alu consensus sequence were used to enrich for Alu sequences (see Supplementary Methods). Three primers were designed to amplify ‘upstream’ of the SmaI site (towards ALU promoter): BAu-TT, BAu-TTCA, BAu-TTCAAGC. Three other primers were designed to amplify ‘downstream’ (towards ALU poly-A): BAd-AG, BAd-AGGC, BAd-AGGCGGA. Letters after the dash correspond to the 3′ sequence of the primer (see Supplementary Data). Data reported here were obtained by using the BAu-TT primer.
In each PCR reaction only one primer was used at a time. Products were resolved on denaturing sequencing gels. Although bands can be visualized by silver staining of the gels, radioactive AUMA's were performed for normal–tumor comparisons. A more detailed description of the PCR and the visualization of the bands are given as Supplementary Data.
Only sharp bands that were reproducible and clearly distinguishable from the background were tagged and included in the analysis. Faint bands with inconsistent display due to small variations in gel electrophoresis resolution were not considered. Band reproducibility was assessed with the analysis of PCR duplicates of three independent sample digests from two different samples and PCR replicates from the same digest from four paired tumor–normal samples. AUMA fingerprints were visually checked for methylation differences between bands in the tumor with regard to its paired normal mucosa. Under these premises, a given band was scored according to three possible behaviors: hypomethylation (increased intensity in the tumor), hypermethylation (decreased intensity in the tumor) and no change (no substantial difference in intensity between normal and tumor samples) (Figure 2). Only those bands showing clear changes in their intensities in the fingerprint were considered to represent methylation changes. This is consistent with previous studies done using a related technique (42,43).
Competitive hybridization of AUMA products to metaphase chromosomes and BAC arrays
The origin and chromosomal distribution of sequences generated by AUMA was analyzed using procedures analogous to CGH. Briefly, an AUMA product obtained from a normal tissue DNA was purified using Jet quick PCR product purification kit (Genomed, Löhne, Germany) and labeled with SpectrumRed dUTP (Vysis, Downers Grove, IL, USA) using a Nick Translation kit (Vysis). Similarly, genomic DNA of the same normal sample was labeled with SpectrumGreen dUTP (Vysis) and both probes were cohybridized to metaphase chromosomes. Procedures and image analysis were performed as described (44).
Differential normal–tumor representation of AUMA at the genomic scale was performed by competitive hybridization of AUMA products to BAC arrays. AUMA products from two normal–tumor pairs were purified using Jet quick PCR product purification kit (Genomed, Löhne, Germany) and 1 μg was labeled with dCTP-Cy3 or dCTP-Cy5 (Amersham Biosciences, UK) by use of the Bioprime DNA Labeling System (Invitrogen, Carlsbad, CA, USA). Probes were hybridized to SpectralChip 2600 BAC arrays (Spectral Genomics, Houston, TX, USA) following the manufacturer's instructions. Arrays were scanned with a ScanArray 4000 (GSI Lumonics, Watertown, MA, USA) and processed with GenePix software (Axon Instruments, Union City, CA, USA). The resulting data were processed to filter out low-quality spots based on spot area and similarity of readings between the two replicates of each BAC. Data manipulation was performed using Excel spreadsheets. Because AUMA products are not evenly distributed along chromosomes, only BACs with intensities above the 10% of maximum intensity in at least one of the two channels were considered for ratio calculations. The pattern of chromosomal alterations in these two tumors was determined by conventional CGH as described (44).
Isolation and cloning of AUMA tagged bands
DNA excised from gels was directly amplified with the same primer used in AUMA (BAu-TT) (Supplementary Figure 2). The amplified product was cloned into plasmid vectors using the pGEM-T easy vector System I cloning kit (Promega, Madison, WI, USA). Automated sequencing of multiple colonies was performed using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) to ascertain the unique identity of the isolated band. Sequence homologies were searched for using the Blat engine (http://genome.ucsc.edu/). Selected clones corresponding to AUMA isolated bands were radioactively labeled and used as a probe to confirm the identity of the excised band by hybridization to AUMA fingerprints as previously described (45).
Bisulfite genomic sequencing
Differential methylation observed in some AUMA tagged bands was confirmed by direct sequencing of bisulfite treated normal and tumor DNA as previously described (46). Prior to sequencing, DNA was amplified using a nested or semi-nested PCR approach, as appropriate. Three independent PCRs were done and products were pooled to ensure a representative sequencing. The sequence of PCR primers is described in Supplementary Data.
Histone modification analysis by chromatin immunoprecipitation (ChIP)
Briefly, 6 × 106 cells were washed twice with PBS and cross-linked on the culture plate for 15 min at room temperature in the presence of 0.5% formaldehyde. Cross-linking reaction was stopped by adding 0.125 M glycine. All subsequent steps were carried out at 4°C. All buffers were pre-chilled and contained protease inhibitors (Complete Mini, Roche). Cells were washed twice with PBS and then scraped. Collected pellets were dissolved in 1 ml lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris pH 8) and were sonicated in a cold ethanol bath for 10 cycles at 100% amplitude using a UP50H sonicator (Hielscher, Teltow, Germany). Chromatin fragmentation was visualized in 1% agarose gel. Obtained fragments were in the 200–500 pb range. Soluble chromatin was obtained by centrifuging the sonicated samples at 14 000g for 10 min at 4°C. The soluble fraction was diluted 1/10 in dilution buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris pH 8, 150 mM NaCl) then aliquoted and stored at −80°C until use.
Immunoprecipitation was carried out at 4°C by adding 5–10 μg of the desired antibody to 1 ml of chromatin. Chromatin–antibody complexes were immunoprecipitated with specific antibodies using a protein A/G 50% slurry (Upstate, Millipore, Billerica, MA, USA) and subsequently washed and eluted according to the manufacturer's instructions. Antibodies against acetylated H3 K9/K14 (Upstate), dimethylated H3 K79 and trimethylated H3 K9 (Abcam, Cambridge, UK) were used. Enrichment for a given chromatin modification was quantified as a fold enrichment over the input using quantitative real-time PCR (Roche). For every PCR, a standard curve was obtained to assess amplification efficiency. All quantifications were performed in duplicate.
RESULTS
Genomic estimation of the targets and evaluation of the adequacy of the approach by computational analysis
The availability of the human genome map has allowed us to make a detailed estimation of the frequency and distribution of the sites targeted by our approaches on the genomic scale. A Perl routine was used to score all positions containing the target sequences in all chromosomes and was also applied to perform a virtual AUMA (see Material and Methods section). Some of the most important data derived from the bioinformatic analysis are shown in Table 1 and Figure 3.
Table 1.
Content and distribution of QUMA and AUMA hits in the human genome
| Sequence | Mba | Number of elementsb | SmaI sites (CCCGGG) | AACCCGGG hitsc | Virtual AUMA hitsd | AUMA hitse | Unmethylated hitsf | Unmethylated hits (%)g |
|---|---|---|---|---|---|---|---|---|
| Total | 3080.4 | 1 118 195 | 486 835 | 168 309 | 5498 | 201 | 14332 ± 2418 | 8.52 ± 1.4% |
| Alu (S+J+Y) | 227.3 | 1 091 110 | 198 201 | 155 226 | 5109 | 59 (29.3%) | 4104 ± 688 | 2.64 ± 0.44% |
| AluS | 141.2 | 660 415 | 122 459 | 97 951 | 3382 | 45 (22.4%) | 3028 ± 510 | 3.09 ± 0.51% |
| AluJ | 54.0 | 283 104 | 14 017 | 1235 | 38 | 2 (1.0%) | 151 ± 25 | 12.25 ± 1.97% |
| AluY | 32.1 | 147 591 | 61 725 | 56 040 | 1689 | 12 (6.0%) | 925 ± 156 | 1.65 ± 0.27% |
| CpG islands | 16.2 | 27 085 | 49 430 | 1673 | 63 | 55 (27.4%) | 1501 ± 97 | 90.5 ± 5.79% |
| Rest | 2836.9 | – | 239 204 | 11 410 | 326 | 87 (43.3%) | 8530 ± 1650 | 75.9 ± 14.63% |
aGenome Mb represented by each type of element. Total number corresponds to the number of megabases analyzed for the presence of hits. Only assembled chromosome fragments were considered.
bElements considered in the analysis as obtained from the Repbase and the Genome Browser Databases (see Material and Methods section).
cNumber of occurrences of the sequence AACCCGGG (or CCCGGGTT) within each type of element.
dNumber of AUMA hits present in virtual PCR products of up to 1000 bp.
eHits of actual AUMA products. Only bands appearing in normal tissue were considered. Eighty-seven bands contributed two hits each (174 hits) and 27 bands contributed only one due to poor sequence or incomplete homology with the NCBI Build 36.1 of the human genome (hg18 assembly, March 2006). Twenty-three additional bands were detected mainly in tumor tissue and were not considered to perform calculations.
fEstimated number of unmethylated sites using Monte Carlo simulations (Material and Methods section).
gIn respect to the total number of AACCCGGG (or CCCGGGTT) hits.
Figure 3.
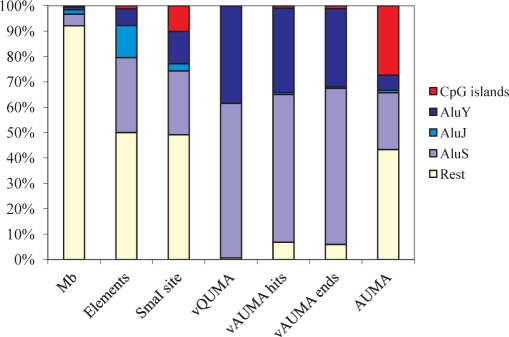
Relative distribution the Alu elements and sequence targets considered in bioinformatic and experimental QUMA and AUMA. Mb: number of megabases occupied by each type of element; elements: number of elements considered (‘Rest’ has been set arbitrarily to 50%); SmaI site: CCCGGG sequence; vQUMA hits: AACCCGGG (or GGGCCCTT) sites in Alu elements; vAUMA hits: AACCCGGG (or GGGCCCTT) sites; vAUMA ends: vAUMA hits considering only putative AUMA products of <1 kb (see Material and Methods section); AUMA: elements at each one of the two ends of actual AUMA products.
Because of the C to T mutational bias at CpG sites (47), any amplification method relying on the consensus sequence (see Supplementary Data) will only cover a fraction of all the Alu's. Therefore it is important to estimate the degree of representativity of the methods used here if genome-wide estimations are to be made. Alu repeats constitute 7.4% of the human genome but accumulate 40.7% of all SmaI sites (Table 1). Nearly 200 000 Alu's (18% of all Alu's) contain a SmaI site and 155 000 retain the AACCCGGG consensus sequence (Table 1 and Figure 3) and are therefore potential targets of QUMA and AUMA. While 38.0% of the youngest AluY elements contain this sequence, the proportions drop to 14.8 and 0.4% in AluS and AluJ families, respectively (Table 1 and Figure 3). These frequencies are consistent with a higher C to T transition trend at CpG sites in older Alu's (47).
The representativity of AUMA was analyzed by a virtual bioinformatic assay of the human genome sequence. A total of 168 309 AACCCGGG (or CCCGGGTT) hits were identified throughout the genome, with 92.9% of all hits within Alu elements (Table 1 and Figure 3). This implies that 14.2% of all Alu elements contained the AACCCGGG sequence. Another 1.0% of the hits were in CpG islands and 6.8% in the rest of the genome (including unique sequences and other repeats) (Table 1 and Figure 3). As expected, Alu elements containing the target sequence mostly belonged to the AluS (2/3) and AluY families (1/3), with a minimal representation of the older family AluJ (<1%). Virtual AUMA determined the presence of 5498 putative products of <1 Kb (the sequence AACCCGGG in the up strand and the sequence CCCGGGTT in the down strand at a distance of <1 kb). Although actual AUMA PCR products may reach 2 kb length (see below), we used the 1 kb limit to compare with AUMA-isolated bands, which were shorter than 1 kb. Most virtual AUMA products contained an Alu element at one of the ends at least (93%).
Quantification of unmethylated Alu in normal and tumor tissues
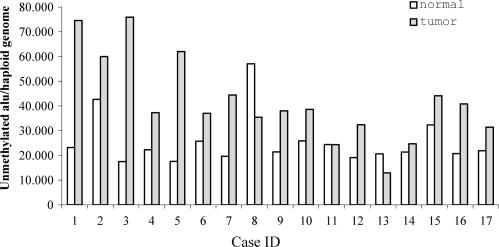
The QUMA approach was applied to quantify unmethylated Alu's in a series of 18 colorectal carcinomas and their paired normal colonic mucosa. An external DNA fragment containing an AluSx element was used as a standard (see Materials and Methods section and Supplementary Data) in order to make an absolute quantification of the number of unmethylated Alu's. Replicates and dilution curves of the samples and standard were performed to assess reproducibility, sensitivity and accuracy (Supplementary Figure 1). Results were normalized by assessment of the number of haploid genomes per test tube (see Material and Methods section). The average number of unmethylated Alu's per haploid genome was 25 486 ± 10 157 in normal mucosa, and 41 995 ± 17 187 in tumor samples (P = 0.004, paired t-test) (Figure 4). We computationally identified a total of 168 309 Alu elements containing the AACCCGGG sequence (potential targets of QUMA) (Table 1). Therefore we estimate that 15.1% of Alu repeats with a AACCCGGG site are unmethylated in the average normal colonic mucosa cell, while this figure is 24.9% in the cancer cell. Considering that the human genome contains ∼1.1 million Alu elements, these estimates indicate that unmethylated Alu's constitute the 2.3 and 3.8% of all Alu's in the normal and tumor tissues, respectively.
Figure 4.
Quantitation of unmethylated Alu's in 17 paired normal mucosa and colorectal carcinoma by QUMA. The values represent the estimated number of unmethylated Alu's per haploid genome. Most tumors exhibited a higher level of hypomethylation when compared with the respective normal.
Set-up and optimization of AUMA fingerprinting
Because QUMA products are fully contained within the Alu sequence, it is not possible to identify and position in the genome the unmethylated Alu elements. To achieve this it is necessary to amplify the targeted unmethylated Alu element together with an adjacent unique sequence. This was attained through the use of the second method, the Amplification of UnMethylated Alus (AUMA). AUMA also targets the unmethylated AACCCGGG sequence, as in QUMA, but in this case a single primer is used in the PCR (BAu-TT) (see Figure 1 and Supplementary Data). Moreover, the product is resolved on a high-resolution gel electrophoresis resulting in a band-rich fingerprint. AUMA bands correspond to sequences flanked by two unmethylated target sequences in opposite strands and sufficiently close to allow PCR amplification (Figure 1 and Supplementary Data). Since 92% of the AACCCGGG occurrences in the human genome are in Alu's (Table 1), the approach is largely biased towards the amplification of unmethylated Alu's. The presence of non-Alu sequences at one of the ends or between two repetitive elements allows the positioning within the genome map of all products.
AUMA products generated using the BAu-TT primer produced highly reproducible fingerprints consisting of bands ranging from ∼100 to ∼2000 bp when resolved in high-resolution sequencing gels (Figure 2 and Supplementary Figure 2). Some well-identifiable bands (up to 5 per experiment) showed random display in both intra-assay and inter-assay replicates (Supplementary Figure 2) and were not considered for analysis. A subset of 110 bands with consistent display among all the experiments were tagged and selected for comparative analysis between samples (see Materials and Methods section).
It should be noted that different fingerprints containing alternative representations may be obtained by AUMA just by using primers that either amplify from the SmaI site towards the Alu promoter (upstream Alu amplification) or towards the Alu poly-A tail (downstream Alu amplification). Also the stringency of the Alu selection may be increased by using longer primers containing additional nucleotides corresponding to the Alu consensus sequence (see Material and Methods section). An illustrative example of AUMA fingerprints generated with different Alu-upstream and Alu-downstream primers is shown in Supplementary Figure 2. All the data reported in this article regarding AUMA were obtained using the BAu-TT primer.
Chromosomal origin of AUMA products
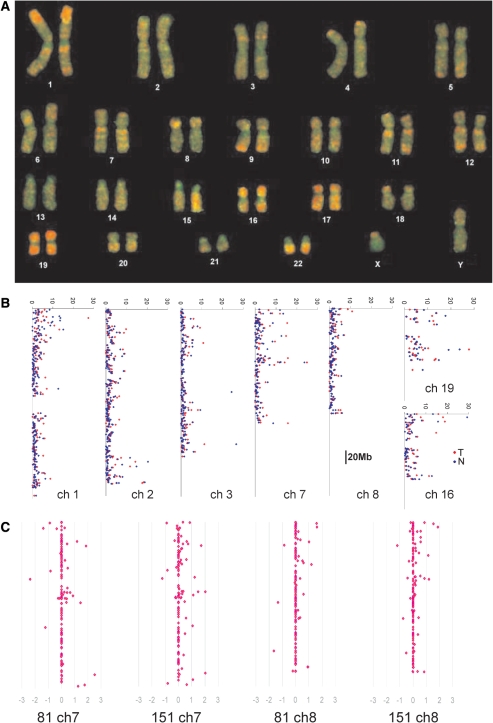
Competitive hybridization between AUMA products and genomic DNA on metaphase chromosomes yielded a characteristic hybridization pattern demonstrating the unequal distribution of AUMA products along the human genome (Figure 5A). Competitive hybridization of AUMA products to BAC arrays showed profiles consistent with those obtained on metaphase chromosomes (Figure 5B). The highest AUMA signal was detected in whole chromosomes 16, 17 and 19 in contrast with chromosomes 2, 13, 18 and X which were mainly labeled by genomic DNA. Other chromosomes showed a discrete pattern of AUMA product hybridization, in which telomeric bands in chromosomes 1, 4, 5, 9, 12 and X, and interstitial bands in chromosomes 1, 3, 7, 11 and 12 are the most prominent examples.
Figure 5.
(A) Chromosomal origin of AUMA products. A competitive hybridization of AUMA product obtained from normal tissue DNA (red) and genomic DNA (green) to metaphase chromosomes was performed. AUMA products showed an unequal distribution along chromosomes, displaying highest densities at most telomeric regions and some interstitial bands. Chromosomes 16, 17 and 19 yielded the highest AUMA density. (B) Intensity distribution of AUMA products hybridized to BAC arrays in selected chromosomes. The average intensity (X-axis) of the two normal (blue) and tumor samples analyzed (red) for each BAC is shown. BACs are arranged along the Y-axis according to its position in the chromosome. (C) Differential methylation profiles determined by competitive hybridization of AUMA products from normal and tumor tissue to BAC arrays. Illustrative examples are shown for chromosomes 7 and 8 from the two cases analyzed (81 and 151). X-axis indicates log2 ratio of tumor/normal intensities. Positive values (to the right) indicate hypomethylations, negative values (to the left) indicate hypermethylations. Additional examples are shown in Supplementary Figure 5.
Identification of AUMA amplified DNA products
To determine the identity of bands displayed by AUMA, 38 tagged bands were isolated and cloned. Multiple clones from each band were sequenced, resulting in a total of 49 different sequences due to the coincidence of more than one sequence in some bands. Characterized bands included bands displaying no changes in the normal–tumor comparisons and bands recurrently altered in the tumor. Table 2 summarizes the main features of a subset of the bands showing recurrent alterations. A list of all the sequences isolated from AUMA fingerprints is provided as Supplementary Table 1. All sequenced bands contained a region of non-repetitive sequence and matched with the BLAST reference sequence, allowing the assignment of a unique chromosomal localization. The BLAST reference sequence corresponding to the 49 sequences isolated from the AUMA fingerprint presented the target sequence CCCGGGTT including the SmaI at both ends. Southern blot analysis of selected cloned sequences showing coincidental size was performed to confirm its correspondence with the band displayed in AUMA fingerprints (Supplementary Figure 4).
Table 2.
A selection of characterized AUMA bands
| Band ID | Size (bp) | % GC | Chromosome map (Locationa) | Gene | CpG islandb | Repetitive elements in band ends (5′/3′) | Methylation status in tumorc |
|---|---|---|---|---|---|---|---|
| Ai1 c3 | 509 | 55 | 17p11.2 (18206453–18206961) | SHMT1 | Yes | Alu Sx/MIR | Hypermethylated |
| Aj2 c1 | 458 | 49 | 1q32.2 (206389082–206389539) | MGC29875 | Yes | Alu Sq/None | Hypomethylated |
| Ao1 c4 | 365 | 50 | 19q13.32 (53550179–53550543) | AK001784 | No | Alu Sx/MIRb | Hypomethylated |
| Ap1 c6 | 358 | 56 | 5q35.2 (175157321–175157674) | CPLX2 | Yes | None/MIR | Hypermethylated |
| Aq3 c6 | 339 | 58 | 8p23.3 (2007343–2007682) | MYOM2 | No | LTR/Alu Y | Hypomethylated |
| Ar3 c3 | 329 | 57 | 2q14.3 (127875178–127875506) | AF370412 | Yes | None/MIRb | Hypomethylated |
| As3 c6 | 306 | 63 | 16p13.3 (3160477–3160782) | None | Yes | None/None | Hypermethylated |
| Au4 c1 | 268 | 57 | 16p13.3 (3162099–3162366) | None | No | tRNA/None | Hypermethylated |
aNucleotide position within the contig (strand +). NCBI Build 36.1 of the human genome.
bThe whole sequence or a fragment of the sequence lays not further than 200 bp of a predicted CpG island.
cAs compared to the paired normal tissue.
To obtain a more representative collection of AUMA bands, 200 clones obtained from normal tissue AUMA products were sequenced. The analysis revealed 88 additional sequences. This resulted in a total of 137 different loci represented in AUMA (Supplementary Table 1). Most sequences obtained by random cloning were also flanked by two AACCCGGG sequences in opposite DNA strands. Nevertheless, in 27 sequences the AACCCGGG site was only present at one of the ends, with the other end showing high homology with the primer although it was not a perfect match. The presence of these sequences suggests that, in some instances, a single cut in the sequence may be enough to produce an amplifiable fragment. This is not considered an artifact since these bands still represent an unmethylated AACCCGGG site.
Genome-wide estimations of unmethylation in Alu's and distribution by subfamily
Of the 137 identified loci represented in AUMA, 114 were isolated from normal tissue DNA and 23 from tumor DNA. Half of the sequences contained an Alu sequence at one of the ends and two were flanked by two inverted Alu's. AUMA sequences isolated from tumor tissue and not present in normal tissue (this corresponds to a tumor-specific hypomethylation) showed a higher proportion of Alu elements (16 out of 23, 70%), and included one sequence flanked by two inverted Alu's. Globally, 78 unmethylated Alu elements were identified and positioned in the human genome map.
To study the genomic distribution of unmethylated sequences in normal colon mucosa, we only considered the 114 sequences obtained from normal tissue. This resulted in a total of 201 unmethylated hits characterized throughout the genome. The nature of the sequences represented in actual AUMA showed striking differences with the distribution expected from the virtual AUMA analysis. The methylation status of the sequence is likely to be the main (if not the only) source of these differences because the virtual AUMA did not consider this state. Therefore we can use these differences to estimate the degree of unmethylation of the Alu repeats. Only 29.4% of the AUMA ends consisted of Alu's, as compared with the expected 92.9% resulting from the bioinformatic analysis. The highest downrepresentation corresponded to the youngest AluY family, which was present in 6.0% of the AUMA ends, while it was expected to add up to 33.1% in virtual AUMA. AluS representation in actual and virtual AUMA was 22.3% and 60%, respectively. Interestingly, AluJ representations, in both actual and virtual AUMA, were closer (1.0% and 0.7%, respectively) (Figure 3 and Table 1). These results suggest that there is a stronger pressure to methylate younger Alus. Alternatively, the hits corresponding to CpG islands were overrepresented in actual versus virtual AUMA by a factor of nearly 25-fold (27.4% versus 1.1%), consistently with the unmethylated status of most CpG islands (Figure 3 and Table 1). The rest of the hits were located in different types of repetitive elements (MIR, MER, LTR, LINE, etc.) and unique sequences (Supplementary Table 1). The miscellaneous collection of sequences (‘Rest’) was over-represented by about 7-fold (observed hits: 43.3%; expected hits: 5.9%, Table 1). The 46 AUMA hits represented by the 23 bands specific of tumor tissue showed a higher proportion of Alu's compared with those obtained from normal tissue (41% versus 29%, respectively), but similar distribution by Alu family (10 AluS, 5 AluY, 1 AluJ, 4 in CpG islands and 26 in other sequences).
To calculate the proportion and distribution of unmethylated Alu elements on a genomic scale, we performed Monte Carlo simulations taking into account the observed and expected distribution of hits in each Alu family and CpG islands and the rest of sequences (see Material and Methods section). We estimate that at least 4104 Alu elements are unmethylated or partially unmethylated in normal colonic mucosa. This corresponds to 2.64% of all Alu elements containing the target sequence AACCCGGG (Table 1). Although AluS and AluY represent the majority of these sequences it should be noted that the methylation pressure is inverse to the conservation of the SmaI site. That is, the most conserved and younger AluY family shows the lowest relative rate of unmethylation; and the older and more degenerated AluJ family exhibits the highest unmethylation.
Application of AUMA to detect differential DNA methylation in colorectal carcinomas
In order to test the usefulness of the method for the detection of new altered methylation targets, we applied AUMA to a series of 50 colorectal carcinomas and their paired normal mucosa. Two cases were excluded from the analysis due to recurring experimental failure of the normal or tumor tissue DNA. For the rest of 48 normal–tumor pairs, consistent and fully readable fingerprints were generated and evaluated for normal–tumor differential representation. A given case presented, on average, 107 ± 2.9 informative bands (range 98–110). The variation was due to polymorphic display or variable resolution power of gel electrophoresis.
In this study, only those bands showing clear intensity differences between normal and tumor tissue fingerprints (Figure 2) have been scored as methylation changes since they are more likely to reflect tumor-wide alterations. Because the fingerprints represent sequences flanked by two unmethylated sites, a decreased intensity in a given band in the tumor in regard to the paired normal tissue is indicative of hypermethylation, while an increased intensity corresponds to hypomethylation (Figure 2). All tumors displayed changes in regard to the paired normal tissue. The average tumor showed 19 ± 7 (range 6–37) hypomethylations and 22 ± 10 (range 1–39) hypermethylations. It is of note that hypomethylations could either be seen as an increase in the intensity of a pre-existing band in the normal tissue or as the appearance of an non-existent band in the normal tissue. This contrasts with hypermethylation events, which rarely showed the complete loss of a band in the tumor sample, most likely due to the unavoidable contamination of normal tissue.
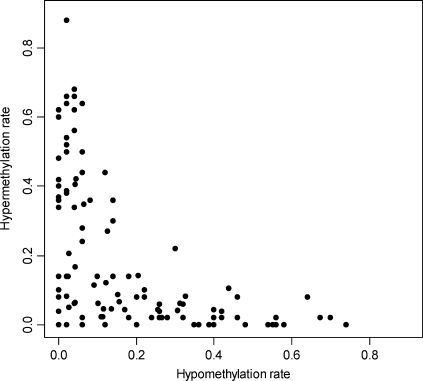
Virtually, all tagged bands (109 out of 110) were found to be altered in at least one tumor when compared to its normal paired mucosa. AUMA tagged bands presented a wide distribution in the hypomethylation/hypermethylation rates (proportion of tumors showing differential display compared to the paired normal tissue) (Figure 6). Hypomethylation and hypermethylation showed a strong negative correlation (r = −0.55 and P < 0.0001), indicating that most bands tended to be either hypomethylated or hypermethylated. A large proportion of tagged bands (78 bands) were recurrently altered in over 25% of the cases included in this series.
Figure 6.
Distribution of hypermethylation and hypomethylation rates in the 110 AUMA tagged bands. Rates were obtained by comparison of the AUMA fingerprints obtained in 50 colorectal tumors as compared to their respective matched normal tissue.
In order to determine whether normal–tumor differences were limited to isolated independent loci or changes that might affect larger chromosomal regions, we compared the distribution of AUMA products generated from two paired normal and tumor tissues and hybridized to BAC arrays. Differential hybridization was observed in many BACs, suggesting that relatively large regions encompassing from several hundred Kbs to a few Mbs may undergo concurrent hypomethylation or hypermethylation. Telomeric regions of many chromosomes contained most of the differential display (Figure 5C). The differential methylation profiles were unaffected by chromosomal dosage as demonstrated by its independence of chromosomal losses and gains (as detected by Comparative Genomic Hybridization (CGH) (Supplementary Figure 5).
Validation of methylation changes detected by AUMA
To confirm that the changes observed in AUMA fingerprints corresponded to actual changes in the methylation status of the sequence, eight different sequences obtained from AUMA fingerprints were analyzed in normal and tumor tissues by direct sequencing of sodium bisulfite-treated DNAs (Table 2). Moreover, it was demonstrated that methylation changes affected not only the CpG in at least one of the two flanking SmaI sites (whose methylation prevents AUMA representation) but also neighboring CpGs (Supplementary Figure 6). In two samples, hypermethylations/hypomethylations detected by AUMA could not be confirmed by bisulfite sequencing, suggesting that the change could affect only a small fraction of tumor cells and that both methods may exhibit different sensitivities. The presence of minor subpopulations can be detected using more sensitive techniques, i.e. the Methylation Specific PCR or by sequencing of multiples clones.
Functional implications of changes detected by AUMA
Next, we wondered if DNA methylation changes detected by AUMA may have any functional consequences. We chose one of the most recurrent hypomethylated AUMA sequences (Aq3) and performed an insightful epigenetic characterization of the region in a series of normal–tumor pairs and in colon cancer cell lines.
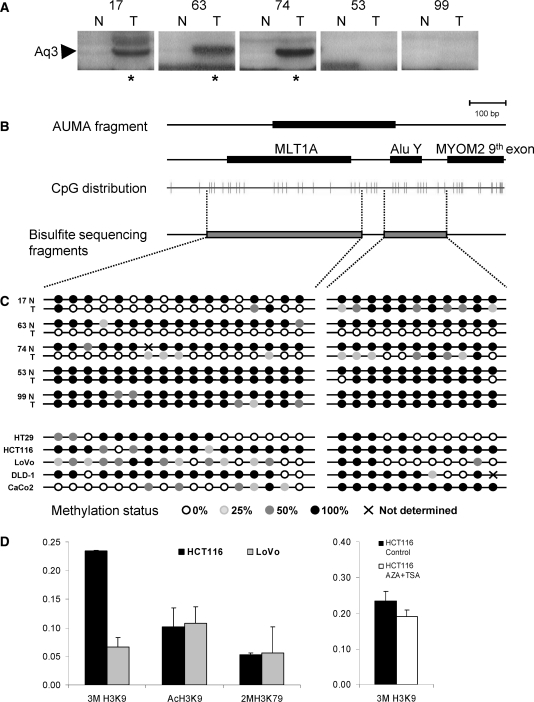
Aq3 band is recurrently hypomethylated in tumors according to AUMA fingerprints (Figure 7A). It represents a sequence situated in the eighth intron of the MYOM2 gene (Table 2) and does not fall inside or close to any CpG island. The SmaI sites are located in a MLT1A repeat and an AluYd3 element. The methylation status the two flanking regions of the AUMA band (465 bp and 213 bp long spanning 20 and 11 CpGs, respectively) was analyzed by bisulfite direct sequencing (Figure 7B). Confirmation of AUMA data was performed in three normal–tumor pairs exhibiting differential display of the Aq3 band in AUMA fingerprints (cases 17, 63 and 74) and two cases lacking this band in both normal and tumor pair (cases 53 and 99) (Figure 7A), as well as five cell lines (HCT116, DLD-1, LoVo, HT29 and CaCo2). All normal tissues as well as tumors 53 and 99 showed heavy methylation of this region (Figure 7C). In contrast to this and in agreement with AUMA results, tumors 17, 63 and 74 exhibited hypomethylation at most CpGs. Cell lines showed variable profiles of DNA methylation, with CaCo2 exhibiting unmethylation of the MLT1A element but heavy methylation of the AluYd3 element, which was also heavily methylated in HCT116 cells but not in the rest of the cell lines tested. MYOM2 expression levels analyzed by real-time RT-PCR were not affected by the methylation status of this sequence (data not shown). Further 45 normal–tumor pairs were analyzed for methylation of the AluYd3 element by real-time dissociation analysis (Supplementary Figure 7) and it was found hypomethylated in 26 tumors (58%).
Figure 7.
(A) Detail of the AUMA fingerprints generated from five normal–tumor sample pairs. The presence of the Aq3 band is indicated by an asterisk under the three Aq3 positive cases. (B) The relative position of the AUMA Aq3 band, MLT1A and Alu Y repetitive elements, as well as MYOM2 ninth exon are shown. Each vertical line in the CpG distribution represents a CpG dinucleotide along the DNA sequence. Two different fragments were amplified for the bisulfite sequencing analysis (gray boxes). Sequence is oriented 5′–3′ in regard to MYOM2 3′ end. (C) Methylation status of the CpG nucleotides in the two fragments amplified were ascertained by direct sequencing of bisulfite-treated DNAs of 5 normal–tumor pairs and 5 colon cancer cell lines. (D) ChIP analysis of the AluY element frequently hypomethylated in cancer revealed loss of trimethylation in histone 3 lysine 9 residue (3mH3K9) in LoVo cells (unmethylated at DNA level) as compared to HCT116 (methylated at DNA level). Treatment of HCT116 cells with 5AzaC and TSA produced a moderate decrease in the levels of trimethylation in H3K9.
Next, we wondered whether the DNA methylation status of the AluYd3 element was associated with alternative chromatin states. We performed Chromatin ImmunoPrecipitation (ChIP) analysis of histone 3 (H3) modifications indicative of active chromatin: acetylation of lysines 9 and 14 (AcH3K9/K14), and dimethylation of lysine 79 (2mH3K79); and silent chromatin: trimethylation of lysine 9 (3mH3K9). These histone marks were compared between cell lines HCT116 and LoVo (with 100% and 30% methylation of the AluY element, respectively). The silencing mark 3mH3K9 was 3.5-fold higher in HCT116 cells compared to the LoVo cell line (Figure 7D). No differences in active marks were observed and these were significantly lower than the silencing mark 3mH3K9. When HCT116 cells were treated with the demethylating agent 5-aza-2′-deoxycytidine (5AzaC) and the inhibitor of histone deacetylase trichostatin A (TSA), a moderate decrease in the amount of the 3mH3K9 mark was observed (Figure 7E). As a whole, these data suggest that DNA methylation changes in this AluYd3 element are accompanied by alternated chromatin states. The molecular consequences of such epigenetic changes remain to be identified.
DISCUSSION
Epigenetic states of Alu elements
Full genome sequencing has provided precise maps of repetitive elements, and several studies have investigated their distribution and relationship with genome structure (48–51). More recently, a few studies have explored sequence-dependent associations between repetitive elements and the epigenetic landscape. There is a characteristic distribution of interspersed elements along methylated and unmethylated domains, with most elements in the methylated compartment of the genome (21). Nevertheless, SINEs, which include Alu elements, are the repetitive sequences most commonly found in unmethylated domains (21) and some Alu elements may contain discriminatory motifs associated with methylation-resistant CpG islands (52). Somatic cells show unstable epigenetic profiles in repetitive elements as demonstrated by global measurements of either DNA methylation (18,20,40,41) or histone modifications (53,54). Recent studies have revealed interindividual variability in DNA methylation profiles at specific Alu elements (55), and Fraga and colleagues detected epigenetic changes arising during the lifetime of monozygotic twins in Alu elements and other sequences (56).
Beyond these few studies, the extension and nature of the epigenetic state of interspersed elements is largely unknown. Global estimates of DNA methylation in repetitive elements have been obtained by Southern blot analyses (30) and, more recently, by using approaches based on bisulfite conversion of the unmethylated cytosine (40,41,57). These studies have confirmed the global hypomethylation of most tumors but they do not provide detailed information on the nature and localization of the unmethylated elements. In silico analysis has revealed that a number of Alu elements close to CpG islands retain a high proportion of CpG sites, and this is presumed to be a sign of unmethylation (58), but no experimental proof has been provided. In our point of view, the lack of simple, specific and sensitive methodologies to screen for epigenetic changes in repetitive elements on the genomic scale has precluded a clearer understanding of the nature and implications of these sequences in cell biology.
Properties of QUMA and AUMA
Here we report a systematic screening of unmethylated Alus as a tool to determine the extent of DNA hypomethylation, to identify specifically unmethylated elements and to detect epigenetic alterations in cancer cells. QUMA is a very simple and specific method and provides accurate relative estimates of the number of unmethylated elements. QUMA is specially appropriate for comparative studies, but also provides a raw quantitation of the number of unmethylated elements per haploid genome, outlining the extent of hypomethylated Alu's in normal and pathologic cells. QUMA analysis indicates that about 1 out of 6 Alu elements containing the AACCCGGG site are unmethylated, while in tumors, this figure nearly doubles in agreement with previous studies (23). Although these analyses are likely to generate good estimates at the comparative level (between samples), absolute values should be treated with caution because the determination refers to a single CpG site within the Alu element.
To date there is still a lack of proper methodologies allowing genome-wide screenings for recurrent hypomethylated regions that may have some impact on tumor biology. Even though QUMA and other methodologies (41,59) allow quantitation of unmethylated repeats, they do not provide a straightforward approach to identify and map the amplified targets. At this point, AUMA takes us a step further, allowing the undoubtful identification of hypomethylated sequences, in addition to hypermethylated targets. Although AUMA is specially suited to determine the nature of the unmethylated elements, it also allows the calculation of global unmethylation in Alu elements. Nevertheless, it should be taken into account that this is an indirect measure, because it relies in the extent of methylation in CpG islands and other sequences. Moreover, unmethylation of a second SmaI site near the Alu is also required to generate the AUMA band and hence to be detected. While AUMA shares many technical steps with other techniques, namely MCA (60) and AIMS (43), its design is conceptually unique since AUMA scans for the atypically unmethylated Alu sequences, unlike the other approaches that are enriched for typical methylated sequences.
Due to sequence degeneration, both QUMA and AUMA are more effective in screening for unmethylation in younger elements. This trend is more clearly seen in AUMA, with only 9% of the Alu elements of the old J subfamily containing the SmaI site retain the AA dinucleotide needed for their amplification, while this figure is 91 and 80% in the younger AluY and AluS subfamilies, respectively (Table 1), making clear that younger Alu elements tend to retain the SmaI site nearly as much as they retain the AA dinucleotide required for their amplification. This bias is not a handicap, since unmethylated Alu sequences revealed by AUMA are likely to represent the most relevant events of this kind, because spurious unmethylation of old Alu elements retaining a single or a few CpG sites is expected to have less biological significance than unmethylation of younger Alu elements that are usually closer to active chromatin regions (21) and retain more CpGs. The stronger methylation pressure observed in the AluY class is consistent with this postulate.
AUMA was designed to amplify DNA fragments containing the target sequence (AACCCGGG), which is present in Alu and other repetitive elements. Because a single primer was used for PCR amplification, the target sequence must appear in both strands of the DNA at relatively nearby positions. As expected, Alu elements, with more than one million copies per human genome (15), were the most frequent repeat in AUMA bands (50% in sequences isolated from non-tumor tissue), but only two sequenced bands contained two inverted Alu repeats (Supplementary Table 1). This observation is in concordance with previous works reporting on the instability of this inverted repeats, which might have caused their exclusion from the human genome (61,62). More restrictive conditions to select for Alu, or any other repeat of interest, may be achieved by extending the 3′ end of the primer specific sequence (see Supplementary Data); however, the number of sequences we obtained was considered appropriate to accomplish the original aim of the study which is to screen for differentially methylated repetitive elements in colorectal cancer.
It is worth noting that AUMA patterns are highly reproducible not only in replicates but also among different samples, which indicates that the unmethylated status of these repeats is tightly controlled, probably by the epigenetic status of nearby regions. This is strengthened by the confirmation that unmethylation extends many CpG sites beyond the SmaI cut site. Moreover, about 50% of the bands tagged in AUMA fingerprints exhibited variable display among normal tissues (data not shown), suggesting the usefulness of this technique to investigate epigenetic polymorphisms.
Alu's and other repetitive elements tend to be highly methylated in most somatic tissues (8,9,40,63). Here we have identified 78 ‘atypical’ Alu elements exhibiting full or partial unmethylation in normal colonic mucosa cells. Different evidences underscore the adequacy of this approach to track changes with possible functional implications: (i) a significant portion of the characterized bands are located inside or nearby CpG islands and genes; (ii) AUMA products show a characteristic distribution in R bands, coincidentally with the distribution of Alu sequences (15), indicating a bias toward the gene-richest portion of the genome known as the H3 isochore (64).
DNA methylation along Alu families
Alu families showed striking differences in their methylation level. Most of the Alu elements characterized here are from the younger families AluS and AluY (74% and 22%, respectively). Nevertheless, this observation is mainly due to the depletion of CpG sites in older Alu elements. Hence, only 1 out 230 AluJ elements maintains the AUMA target site (AACCCGGG), while younger elements show higher rates of maintenance in accordance with their age (AluS: 1 out of 7; AluY: 2 out of 5) (Table 1). Interestingly, the rate of unmethylation is higher in older elements (AluJ: 12.2%; AluS: 3.1%; AluY: 1.6%) (Table 1). As noted by Rollins et al. (21), the boundaries of unmethylated domains tend to be occupied by methylated Alu transposons of the younger AluS and AluY families. Other studies have noted that CpG island-associated Alu's retain a higher proportion of CpG sites, suggesting that these elements are unmethylated in the germ line (58). These unmethylated elements that can be easily revealed by AUMA are likely to play a regulatory role in a significant number of genes (65). AUMA of tumors was enriched in Alu sequences as compared with normal tissue (41% versus 29%, respectively), suggesting a favored hypomethylation of Alu elements within the global genomic hypomethylation associated with tumorigenesis (23).
Application of AUMA to detect epigenetic changes in cancer cells
Cancer-related hypomethylation is well documented (23) and different studies have demonstrated the demethylation of Alu's and other repetitive elements in different types of neoplasias (66). Our data are consistent with previous estimates and go one step forward in the characterization of unmethylated repeats. The AUMA approach was conceived as a straightforward DNA methylation screening strategy targeting specific interspersed repeats and suitable to be applied to large series of samples or experimental conditions as reported here.
The application of AUMA to a series of colorectal carcinomas and their paired matched normal tissue has revealed a high rate of alterations. This indicates the plasticity of epigenetic control of the elements screened by AUMA in colorectal carcinogenesis. Although some bands show bidirectional changes (hypomethylations and hypermethylations), which have been also reported in other sequences (42,67), most bands display an alternative trend either toward hypermethylations or hypomethylations. Some of these changes are highly recurrent (in more than 50% of tumors), suggesting that they may represent relevant alterations related to mechanisms frequently disturbed in colon cancer. Because the default status of repetitive elements is methylation, hypomethylations are readily detected as the emergence of a new band in the tumor AUMA fingerprint. Since all amplified bands include a unique sequence, it has been possible to identify all of the isolated bands and pinpoint them in the genomic map.
As an example, we have investigated the Aq3 sequence, one of the most recurrent hypomethylations in this study. Aq3 band is flanked by two repeats, a LTR and an AluY, which map within an intron between exons 8 and 9 of the MYOM2 gene at 8p23.3. Both elements are heavily methylated in normal tissue and partially to fully unmethylated in tumor tissue. Interestingly, we have found moderately high levels of the heterochromatin associated mark 3mH3K9 in the fully methylated AluYd3 element in the HCT116 cell line, while the levels were significantly lower (3.5-fold) in the partially demethylated LoVo cell line. Furthermore, none of the classical active marks AcH3K9/K14 and 2mH3K79, have been found enriched in the LoVo cell line. These data are in concordance with preliminary data showing that the hypomethylation does not affect the expression of MYOM2 (data not shown) but could rather affect chromatin structure in the region. In agreement with these observations, this genomic region undergoes frequent losses (68–70) and is rearranged in many different types of tumors (71,72), which hints at a role for DNA hypomethylation in genomic instability (24–27,73). The specific functional consequences of this hypomethylation deserve further investigations.
Another application of AUMA is the detection of genomic regions that have been silenced in cancer. Interspersed elements are concentrated in gene-rich regions and due to the intended selection of unmethylated repetitive elements in AUMA, it appears reasonable to postulate that normally unmethylated sequences are likely to pinpoint active genomic regions. In this context, AUMA provides a large collection of genomic regions undergoing hypermethylation, which are readily seen as bands recurrently loss in the fingerprints. DNA methylation associated epigenetic silencing is probably one of the most prevalent mechanisms of tumor suppression inactivation in cancer (33,37,74). Therefore, AUMA can also be used to screen for differential methylation not only in Alu elements but also in unique sequences and repetitive elements other than Alu.
In summary, QUMA and AUMA methodologies are a simple and novel approach to explore and gain insights into the functional significance of interspersed genomic elements and neighboring sequences. Due to its distinctive features (bias for unmethylated elements in gene-rich regions and detection of both hypomethylation and hypermethylation) we think that these techniques constitute a new and unique tool that should complement global determinations and high-resolution genome-wide scanning strategies. Beyond unmethylated repetitive elements, AUMA can be also used to detect recurrent epigenetic changes associated with tumorigenesis including gene epigenetic inactivation.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Gemma Aiza for technical support and Jessica Halow for critical review of the manuscript. J.R. was a fellow of the Generalitat de Catalunya; E.V. was a fellow of the Fondo de Investigación Sanitaria (FIS). This work was supported by a grant from the Ministry of Education and Science (SAF2006/351) and the Consolider-Ingenio 2010 Program (CSD2006-49). Funding to pay the Open Access publication charges for this article was provided by the Institute of Predictive and Personalized Medicine of Cancer (IMPPC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 2.Zuckerkandl E. Why so many noncoding nucleotides? The eukaryote genome as an epigenetic machine. Genetica. 2002;115:105–129. doi: 10.1023/a:1016080316076. [DOI] [PubMed] [Google Scholar]
- 3.Kreahling J, Graveley BR. The origins and implications of Aluternative splicing. Trends Genet. 2004;20:1–4. doi: 10.1016/j.tig.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Jasinska A, Krzyzosiak WJ. Repetitive sequences that shape the human transcriptome. FEBS Lett. 2004;567:136–141. doi: 10.1016/j.febslet.2004.03.109. [DOI] [PubMed] [Google Scholar]
- 5.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 6.Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 8.Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 10.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 11.Weissmann F, Lyko F. Cooperative interactions between epigenetic modifications and their function in the regulation of chromosome architecture. Bioessays. 2003;25:792–797. doi: 10.1002/bies.10314. [DOI] [PubMed] [Google Scholar]
- 12.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 13.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nat. Rev. Genet. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 14.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 16.Korenberg JR, Rykowski MC. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988;53:391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Gentles AJ, Jurka J, Karlin S. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:2930–2935. doi: 10.1073/pnas.052692099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmid CW. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991;19:5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gama-Sosa MA, Wang RY, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983;11:3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kochanek S, Renz D, Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993;12:1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 23.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 24.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 25.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capella G, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 28.Liu WM, Maraia RJ, Rubin CM, Schmid CW. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994;22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 30.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr. Top. Microbiol. Immunol. 2006;310:251–274. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 31.Plass C. Cancer epigenomics. Hum. Mol. Genet. 2002;11:2479–2488. doi: 10.1093/hmg/11.20.2479. [DOI] [PubMed] [Google Scholar]
- 32.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 33.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 34.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 35.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 36.Laird PW. Cancer epigenetics. Hum. Mol. Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;6:6. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 38.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 39.Cottrell SE. Molecular diagnostic applications of DNA methylation technology. Clin. Biochem. 2004;37:595–604. doi: 10.1016/j.clinbiochem.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frigola J, Sole X, Paz MF, Moreno V, Esteller M, Capella G, Peinado MA. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum. Mol. Genet. 2005;14:319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 43.Frigola J, Ribas M, Risques RA, Peinado MA. Methylome profiling of cancer cells by amplification of inter- methylated sites (AIMS) Nucleic Acids Res. 2002;30:e28. doi: 10.1093/nar/30.7.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vendrell E, Ribas M, Valls J, Sole X, Grau M, Moreno V, Capella G, Peinado MA. Genomic and transcriptomic prognostic factors in R0 Dukes B and C colorectal cancer patients. Int. J. Oncol. 2007;30:1099–1107. [PubMed] [Google Scholar]
- 45.Perucho M, Welsh J, Peinado MA, Ionov Y, McClelland M. Fingerprinting of DNA and RNA by arbitrarily primed polymerase chain reaction: applications in cancer research. Methods Enzymol. 1995;254:275–290. doi: 10.1016/0076-6879(95)54020-2. [DOI] [PubMed] [Google Scholar]
- 46.Stirzaker C, Song JZ, Davidson B, Clark SJ. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 2004;64:3871–3877. doi: 10.1158/0008-5472.CAN-03-3690. [DOI] [PubMed] [Google Scholar]
- 47.Xing J, Hedges DJ, Han K, Wang H, Cordaux R, Batzer MA. Alu element mutation spectra: Molecular clocks and the effect of DNA methylation. J. Mol. Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- 48.Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grover D, Majumder PP, Rao CB, Brahmachari SK, Mukerji M. Nonrandom distribution of alu elements in genes of various functional categories: insight from analysis of human chromosomes 21 and 22. Mol. Biol. Evol. 2003;20:1420–1424. doi: 10.1093/molbev/msg153. [DOI] [PubMed] [Google Scholar]
- 50.Grover D, Mukerji M, Bhatnagar P, Kannan K, Brahmachari SK. Alu repeat analysis in the complete human genome: trends and variations with respect to genomic composition. Bioinformatics. 2004;20:813–817. doi: 10.1093/bioinformatics/bth005. [DOI] [PubMed] [Google Scholar]
- 51.Price AL, Eskin E, Pevzner PA. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;15:15. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 53.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;13:13. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 54.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003;278:27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 55.Sandovici I, Kassovska-Bratinova S, Loredo-Osti JC, Leppert M, Suarez A, Stewart R, Bautista FD, Schiraldi M, Sapienza C. Interindividual variability and parent of origin DNA methylation differences at specific human Alu elements. Hum. Mol. Genet. 2005;14:2135–2143. doi: 10.1093/hmg/ddi218. [DOI] [PubMed] [Google Scholar]
- 56.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brohede J, Rand KN. Evolutionary evidence suggests that CpG island-associated Alus are frequently unmethylated in human germline. Hum. Genet. 2006;119:457–458. doi: 10.1007/s00439-006-0161-x. [DOI] [PubMed] [Google Scholar]
- 59.Choi IS, Estecio MR, Nagano Y, Kim do H, White JA, Yao JC, Issa JP, Rashid A. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod. Pathol. 2007;20:802–810. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 60.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 61.Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J, Resnick MA. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 63.Hellmann-Blumberg U, Hintz MF, Gatewood JM, Schmid CW. Developmental differences in methylation of human Alu repeats. Mol. Cell. Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saccone S, Caccio S, Kusuda J, Andreozzi L, Bernardi G. Identification of the gene-richest bands in human chromosomes. Gene. 1996;174:85–94. doi: 10.1016/0378-1119(96)00392-7. [DOI] [PubMed] [Google Scholar]
- 65.Oei SL, Babich VS, Kazakov VI, Usmanova NM, Kropotov AV, Tomilin NV. Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters. Genomics. 2004;83:873–882. doi: 10.1016/j.ygeno.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 66.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim. Biophys. Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 67.Nishiyama R, Qi L, Tsumagari K, Weissbecker K, Dubeau L, Champagne M, Sikka S, Nagai H, Ehrlich M. A DNA Repeat, NBL2, Is Hypermethylated in Some Cancers but Hypomethylated in Others. Cancer Biol. Ther. 2005;4:4. doi: 10.4161/cbt.4.4.1622. [DOI] [PubMed] [Google Scholar]
- 68.Emi M, Fujiwara Y, Nakajima T, Tsuchiya E, Tsuda H, Hirohashi S, Maeda Y, Tsuruta K, Miyaki M, et al. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992;52:5368–5372. [PubMed] [Google Scholar]
- 69.Sunwoo JB, Sun PC, Gupta VK, Schmidt AP, El-Mofty S, Scholnick SB. Localization of a putative tumor suppressor gene in the sub-telomeric region of chromosome 8p. Oncogene. 1999;18:2651–2655. doi: 10.1038/sj.onc.1202838. [DOI] [PubMed] [Google Scholar]
- 70.Muscheck M, Sukosd F, Pesti T, Kovacs G. High density deletion mapping of bladder cancer localizes the putative tumor suppressor gene between loci D8S504 and D8S264 at chromosome 8p23.3. Lab Invest. 2000;80:1089–1093. doi: 10.1038/labinvest.3780114. [DOI] [PubMed] [Google Scholar]
- 71.Wong N, Wong KF, Chan JK, Johnson PJ. Chromosomal translocations are common in natural killer-cell lymphoma/leukemia as shown by spectral karyotyping. Hum. Pathol. 2000;31:771–774. doi: 10.1053/hupa.2000.7625. [DOI] [PubMed] [Google Scholar]
- 72.Jin Y, Jin C, Wennerberg J, Hoglund M, Mertens F. Cytogenetic and fluorescence in situ hybridization characterization of chromosome 8 rearrangements in head and neck squamous cell carcinomas. Cancer Genet. Cytogenet. 2001;130:111–117. doi: 10.1016/s0165-4608(01)00476-9. [DOI] [PubMed] [Google Scholar]
- 73.Karpf AR, Matsui S. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–8639. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 74.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.