Abstract
The poly(A) (pA) signal possesses a dual function in 3′ end processing of pre-mRNA and in transcriptional termination of RNA polymerase II (Pol II) for most eukaryotic protein-coding genes. A key protein factor in yeast and Drosophila Pol II transcriptional termination is the 3′-end processing factor, Pcf11. In vitro studies suggest that Pcf11 is capable of promoting the dissociation of Pol II elongation complexes from DNA. Moreover, several mutant alleles of yeast Pcf11 effect termination in vivo. However, functions of human Pcf11 (hPcf11) in Pol II termination have not been explored. Here we show that depletion of hPcf11 from HeLa cells reduces termination efficiency. Furthermore, we provide evidence that hPcf11 is required for the efficient degradation of the 3′ product of pA site cleavage. Finally, we show that these functions of hPcf11 require an intact pA signal.
INTRODUCTION
Most eukaryotic genes code for proteins and are transcribed by RNA polymerase II (Pol II). Transcriptional termination on these genes generally requires a poly(A) (pA) signal, multiple trans-acting proteins and, in higher eukaryotes, additional downstream sequences (1). Two models have been proposed to explain the process: the allosteric and torpedo hypotheses. The allosteric model predicts that transcription of the pA signal induces a change in the Pol II elongation complex that leads to termination (2). The torpedo model postulates that a 5′→3′ exonuclease degrades the Pol II-associated product of pA site cleavage and promotes termination upon reaching the elongation complex (3,4). Accumulating evidence suggests that Pol II termination incorporates aspects of both of these models (5).
In humans and yeast, the 3′ products of pre-mRNA cleavage at the pA site are degraded by the 5′→3′ exonucleases, Xrn2 and Rat1, respectively, both of which are required for efficient Pol II termination (6–8). In addition to cleavage at the pA site, some higher eukaryotic pre-mRNAs are initially cleaved beyond the pA signal (9,10). The best example of co-transcriptional cleavage (CoTC) of pre-mRNA is in the human β-globin gene (10). Degradation of the 3′ product of CoTC, by Xrn2, is also correlated with transcriptional termination efficiency (11).
In addition to exonucleases, several proteins that play roles in 3′ end processing are also required for termination (12). Of these, Pcf11 has emerged as a key factor in the process. In yeast and Drosophila, Pcf11 is capable of promoting the release of Pol II from DNA in vitro and is also required for termination in vivo (12–15). Human Pcf11 (hPcf11) is a component of cleavage factor II (CFII), which is required for pre-mRNA cleavage during 3′ end processing (16). More recent studies showed that it facilitates premature termination at the HIV-1 promoter (17).
We have investigated the function of hPcf11 in Pol II termination using RNA interference (RNAi) to deplete it from HeLa cells. Depletion of hPcf11 in HeLa cells reduces the efficiency of transcriptional termination on transfected constructs and stabilizes the 3′ products of RNA cleavage at or beyond the pA site. Finally, we show that these functions of hPcf11 require an intact pA signal.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotide sequences are provided in Table 1.
Table 1.
Oligonucleotide sequences
| upA | GGGATATTATGAAGGGCCTTGAC |
| Rzr | GAACTAGCTCTTCATTTCTTTATG |
| Rzf | CCTTGGGAAAATACACTATATC |
| Vf | CAGGAAACTATTACTCAAAGGGTA |
| Vr | TTGAATCCTTTTCTGAGGGATG |
| pAr | AATCCAGATGCTCAAGGCC |
| HHr | CCTGTCACCGGATGTGTTTTCCGGTCT GATGAGTCCGTGAGGACGAAACAGG |
| HHr | CCTGTTTCGTCCTCACGGACTCATCAG ACCGGAAAACACATCCCGGTGACAGG |
| ΔpAf | CTGCCGAATTCAAAACATTTATTTTC |
| RT | AAACCCGACAGGACTATAAAGATAC |
| RTf | CAGGAAACTATTACTCAAAGGGTA |
| RTr | AGAAAATACCGCATCAGGCGC |
| Pf | ACCGCAGCAACAGCATCGATTATTCAA GAGATAATCGATGCTGTTGCTGCTTTTTC |
| Pr | TGCAGAAAAAGCAGCAACAGCATCGAT TATCTCTTGAATAATCGATGCTGTTGCTG |
| Pcf115′ | GAAGATCAAGATGTTCCAGATC |
| Pcf113′ | GTTCTTCCAGATCAGCTATCTC |
| Drosha5′ | GAGACCTAGCCTAGTTTTCCTG |
| Drosha3′ | AATGCACATTCACCAAAGTCAAG |
Constructs
pMAZ4 (8), pΔterm (previously called pΔCoTC) (11), pCoTC (11), pΔpA (previously called pCoTCΔpA) (11), Tat (18) and pRZ (previously called pHH) (11) have been described. przMAZ4 was made by inserting the annealed HHf/HHr primer pair into a vector prepared by Rzr/Rzf PCR amplification of pMAZ4. przMAZ4ΔpA was made by ligation of a pAr/ΔpAf PCR product obtained from przMAZ4. The hPcf11 shRNA expression construct was generated by inserting the annealed Pf/Pr primers into the siSTRIKE vector (Promega). This vector is provided pre-linearized.
Single-stranded M13 probes
The P and U3 (19), B3 and B4 (20) and A probes (11) have been described previously. M is empty M13 vector.
Transfection
Sub-confluent HeLa cells were transfected with 10 μg of reporter plasmid and 1 μg of Tat, using 20 μl of Lipofectamine 2000 (Invitrogen). RNA was isolated 12–24 h post-transfection.
RNA isolation
To isolate nuclear RNA, HeLa cell pellets were re-suspended in 0.5 ml of lysis buffer (10 mM Tris–HCl, 140 mM NaCl, 1.5 mM MgCl2 and 0.5% NP-40). After a 5-min incubation on ice, the suspension was under layered with 0.5 ml of lysis buffer containing 24% (w/v) sucrose. Tubes were spun at 13 000 r.p.m. in a bench-top centrifuge for 10 min. RNA was isolated from pelleted nuclei using Trizol (Invitrogen), following the manufacturers instructions. When required, total RNA was also isolated using Trizol. When analysing RZ cleavage by hsNRO, RNA was also isolated under denaturing conditions, using Trizol, in the absence of any divalent cations. This was to prevent in vitro RZ cleavage.
RT-PCR
Reverse transcription was performed using SuperScript III (Invitrogen) following the manufacturers guidelines. Extension temperatures were 37°C for oligo-dT and 55°C for all other primers. Real-time PCR analysis was performed using 10 pmol of each oligonucleotide, 7.5 μl of SYBR green mix (Qiagen) and 1/20th of the cDNA from reverse transcription. All of this was in a 15-μl final volume. For each sample, a control experiment was performed in the absence of reverse transcriptase to test for any DNA contamination. The value obtained for the minus reverse transcriptase control was deducted from that obtained in the presence of reverse transcriptase in order to obtain the RNA specific signal.
RNA interference
On Day 1, HeLa cells were transfected with 10 μg of plasmid expressing hPcf11-specific shRNA (described above) or a scrambled siRNA oligonucleotide (siCONTROL1 from dharmacon). On day 3, cells were transfected with the appropriate β-globin reporter plasmid, the Tat expression plasmid and, where required, the VA plasmid. Assays were performed on Day 4. hPcf11 and drosha mRNA were detected by PCR using primers Pcf115′/Pcf113′ and Drosha5′/Drosha3′, respectively, following cDNA synthesis with oligo-dT.
Hybrid selection
The hybrid selection procedure is described elsewhere, including the exon 3 biotinylated anti-sense probe that was used (19).
Nuclear run on (NRO) analysis
NRO analysis was performed as described in Ref. (20).
S1 nuclease analysis (S1A)
The probe to detect β-globin mRNA was prepared by digesting the relevant β-globin reporter plasmid with EcoR1, whilst the VA probe was prepared by digesting the VA plasmid with BamH1. The procedure for radiolabelling each probe and the protocol for the assay is taken from Ref. (21).
Western blotting
Western blotting was performed as described by Ref. (17). Blots were probed with anti-Pcf11 polyclonal anti-sera (1:500 dilution), which was a kind gift from David Gilmour's lab and anti-actin (1:2000 dilution), which is from SIGMA. Proteins were detected using an ECL kit (GE healthcare).
Quantitation
S1A and NRO experiments were quantitated using Microsoft imagequant software. Error bars represent the standard error obtained from a minimum of three repeats.
RESULTS
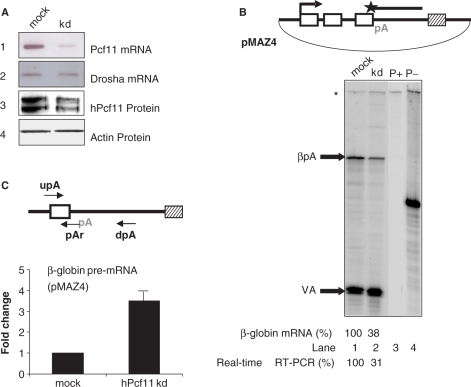
To investigate the functions of hPcf11 in vivo, we used RNAi to deplete it from HeLa cells. Depletion levels were measured using RT-PCR and western blotting to analyse hPcf11 mRNA and protein, respectively (Figure 1A). As compared to the mock-treated sample, hPcf11 mRNA was reduced in the shRNA-treated sample (kd, panel 1). This reduction varied between 2- and 3-fold between replicate experiments, as determined by real-time RT-PCR (data not shown). However, levels of drosha mRNA (panel 2), which was used as a control, remained similar. hPcf11 protein was also depleted by the shRNA treatment (panel 3). The multiple bands may correspond to different isoforms, previously seen by mass spectrometry (16). In contrast, actin protein levels were unchanged (panel 4). These data show that shRNA treatment causes specific partial depletion hPcf11 from HeLa cells. We predict that the essential nature of hPcf11 function may preclude more efficient depletion, since this would result in cell death.
Figure 1.
hPcf11 depletion and its effect on 3′ end processing. (A) Analysis of the efficiency of hPcf11 RNAi treatment. Panel 1; hPcf11 mRNA in mock cells or hPcf11-depleted cells (kd). Panel 2; drosha mRNA. Panel 3; hPcf11 protein. Panel 4; actin protein. (B) S1 analysis of β-globin mRNA levels in mock- and hPcf11-depleted cells (kd). The diagram shows the pMAZ4 plasmid, depicting β-globin exons (white boxes), pA signal (pA), MAZ4 termination element (hatched box) and S1 probe (shown above plasmid). Bands for the VA transfection control (VA) and β-globin mRNA (βpA) are indicated with arrows. Undigested probe is indicated with an asterisk. Lanes 3 and 4 show the β-globin and VA probes in the presence (P+) and absence (P–) of S1 nuclease, respectively. Note, that the P– lane only contains 20% of the probe used in the analysis to prevent it appearing overexposed on the gel. Quantitation measures the percentage of β-globin mRNA in kd cells as compared to mock cells (value fixed at 100%). Quantitation of a real-time RT-PCR experiment used to detect β-globin mRNA is also shown. (C) Real-time RT-PCR analysis of β-globin pre-mRNA, not cleaved at the pA site, in mock-treated and hPcf11-depleted cells (kd). The diagram shows primers used for reverse transcription (dpA) and for PCR (upA/pAr). Graph shows fold change in the abundance of this species. The value obtained in mock-treated cells is set to 1.
Previous results show that hPcf11 is required for pA site cleavage in vitro (16). To test this in vivo, mock- or shRNA-treated HeLa cells were transfected with pMAZ4 and the VA co-transfection control plasmid. pMAZ4 contains the β-globin gene, with transcription driven by the HIV promoter, followed by the MAZ4 termination element (Figure 1B). Levels of β-globin mRNA were determined using S1A with probes specific to the β-globin and VA transcripts (Figure 1B). After normalizing to the VA control signal, we observed that β-globin mRNA levels were reduced to ∼40% of the mock levels (lane 1) after depletion of hPcf11 (lane 2). These bands are specific to β-globin since they are not present in the control lane, which displays the probes and tRNA treated with S1 nuclease (lane 3). Lane 4 shows the undigested probes. This experiment was also validated using real-time RT-PCR to detect β-globin mRNA, which again revealed a reduction (to 31%) of the value for mock-treated HeLa cells (quantitation is shown underneath the S1A gel). No above-background signals were observed that represent unprocessed pre-mRNA, presumably because of its instability. Therefore, although this experiment strongly suggests that full levels of hPcf11 are required for 3′ processing in vivo, changes in transcription and/or mRNA turnover rates may also be responsible.
To rule out effects of hPcf11 on transcription and/or pre-mRNA turnover, mock-treated or hPcf11-depleted HeLa cells were transfected with pMAZ4 and the VA plasmid and nuclear RNA was isolated. This was reverse transcribed using the dpA primer and cDNA was real-time PCR amplified with pAR and upA to detect β-globin pre-mRNA, not cleaved at the pA site (Figure 1C). hPcf11 depletion resulted in a 3.5-fold increase in pre-mRNA levels as compared to mock treatment. The enhanced pre-mRNA accumulation indicates that transcription is not reduced by hPcf11 depletion nor is RNA turnover enhanced (these defects would most likely reduce levels of pre-mRNA). Moreover, the enhanced accumulation of pre-mRNA is consistent with the reduction in mRNA seen in Figure 1B. Taken together, results in Figure 1B and C indicate that hPcf11 is required for efficient 3′ end processing in vivo.
hPcf11 is required for efficient Pol II termination
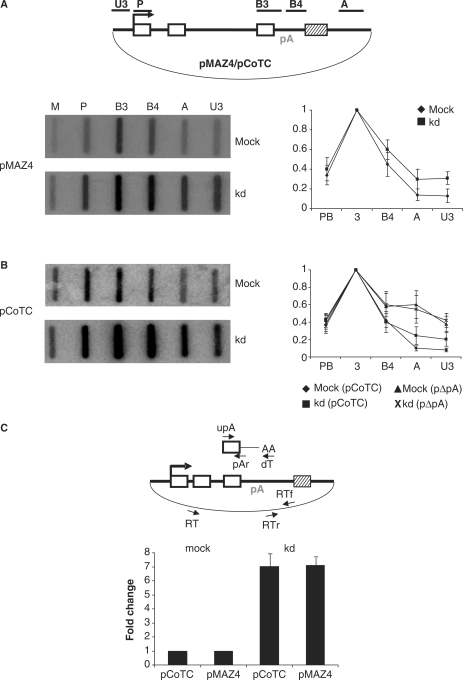
We next examined the potential role of hPcf11 in Pol II transcriptional termination by performing NRO. NRO measures the position and abundance of elongating Pol II via hybridization of de novo radiolabelled transcripts to complementary probes that are immobilized to a filter. We performed NRO on mock- and hPcf11-depleted HeLa cells transfected with pMAZ4 (Figure 2A). As we have previously observed, termination on pMAZ4 was efficient in mock-treated cells. This is shown by the low signals over probes downstream of the MAZ4 termination element (A and U3) compared to those upstream (P-B4). Depletion of hPcf11 reduces the efficiency of transcriptional termination on pMAZ4, causing a ∼2-fold increase in signals over read-through probes A and U3. Quantitation is shown in the accompanying graph.
Figure 2.
hPcf11 is required for efficient Pol II transcriptional termination. (A) Top diagram: plasmid used for NRO. HIV promoter (arrow), exons (white boxes), pA signal (pA) and MAZ4/CoTC termination element (hatched box) are shown. Data panel: NRO analysis of transcriptional termination on pMAZ4 in mock-treated and hPcf11-depleted (kd) cells. NRO probes are indicated above each slot and their positions are shown in the diagram. M is an empty M13 vector and controls for background hybridization. Quantitation is shown in the accompanying graphs. All probes values are relative to the B3 signal (set at 1). (B) NRO analysis of pCoTC in mock-treated and hPcf11-depleted (kd) HeLa cells. Probes are indicated above each slot and positions are as in A. The graph shows quantitation of the experiment and of a NRO that was performed on mock-treated and hPcf11kd cells transfected with pΔpA, which has a mutated pA signal and does not support termination. (C) Real-time RT-PCR analysis of transcriptional read-through on the pCoTC and pMAZ4 constructs in mock-treated and hPcf11-depleted (kd) cells. Primers for reverse transcription (oligo-dT and RT) and PCR (pAr/upA and RTr/RTf) are shown on the accompanying diagrams. Quantitation was determined by dividing the RTr/RTf signal by the pAr/upA signal. The value obtained in mock-treated cells was set to 1.
In addition to the MAZ4 termination sequence, work from our lab identified a sequence in the 3′ flank of the human β-globin gene, called the CoTC element, as an efficient transcriptional terminator (10). The CoTC element functions differently to the MAZ4 element, at least in part because the transcripts produced from it are co-transcriptionally cleaved. We were interested to determine how this alternative mode of termination was affected by hPcf11 depletion. NRO was performed on mock- and hPcf11-depleted HeLa cells transfected with pCoTC, which contains the CoTC element instead of the MAZ4 sequence (Figure 2B). Efficient termination was observed in mock-treated cells, since signal levels over read-through probes A and U3 are similar to those over the background M probe. However, depletion of hPcf11 resulted in elevated A and U3 signals, showing that efficient termination over the CoTC sequence also requires hPcf11. In contrast, very little change was observed over probes P-B4 as expected. Quantitation of this experiment is shown in the accompanying graph. Quantitation of a control NRO carried out on a construct with a mutated pA signal (pΔpA) is also shown. Since loss of pA signal function also promotes loss of Pol II termination, hPcf11 depletion causes no further effect.
Although NRO provides the best measure of termination efficiency, we sought to confirm that hPcf11 is required for termination using RT-PCR (Figure 2C). Nuclear RNA was isolated from mock-treated and hPcf11-depleted HeLa cells transfected with pCoTC or pMAZ4. cDNA was made with either oligo-dT or primer RT and then real-time PCR amplified with the pAr/upA primer pair (to detect β-globin mRNA) or the RTf/RTr primer pair (to detect read-through transcripts). Read-through was calculated as a ratio of these two products. For both the pMAZ4 and pCoTC, we observed 7-fold more read-through RNA in the hPcf11-depleted cells than in the mock-treated cells. This is an enhanced effect when compared to the above NRO, possibly due to the accumulation of read-through product over time. It is important to note that RT-PCR measures the steady-state RNA levels that accumulate over time, whereas NRO provides a ‘snap-shot’ of the consequences of shRNA treatment. Based on both NRO and RT-PCR experiments, we conclude that hPcf11 is required for efficient transcriptional termination on both pCoTC and pMAZ4.
hPcf11 is required to degrade the 3′ product of pA site cleavage
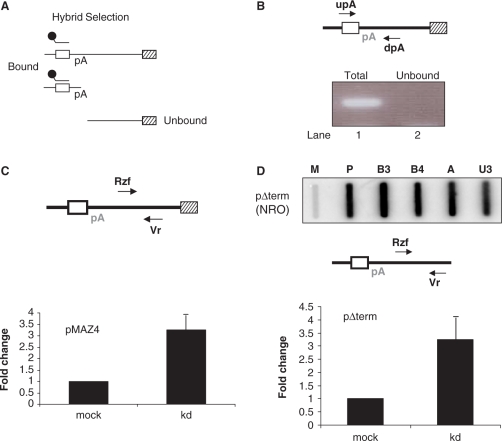
Recent studies show that yeast Pcf11 and Rat1 co-recruit each other to the 3′ end of genes (6). This indicates that they may cooperate in the transcriptional termination process. We next investigated potential hPcf11 effects on the function of Xrn2 (the human homologue of Rat1) in termination. We have shown that Xrn2 is required for degradation of the 3′ pA site cleavage product (8). We therefore tested the effect of hPcf11 on this degradation process using the hybrid selection technique (19). In brief, a biotinylated anti-sense RNA probe was hybridized to β-globin exon 3 and the resulting hybrids were bound to streptavidin-coated magnetic beads (Figure 3A). The selected bound RNA fraction includes β-globin mRNA and pre-mRNA not cleaved at the pA site. However, the 3′ product of pA site cleavage remains unbound because it is not continuous with exon 3. It should be noted that results in Figure 1B show hPcf11 to be required for full 3′-end processing efficiency. Therefore, depletion of hPcf11 would be anticipated to yield less of the 3′ product of pA site cleavage. Thus, these experiments were equalized to the signal obtained for β-globin mRNA (data not shown), which was normalized against the VA co-transfection control plasmid transcripts.
Although previous NRO experiments have shown that hybrid selection is very efficient (10), we performed a control RT-PCR experiment to establish this for our particular experiment. HeLa cells were transfected with pMAZ4 and nuclear RNA was isolated. The RNA was then reverse transcribed using primer dpA and the resulting cDNA was PCR amplified using the upA/dpA primer pair in order to detect RNA not cleaved at the pA site. A strong band was observed that represents this species (lane 1, Figure 3B). Next, an equivalent amount of the isolated RNA was selected with the biotinylated probe specific to exon 3. The unbound fraction of selected RNA was then analysed by RT-PCR under the same conditions. In this case, no band was detected (lane 2). The absence of un-cleaved β-globin pre-mRNA in the unbound fraction clearly shows that the hybrid selection technique is close to 100% efficient.
Figure 3.
hPcf11 is required to degrade the 3′ product of pA site/RZ cleavage. (A) Hybrid selection technique. β-globin RNA that is continuous with exon 3 (white box) is selected by the biotinylated probe (shown above exon 3) and bound by the streptavidin-coated magnetic beads. The 3′ product of pA site cleavage (bottom species) remains unbound. (B) RT-PCR analysis of pMAZ4 RNA from before hybrid selection (lane 1: total) and from the unbound RNA fraction obtained after selection (lane 2: unbound). Primers used for reverse transcription (dpA) and PCR (upA/dpA) are shown in the diagram. (C) Real-time RT-PCR analysis of the 3′ pA site cleavage product (obtained from the unbound fraction of selected RNA) in mock-treated or hPcf11-depleted (kd) cells transfected with pMAZ4 and VA. The β-globin primers used for cDNA synthesis (Vr) and PCR (Rzf and Vr) are indicated in the diagram. Quantitiation is normalized to the VA signal. The mock sample is given a value of 1. (D) Top data panel: NRO analysis of pΔterm. Probes are the same as in Figure 2A. Lower data panel: real-time RT-PCR analysis of the 3′ pA site cleavage product in mock-treated or hPcf11-depleted (kd) cells transfected with pΔterm. Primers used for cDNA synthesis (Vr) and PCR (Rzf and Vr) are shown in the diagram. Quantitiation is normalized to the VA signal. The mock sample is given a value of 1.
Next, we used hybrid selection to isolate the 3′ product of β-globin pA site cleavage. Mock-treated and hPcf11-depleted HeLa cells were transfected with pMAZ4 and the VA control plasmid and the unbound fraction of hybrid selected nuclear RNA was analysed using RT-PCR (Figure 3C). To detect the 3′ pA site cleavage product, cDNA was made with the Vr primer and was then PCR amplified using Vr and Rzf primers. After normalizing to the VA signal and levels of cleaved mRNA, we observed over 3-fold more of this RNA in the hPcf11-depleted sample as compared to the mock-treated sample. Data in Figure 3B show that this signal does not result from contaminating un-processed RNA, since this is efficiently removed by hybrid selection. Moreover, even when hybrid selection was not used to purify the 3′ pA site cleavage product, we observed enhanced accumulation of RNA beyond the pA site in hPcf11-depleted samples (data not shown). Therefore, these data indicate that hPcf11 is in some way required to degrade the 3′ product of pA site cleavage.
We next asked if the hPcf11-dependent degradation occurred even where transcriptional termination was inefficient. To do this, we repeated the above experiment on a construct (pΔterm) that contains the human β-globin gene without any downstream termination elements. The NRO in Figure 3D confirms that termination is very inefficient on pΔterm, as shown by the high A and U3 signals from read-through Pol II. To analyse the stability of the 3′ pA site cleavage product, pΔterm and VA were transfected into mock-treated and hPcf11-depleted HeLa cells and nuclear RNA was isolated and hybrid selected as described above. The unbound fraction of selected RNA was then used for RT-PCR analysis (Figure 3D). cDNA was made using the Vr primer and then PCR amplified using the Vr/Rzf primer pair. After normalizing to the VA and β-globin mRNA signals, we found ∼3.5-fold more of the Vr/Rzf product in the hPcf11-depleted sample as compared to the mock sample. This shows that degradation of the 3′ pA site cleavage product requires hPcf11, even in the absence of transcriptional termination. This also demonstrates that hPcf11 and RNA degradation are not sufficient for termination. These results therefore serve to highlight the complexity of Pol II termination in humans and the role that specific sequences, beyond the pA signal, play in the process.
The pA signal is required for hPcf11-dependent degradation of RNA
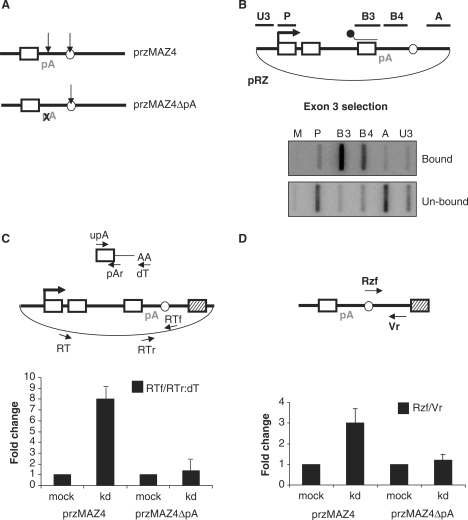
We next investigated the potential role of the pA signal in hPcf11-dependent RNA degradation. It should be noted that a construct containing a mutated pA signal would not provide this information since such a mutation abolishes cleavage at the pA site and so no ‘exposed’ 5′ RNA end would be presented to the degradation machinery. It was therefore necessary to create a situation where a degradable Pol II-associated RNA was produced in the absence of a pA signal. To achieve this, we created a further two constructs: przMAZ4, where a hammerhead ribozyme (RZ) was positioned between the pA signal and MAZ4 sequence (see diagram in Figure 4A) and przMAZ4ΔpA, in which the pA signal of przMAZ4 was inactivated by mutation. In each situation, RZ cleavage severs the RNA independently of a pA signal, which results in an exposed 5′ RNA end in the absence of pA site cleavage. We previously noted that RZ cleavage was not sufficient to promote transcriptional termination (11). However, some recent experiments have shown that positioning the MAZ4 element beyond the RZ promotes efficient termination and that this was preceded by 5′→3′ degradation of the Pol II-associated RZ cleavage product (West,S., Proudfoot,N.J. and Dye,M.J. submitted for publication). These same studies revealed that termination on przMAZ4 is just as efficient as on pMAZ4 and that a construct containing a RZ-inactivating point mutation also behaves like pMAZ4.
Figure 4.
hPcf11 function requires a functional pA signal. (A) Diagram of przMAZ4 and przMAZ4ΔpA. Final exon (white box), pA site (pA), RZ (white circle) and MAZ4 element (hatched box) are shown. pA site mutation is denoted by an X. Arrows represent sites of transcript cleavage. (B) hsNRO analysis of pRZ (shown in diagram). Transcripts were selected with the B3 probe (shown in the diagram above the final exon). Top panel shows selected transcripts (bound) and the bottom panel shows unselected transcripts (un-bound). NRO probes are shown in the diagram. (C) Real-time RT-PCR analysis of transcriptional termination on przMAZ4 or przMAZ4ΔpA in mock-treated or hPcf11-depleted (kd) cells. The plasmid is przMAZ4, where the RZ is shown by a white circle. The primers used for reverse transcription (RT) and PCR (RTf/RTr) are shown. The top diagram depicts the 3′ end of the mRNA (from the final exon to the poly(A) tail). Primers used for reverse transcription (oligo-dT) and PCR (upA/pAr) are indicated. Read-through was calculated as a ratio of the two PCR products and set at 1 in mock-treated cells. (D) Real-time RT-PCR analysis of the 3′ RZ cleavage product in mock-treated and hPcf11-depleted (kd) cells. Primers used for cDNA synthesis (Vr) and subsequent PCR (Rzf/Vr) are shown in the diagram. The RZ is again depicted by a white circle. Graph shows fold change where the value for mock-treated cells is given a value of 1.
We previously showed that this RZ cleaves co-transcriptionally in vivo (11). However, we needed to establish whether RZ cleavage was faster than β-globin pA site cleavage to know which cleavage event generated the initial Pol II-associated RNA on przMAZ4. To do this, we inserted the RZ beyond the β-globin pA signal, in pΔterm, to form pRZ. HeLa cells were transfected with pRZ and hybrid selection NRO (hsNRO) was performed. For hsNRO, radiolabelled nascent transcripts are selected with biotinylated probes (in the same way described in Figure 3). Both the selected RNA and the RNA that escaped selection were retained and hybridized to separate filters containing anti-sense M13 probes. It is important to note that the possibility of in vitro RZ cleavage during this experiment was minimized (see Materials and Methods section). The top panel in Figure 4B shows the profile of nascent pRZ RNA that was selected with the biotinylated probe. Strikingly, the only above-background signals (higher than probe M) in the selected fraction were over regions B3 and B4, which cover the region spanned by the selection probe and the downstream region, between the pA signal and RZ. Strikingly, transcripts beyond the RZ (that would hybridize to probes A and U3) are present exclusively in the unselected RNA (lower panel, Figure 4B). The absence of B3 signal in this fraction illustrates that selection of this RNA was close to 100% efficient. Furthermore, the absence of B4 signal in the unselected fraction demonstrates that cleavage at the pA site has not occurred. This result shows that transcripts continuous with exon 3 extend up to the RZ but not into region A, which lies only 350-bp downstream. Previous data from our lab showed that cleavage of the β-globin pA site does not take place until Pol II has transcribed at least 850 bp (19). Therefore, RZ cleavage is faster than β-globin pA site cleavage. These data show that on przMAZ4, the initial Pol II-associated RNA is produced by RZ cleavage.
We compared transcriptional termination efficiency on przMAZ4, in both mock- and hPcf11-depleted cells, using the RT-PCR assay described in Figure 2C (Figure 4C). cDNA was made with either oligo-dT or primer RT and then PCR amplified using the pAr/upA or the RTf/RTr primer pairs to detect β-globin mRNA or read-through RNA, respectively. There was ∼8-fold more read-through RNA in the hPcf11-depleted cells than in the mock-treated cells. This shows that hPcf11 is required for efficient termination on this construct. We next performed the same assay on mock- and hPcf11-depleted cells transfected with przMAZ4ΔpA. Mutation of the pA signal inhibits Pol II termination (1) and so high levels of read-through RNA are produced, even in mock-treated cells. Consequently, hPcf11 depletion had very little effect on the level of read-through RNA produced from this construct, which is in agreement with data on pΔpA, which is presented in Figure 2B. Although these data were predicted, given the anticipated role of hPcf11 in termination of Pol II, they further support conclusions drawn from experiments presented in Figures 1–3.
We next examined the effect of hPcf11 depletion on the stability of the 3′ RZ cleavage product to see if the reduced termination efficiency on przMAZ4 was correlated with enhanced stability of this species. Mock- and hPcf11-depleted HeLa cells were transfected with przMAZ4 together with the VA plasmid and nuclear RNA was isolated for RT-PCR analysis (Figure 4D). cDNA was made using the Vr primer and then PCR amplified with the Vr/Rzf primer pair in order to detect the 3′ RZ cleavage product. After normalizing to the VA signal, we observed ∼4-fold more of this species in hPcf11-depleted cells than in mock-treated cells. This indicates that the 3′ RZ product is a substrate for hPcf11-dependent degradation just as the 3′pA site cleavage product is. We finally repeated the experiment on przMAZ4ΔpA. In this case, hPcf11 depletion did not change the amount of 3′ RZ cleavage product. This is in spite of RZ cleavage creating a degradable substrate. Consequently, these results indicate that hPcf11-dependent degradation of cleaved RNA is completely coupled to transcription of a pA signal. This result further highlights the interconnections between 3′ end processing and transcriptional termination.
DISCUSSION
5′→3′ degradation of Pol II-associated RNA, following transcript cleavage, has been shown to be required for termination on the protein-coding genes so far tested (6,7,11,22). How this leads to Pol II release is unclear. Since digestion of Pol II-associated RNA in vitro does not cause disassembly of the ternary complex (23), other factors may be required. Recent studies on yeast and Drosophila Pcf11 show that it has the capability to dismantle the ternary complex in vitro (13,15). Moreover, it may aid recruitment of the Rat1 exonuclease to the 3′ end of genes in vivo (6). These observations have lead to a view that Rat1 and Pcf11 may cooperate to promote transcriptional termination as part of a mechanism that incorporates aspects of both the torpedo and allosteric models.
It was recently shown that hPcf11 affects transcription from the HIV promoter in the absence of other activating factors (17). This leads to the conclusion that hPcf11 may normally repress HIV gene expression in the absence of such factors. It is important to note that the constructs that we use rely on the HIV promoter for transcription. However, we do not anticipate that this influences our conclusions, since our experiments are performed in the presence of the trans-activating factor, Tat, which promotes high levels of transcription.
In vitro, Pcf11 releases stalled or paused Pol II much more efficiently than the processive elongating form (13,15). It is interesting that the MAZ4 element was originally identified as a Pol II pause element (24) and could therefore fulfil a role of slowing down the post-pA signal polymerase sufficiently for it to become susceptible to hPcf11. This may be why termination is inefficient on pΔterm, despite hPcf11 playing a role in degrading the 3′ pA site cleavage product. It is not known if the CoTC element functions to pause Pol II or whether cleavage of its transcripts is the key to its function in termination. Interestingly, Pol II can be stalled in vitro by digesting its nascent RNA to within 50 nt of the active site (25). This effect may be mimicked in vivo by rapid transcript cleavage, such as that promoted by CoTC.
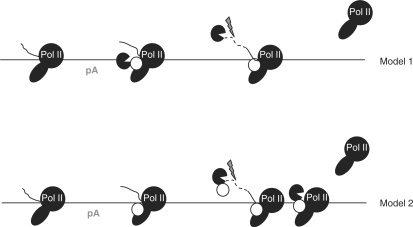
The necessity of hPcf11 for RNA degradation could be conveniently explained by invoking a complex containing Xrn2 and hPcf11, which is recruited to Pol II after transcription of the pA signal (Figure 5, Model 1). Cleavage at the pA site (or at CoTC sites, in the case of pCoTC) would precede degradation of Pol II-associated RNA by Xrn2, assisted by hPcf11. However, it is notable that depletion of Rat1 or Xrn2 does not effect cleavage at the pA site (7,11,22), even though it has been claimed that Rat1 is required for the efficient recruitment of Pcf11 and other factors to genes, which are themselves required for 3′ end processing (6). It is therefore also possible that Xrn2 and hPcf11 are not recruited to genes in a 1:1 ratio (Figure 5, Model 2). In vitro, Pcf11 binds more avidly to Pol II CTD that is phosphorylated on serine 2 than to un-phosphorylated CTD or CTD phosphorylated on serine 5 (26,27). However, deletion of the gene encoding the protein responsible for serine 2 phosphorylation in yeast (CTK1) does not effect either the recruitment of Rat1 to genes nor transcriptional termination (7). Therefore, Xrn2 may recruit further hPcf11 to the cleaved transcript in addition to the hPcf11 involved in 3′ end processing.
Figure 5.
Model. Model 1: after transcription of the pA signal (pA), hPcf11 (white circle) and Xrn2 (black pack-man) are co-recruited to Pol II. Following cleavage at the pA site/RZ/CoTC (shown by grey lightening), Xrn2 degrades Pol II-associated RNA and, together with hPcf11 and a termination element (hatched box) promotes Pol II release. Model 2: hPcf11 is recruited to Pol II upon transcription of the pA signal. However, a further hPcf11 is recruited to the cleaved RNA transcript by Xrn2. Again, degradation of the Pol II-associated transcript, enhanced by hPcf11 function, promotes Pol II release at the termination element.
ACKNOWLEDGEMENTS
We would like to thank Michael Dye for helpful suggestions. We are grateful to Shawn Cowan and David Gilmour for the anti-sera used to detect hPcf11 protein. S.W. is a Junior Research Fellow at Oriel College, Oxford. These studies were supported by a Programme Grant from the Wellcome Trust to N.J.P. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 2.Logan J, Falck-Pedersen E, Darnell JE, Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl Acad. Sci. USA. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proudfoot NJ. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 4.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 5.Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 6.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 8.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West S, Zaret K, Proudfoot NJ. Transcriptional termination sequences in the mouse serum albumin gene. RNA. 2006;12:655–665. doi: 10.1261/rna.2232406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dye MJ, Proudfoot NJ. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 11.West S, Gromak N, Proudfoot NJ. Human 5′ –> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 12.Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Fu J, Gilmour DS. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol. Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 16.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Klatt A, Henderson AJ, Gilmour DS. Transcription termination factor Pcf11 limits the processivity of Pol II on an HIV provirus to repress gene expression. Genes Dev. 2007;21:1609–1614. doi: 10.1101/gad.1542707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams SE, Johnson ID, Braddock M, Kingsman AJ, Kingsman SM, Edwards RM. Synthesis of a gene for the HIV transactivator protein TAT by a novel single stranded approach involving in vivo gap repair. Nucleic Acids Res. 1988;16:4287–4298. doi: 10.1093/nar/16.10.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 20.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashe MP, Griffin P, James W, Proudfoot NJ. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu W, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl Acad. Sci. USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashfield R, Enriquez-Harris P, Proudfoot NJ. Transcriptional termination between the closely linked human complement genes C2 and factor B: common termination factor for C2 and c-myc? EMBO J. 1991;10:4197–4207. doi: 10.1002/j.1460-2075.1991.tb04998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ujvari A, Pal M, Luse DS. RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 2002;277:32527–32537. doi: 10.1074/jbc.M201145200. [DOI] [PubMed] [Google Scholar]
- 26.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 27.Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]