Abstract
The TOPRIM DXDXXG residues of type IA and II topoisomerases are involved in Mg(II) binding and the cleavage-rejoining of DNA. Mutation of the strictly conserved glycine to serine in Yersinia pestis and Escherichia coli topoisomerase I results in bacterial cell killing due to inhibition of DNA religation after DNA cleavage. In this study, all other substitutions at the TOPRIM glycine of Y. pestis topoisomerase I were examined. While the Gly to Ala substitution allowed both DNA cleavage and religation, other mutations abolished DNA cleavage. DNA cleavage activity retained by the Gly to Ser mutant could be significantly enhanced by a second mutation of the methionine residue adjacent to the active site tyrosine. Induction of mutant topoisomerase with both the TOPRIM glycine and active site region methionine mutations resulted in up to 40-fold higher cell killing rate when compared with the single TOPRIM Gly to Ser mutant. Bacterial type IA topoisomerases are potential targets for discovery of novel antibiotics. These results suggest that compounds that interact simultaneously with the TOPRIM motif and the molecular surface around the active site tyrosine could be highly efficient topoisomerase poisons through both enhancement of DNA cleavage and inhibition of DNA rejoining.
INTRODUCTION
DNA topoisomerases can carry out the important functions of DNA supercoiling regulation and DNA untangling because these enzymes can catalyze the interconversion of DNA topological forms by the concerted breaking and rejoining of DNA strands coupled to DNA strand passage through the DNA cleavage sites (1–4). A covalent enzyme–DNA complex is formed after nucleophilic attack of an active site tyrosine on the DNA phophodiester backbone during the DNA cleavage step. Topoisomerase poisons are compounds that interfere with the DNA cleavage-rejoining equilibrium of topoisomerases and increase the physiological concentration or life-time of the covalent complex intermediates formed between topoisomerases and cleaved DNA, resulting in events that lead to cell death (5,6). Many clinically important antibacterial and anticancer drugs are topoisomerase poisons targeting type IB and type IIA topoisomerases (5–10). Both type IA and type IB topoisomerases cut and rejoin a single-strand of DNA during catalysis but type IA topoisomerases are more similar to type IIA topoisomerases in catalytic mechanism of DNA cleavage and rejoining (3).
Based on genome sequences and genetic studies, there is at least one type IA topoisomerase activity required to be present in every organism to resolve entanglement of single-stranded DNA during replication or recombination (2,11). Although topoisomerase poison known to target type IA topoisomerase with high specificity is not available currently, it has been demonstrated that trapping of covalent complex intermediate formed by a mutant form of recombinant Yersinia pestis or Escherichia coli topoisomerase I can result in rapid and extensive cell killing in E. coli (12). This cell killing effect from the stabilization of topoisomerase I cleavage complex validates the potential of bacterial type IA topoisomerase as a target for development of novel antibacterial compounds to combat multi-drug resistant bacterial pathogens (12–14). The cell killing mutation previously characterized was identified by screening a random recombinant Y. pestis topoisomerase I mutant library for induction of cellular SOS response in E. coli, and involves change of the strictly conserved Gly residue in the TOPRIM motif (DXDXXG) to a Ser residue (12). The understanding of how DNA cleavage-rejoining can be interfered with at the molecular level is important for the elucidation of how the enzyme maintains the DNA cleavage-rejoining equilibrium so that the degree of DNA cleavage is sufficient for the required conversion of DNA topology, but avoids toxic level of cleaved DNA complex being accumulated. Such biochemical investigations could also be useful for the design of novel type IA topoisomerase poisons or modification of hit molecules identified via high through-put screening in the development of antibacterial compounds targeting type IA topoisomerases (13,14). In this study, it is shown that the TOPRIM Gly to Ser mutation in subdomain I is unique in its cell killing capability among all other possible substitutions at the conserved Gly residue because it maintains a sufficient level of DNA cleavage activity while DNA religation is abolished. All other possible amino acid substitutions at the conserved glycine residue of the TOPRIM motif were examined and found not to result in cell killing. Every other substitution except Ala had no detectable DNA cleavage or relaxation activity. Unlike the Gly to Ser substitution, DNA religation was not inhibited by the Gly to Ala substitution. We also demonstrated that in a different subdomain of the enzyme (subdomain III), although the conversion of the highly conserved methionine residue adjacent to the active site tyrosine of Y. pestis topoisomerase I did not inhibit relaxation of DNA or affect cell viability by itself, the mutation enhanced DNA cleavage by the enzyme. When this increase in DNA cleavage was combined with the inhibition of DNA rejoining by the TOPRIM Gly to Ser mutation, the level of DNA cleavage product and degree of cell killing of the double mutant were increased significantly over the TOPRIM Gly to Ser single mutant. This demonstrates that molecular interactions near the active site tyrosine of type IA topoisomerases can also affect the DNA cleavage-religation equilibrium and be synergistic with other molecular perturbations elsewhere in the enzyme that inhibit DNA rejoining to achieve higher degree of DNA cleavage product accumulation and overall bactericidal action.
MATERIALS AND METHODS
Site-directed mutagenesis of recombinant topoisomerase I
Site-directed mutagenesis was carried out with using Pfu Ultra DNA polymerase (from Stratagene) with procedures based on the Stratagene QuikChange protocol. Oligonucleotide primers coding for the desired amino acid substitutions were synthesized by Sigma Genosys. Random substitution of Y. pestis topoisomerase I (YTOP)2 at Gly122 was achieved with plasmid pYTOP (12) as template and primers 5′GACCTTGATCGCGAANNNGAGGCTATTGCCTG 3′ and 5′ CAGGCAATAGCCTCNNNTTCGCGATCAAGGTC 3′. YTOP mutants with substitutions of Gly122 with Asn, His, Trp and Tyr or Met326 substitution with Val were created by using oligonucleotides specifying these substitutions. Recombinant E. coli topoisomerase I (ETOP) with Val substitution at Met320 were made by introducing the site-directed mutation into plasmid pETOP or pETOP-G116S (12). In the pYTOP and pETOP plasmids, the synthesis of the recombinant topoisomerase I is under the control of the arabinose-inducible BAD promoter. In addition to ampicillin, the plates and media used for isolation, maintenance and overnight growth of the E. coli strains transformed with plasmids pYTOP or pETOP contained 2% glucose to suppress the expression of the potentially lethal mutant topoisomerases from the BAD promoter.
Identification of SOS inducing YTOP mutants
Escherichia coli strain JD5 with chromosomal dinD1::lacZ fusion (15) was transformed with plasmid pYTOP derivatives encoding mutant YTOP enzymes. Transformations obtained on LB plates with 2% glucose and 100 μg/ml ampicillin were replicated onto plates with ampicillin, 35 μg/ml X-gal and 0.002% arabinose to identify SOS-inducing mutants that gave rise to blue colonies after overnight incubation at 37°C due to induction of the dinD1 promoter (16).
Effect of recombinant topoisomerase expression on viability
The ability of the YTOP mutants with different substitutions at Gly122 to cause bacterial cell death was first evaluated in strain JD5 by inducing their expression with saturating concentration of arabinose (0.2%) for 2 h at early exponential phase (OD600 = 0.4) after dilution of overnight culture in the presence of 2% glucose into fresh LB medium with antibiotics but no glucose. Viable counts were measured by plating of dilutions on LB plates with 2% glucose and ampicillin and compared with viable counts from cultures not induced with arabinose after overnight incubation of the plates at 37°C. The cell killing effects of mutant recombinant topoisomerases with the Gly to Ser substitution were also measured at a range of lower arabinose concentrations (0.00006–0.002%) in E. coli strain BW27784 (from Yale E. coli Genetic Stock Center). In this strain, the control of the arabinose transporter araE gene by a constitutive promoter (17) allows expression from the BAD promoter to be regulated by increasing arabinose concentration instead of the ‘all-or-none’ expression pattern found in strain JD5 (12,18). This enabled comparison of the cell killing efficiency with increasing arabinose concentrations.
Protein purification
Wild-type and mutant YTOP proteins were induced in E. coli JD5 strain after growth in LB medium with ampicillin to exponential phase with either 0.02 or 0.2% arabinose for 4 h at 37°C. The recombinant proteins with thioredoxin N-terminal tag and His6 C-terminal tag were purified with the His SpinTrap column (GE Healthcare) according to manufacturer's protocol. Recombinant ETOP mutant proteins without affinity tags were expressed in E. coli strain GP200 (ΔtopA) and purified by combination of phosphocellulose, hydroxyapitite and DNA affinity chromatography columns as previously described (19,20).
Enzyme activity assays
Each assay was carried out at least thrice, and representative results are shown here. Assays of relaxation of negatively supercoiled DNA by topoisomerase I was assayed in the presence of 6 mM MgCl2 as described previously (20). Cleavage of plasmid DNA was assayed in buffer with either no added Mg2+ or with up to 10 mM MgCl2 added as described (12). A 59-base oligonucleotide 5′-GCCCTGAAAGATTATGCAATGCGCTTTGGGCAAACCAAGAGAGCTAATCTTTCAGGGC-3′ with the preferred cleavage site CAAT↓GC for ETOP (21) was labeled at the 5′-end with T4 polynucleotide kinase and [γ32P-ATP]. Cleavage and religation of the oligonucleotide by topoisomerase I were assayed as described and analyzed by electrophoresis in a 15% DNA sequencing gel (22). The wild-type topoisomerase I religation reaction is complete after 10 s at 37°C so some of the religation reactions were carried out on ice at 0°C. Gel shift assay with the same 5′-end labeled oligonucleotide was used to compare non-covalent binding affinities of wild-type and mutant ETOP enzymes with the DNA substrate (22). To assess the effect of the mutations on Mg2+-binding affinity of ETOP, measurements of change in intrinsic tryptophan fluorescence of ETOP enzymes from binding of Mg2+ (20,23) were carried out as described previously (12). Non-linear regression curve fitting for two binding sites was carried out using the GraphPad Prism program.
RESULTS
Only Ser substitution at Gly122 of YTOP produced the SOS inducing and cell killing phenotypes by inhibiting DNA religation while retaining DNA cleavage activity
To determine if in addition to substitution with Ser, other types of amino acid substitutions at Gly122 of Y. pestis topoisomerase I can have SOS inducing and cell killing properties, random mutations at Gly122 were first created by oligonucleotide-directed mutagenesis with all four possible nucleotides at the codon positions for Gly122 of recombinant YTOP expressed under the tight control of the BAD promoter. After sequencing 60 mutants with random substitutions at Gly122 and screening for SOS induction using the dinD1::lacZ reporter strain JD5, only the Ser substitution mutants were found to have the SOS-inducing phenotype on X-gal plates with low level of arabinose (0.002%). Gly122 substitutions to Asn, His, Trp and Tyr were the only possible substitutions not found among the 60 sequenced YTOP mutants. These substitution mutants were then created by sequence specific oligonucleotide-directed mutagenesis. These four mutants were also found not to induce SOS in strain JD5. The effect of overexpression of 13 of the YTOP Gly122 substitution mutants on cell viability in strain JD5 were measured by determination of viable counts at 2 h after induction with saturating concentration of arabinose (0.2%). The list of mutants analyzed for effect on viability included those substitutions most similar to Gly and Ser in size or having the same functional group in the side chain (Thr and Tyr), as well as some examples of substitutions that are much larger in size and hydrophobic in nature. The results (Table 1) confirmed that these YTOP Gly122 substitution mutants that did not induce the SOS DNA damage response also had relatively small effect on cell viability when compared with the Gly122 to Ser substitution YTOP mutant. The substitution with cysteine had a greater effect on viability (induced/non-induced relative viability = 0.03) than the other substitutions examined (average relative viability = 0.11), but the relative viability of the cysteine substitution is still 100-fold higher than that obtained for the serine substitution.
Table 1.
Effect of overexpression of recombinant wild-type or mutant Y. pestis topoisomerase I with substitutions at Gly122 on the viability of E. coli JD5
| Subsitution at Gly122 | Induced/non-induced relative viability |
|---|---|
| none | 0.17 ± 0.11 |
| Ser | 5.2 × 10−4 ± 1.8 × 10−5 |
| Ala | 0.093 ± 0.015 |
| Asp | 0.19 ± 0.02 |
| Asn | 0.17 ± 0.10 |
| Cys | 0.030 ± 0.004 |
| Glu | 0.092 ± 0.040 |
| Gln | 0.11 ± 0.06 |
| His | 0.14 ± 0.02 |
| Ile | 0.086 ± 0.033 |
| Phe | 0.075 ± 0.035 |
| Thr | 0.087 ± 0.018 |
| Trp | 0.13 ± 0.05 |
| Tyr | 0.088 ± 0.016 |
Relative viability (RV) was measured by the ratio of the viable colonies obtained after induction of the BAD promoter directing the expression of recombinant YTOP with 0.2% arabinose for 2 h in comparison with viable colonies from the culture not treated with arabinose. The results shown represent the average and standard deviation from at least three measurements.
Effect of the Gly122 substitutions on YTOP enzyme activity
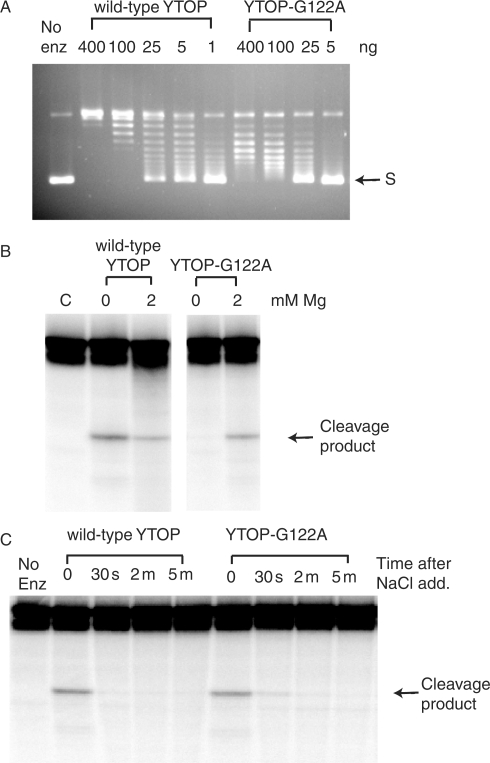
The YTOP mutants with every possible Gly122 substitutions were purified for assay of relaxation activity and DNA cleavage activity. The expression level of each of the mutant proteins in the soluble extract of E. coli JD5 was similar to that of the wild-type YTOP protein. The G122A mutant YTOP was found to retain around 10% of the relaxation activity (Figure 1A). All the other substitution mutants, similar to the G122S mutant (12), had no detectable relaxation activity (data not shown). DNA cleavage activity was assayed both in the absence and presence of up to 10 mM MgCl2 in the reaction. The addition of Mg2+ is not necessary for the DNA cleavage activity of E. coli and Y. pestis topoisomerase I to be observed (12,24), but is necessary for DNA rejoining (12,24,25). For the TOPRIM Gly to Ser mutant, Mg2+-binding affinity has been found to be reduced and it was necessary to have Mg2+ added to the reaction mixture to observe DNA cleavage (12). The YTOP-G122A mutant enzyme was also dependent for DNA cleavage activity and had no detectable DNA cleavage activity in the absence of added Mg2+ (Figure 1B). For the wild-type YTOP enzyme, addition of MgCl2 promotes religation, so the amount of cleavage product in the presence of 2 mM MgCl2 was reduced by 56% (from densitometry analysis of four sets of data) when compared with that observed in the absence of Mg2+ (Figure 1B, left panel). In the presence of 2 mM MgCl2, the amount of cleavage product formed by the G122A mutant was about the same as the wild-type enzyme (122% from three sets of data). While the YTOP-G122S mutant enzyme has no detectable DNA rejoining activity (12), YTOP-G122A mutant enzyme was found to be capable of DNA rejoining (Figure 1C). For both wild-type YTOP and YTOP-G122A, rejoining of cleavage product reached >90% of the maximal level within 30 s after the addition of MgCl2 and high salt at 37°C.
Figure 1.
Enzymatic activities of the YTOP-G122A mutant. (A) Relaxation of supercoiled DNA. The indicated amount of wild-type or G122A mutant YTOP protein was incubated in relaxation buffer containing 6 mM MgCl2 at 37°C for 30 min. S: supercoiled plasmid DNA. (B) Magnesium dependent formation of cleavage product from oligonucleotide DNA substrate by wild-type YTOP and YTOP-G122A mutant enzyme. 400 ng of enzyme was incubated with 0.5 pmole of 5′-end labeled oligonucleotide substrate for 30 min at 37°C. The cleavage product was visualized by PhosphorImager after electrophoresis in a 15% sequencing gel. (C) Religation of DNA cleavage product. DNA cleavage reaction between 400 ng of enzyme and 0.5 pmole of oligonucleotide substrate in the presence of 5 mM MgCl2 was allowed to reach cleavage-religation equilibrium and then treated with 1 M NaCl to promote dissociation of the enzyme from the religated DNA. The religation reactions were stopped after the indicated amount of time by the addition of stop solution containing 0.2 M NaOH and 79% formamide.
DNA cleavage was not observed for any of the other YTOP G122 substitution mutant protein in the absence of added Mg2+ and the addition of up to 10 mM Mg2+ did not restore the DNA cleavage activity of any of the other Gly122 substitution mutants (data not shown). Due to its more significant effect on viability among the mutants examined, DNA cleavage activity of YTOP-G122C was also examined using 5′-end labeled oligonucleotide as substrate. DNA cleavage activity was again not detectable, so the small effect on viability from overexpression of YTOP-G122C was probably unrelated to the effect of the mutation on DNA cleavage-religation. It can be concluded that the Gly to Ser substitution is unique in its cell killing phenotype because it is the only amino acid substitution at this critical position that inhibits DNA religation completely while retaining a high degree of DNA cleavage activity.
Mutation of the conserved Met residue adjacent to the active site tyrosine to Val can enhance the cell killing activity of the TOPRIM Gly to Ser topoisomerase I mutant
The sequence of the SOS-inducing YTOP128 mutant isolated in the original mutant screening had two other mutations, M326V and A383P, in addition to the TOPRIM G122S mutation (12). Site-directed mutagenesis showed YTOP with single M326V or A383P mutation had no SOS-inducing or significant cell killing effect in strain JD5 (12). The cell killing effect from YTOP-G122S was slightly lower than the original YTOP128 mutant in strain JD5 (12). Met326 follows the active site tyrosine residue Tyr325 in YTOP sequence and is conserved in bacterial topoisomerase I sequences, while a proline is often found in other type IA topoisomerases at this position (26). To determine the effect of the additional M326V mutation on the cell killing efficiency of YTOP-G122S, the double mutant YTOPG122S/M326V was created by site-directed mutagenesis on the low copy number expression plasmid pAYTOP derived from the cloning vector pACYC184 (14). In the genetic background of JD5, arabinose induction of the BAD promoter is ‘all or none’ because the araC-PBAD system and the associated L-arabinose transporter AraE are regulated autocatalytically by arabinose (18). The experiments in JD5 therefore utilized the saturating concentration of arabinose (0.2%) for measurement of cell viability. In strain BW27784, the synthesis of AraE is arabinose independent due to its control under a constitutive promoter (17), allowing increasing amount of YTOP proteins to be expressed from the BAD promoter by increasing concentrations of arabinose added to the culture. Strain BW27784 was used to compare the cell killing efficiency of the low copy number plasmid pAYTOP expressing wild-type YTOP, and its derivatives expressing YTOP-Gl22S or YTOP-G122S/M326V, as well as the original YTOP128 mutant over a range of non-saturating concentrations of arabinose (Table 2). The results demonstrated that the addition of the M326V mutation to the YTOP-G122S mutant background enhanced further the cell killing efficiency of the resulting YTOP-G122S/M326V mutant topoisomerase I protein by up to 40-fold. The cell killing efficiency of the YTOP-G122S/M326V double mutant in strain BW27784 was found to be nearly identical to that of the original YTOP128 mutant with in addition to G122S and M326V mutations, the A383P substitution also present.
Table 2.
Effect of arabinose concentration on the viability of E. coli BW27784 expressing different recombinant YTOP proteins under the control of the BAD promoter
| Arabinose concentration (%) | R.V. pAYTOP | R.V. pAYTOP128 | R.V. pAYTOP-G122S | R.V. pAYTOP-G122S/M326V |
|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 1 |
| 0.00006 | 0.84 ± 0.09 | 0.014 ± 0.005 | 0.065 ± 0.007 | 0.016 ± 0.007 |
| 0.0002 | 0.48 ± 0.16 | 0.0014 ± 0.0002 | 0.020 ± 0.003 | 0.0012 ± 0.0009 |
| 0.002 | 0.25 ± 0.11 | 0.00035 ± 0.00008 | 0.0091 ± 0.0006 | 0.00023 ± 0.00014 |
Relative viability was determined by calculating the ratio of the viable colony counts after 2 h of induction with the indicated arabinose concentration versus the viable colony counts from the non-induced culture. The recombinant YTOP proteins were expressed in low copy number plasmids (pAYTOP and its derivatives). The results shown represent the average and standard deviation of at least four different measurements for the mutant YTOP proteins and three different measurements for wild-type YTOP. Mutant pAYTOP128 had a third A383P substitution in addition to G122S and M326V mutations. Single mutation of A383P or M326V in YTOP did not induce SOS response when induced by arabinose and had no significant effect on viability (12).
The Met to Val substitution adjacent to the active site tyrosine enhanced DNA cleavage
The effect of the substitution of Met326 with Val on YTOP protein activity was further analyzed to determine the biochemical basis for this enhancement of cell killing. When the enzyme activity of YTOP-M326V was examined, it was found to have no significant effect on the relaxation activity when compared with wild-type YTOP enzyme (Figure 2A). Although YTOP-M326V had no SOS-inducing or cell killing effect in vivo (12), the amount of cleavage product formed by this mutant was higher than that from wild-type YTOP (Figure 2B). Densitometry analysis of results from three experiments showed that the Met to Val substitution resulted in 1.9-fold higher cleavage product than wild-type YTOP in the absence of Mg2+. However, the percent of decrease of cleavage product upon addition of 2 mM MgCl2 was higher for YTOP-M326V (76%) than wild-type YTOP (53%), so the amount of cleavage product was approximately equal for YTOP-M326V and wild-type YTOP at 2 mM MgCl2. This is in good agreement with the lack of SOS induction or cell killing by the YTOP-M326V mutant in vivo with Mg2+ present.
Figure 2.
Effect of the M326V mutation on the relaxation and cleavage activities of YTOP. (A) YTOP-M326V has similar relaxation activity as wild-type YTOP enzyme. S: supercoiled plasmid DNA. (B) YTOP-M326V DNA cleavage activity was magnesium independent, and was enhanced over the wild-type DNA cleavage activity. Formation of oligonucleotide cleavage product was assayed with 400 ng of enzyme, 0.5 pmole of 5′-end labeled oligonucleotide substrate after incubation for 30 min at 37°C.
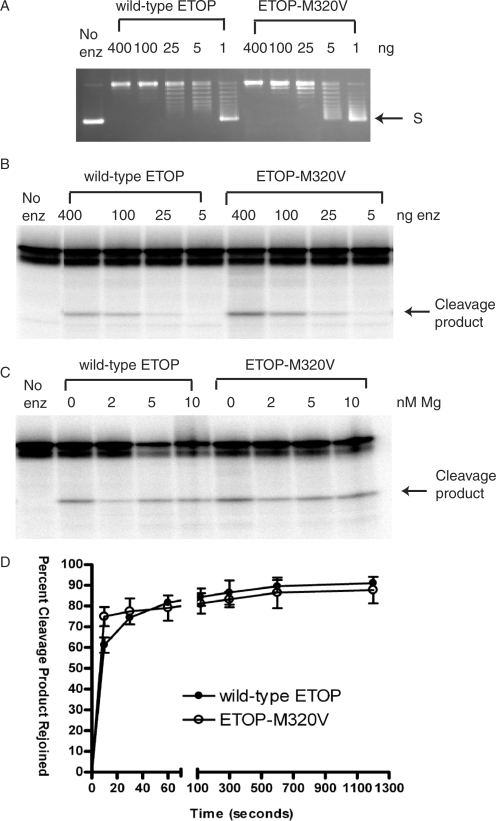
To study the effect of this mutation on the enzymatic activity of type IA topoisomerases with the better characterized system of ETOP, the corresponding M320V and M320V/G116S ETOP mutant enzymes were expressed from the BAD promoter in plasmid pETOP (12). These mutant enzymes without any linked affinity tags were purified by a combination of conventional chromatography procedures and compared with wild-type ETOP and the ETOP-G116S mutant enzyme characterized previously (12). Similar to YTOP-M326V enzyme, ETOP-M320V protein had wild-type relaxation activity (Figure 3A), but enhanced DNA cleavage activity (Figure 3B and C). Densitometry analysis of the cleavage products from three sets of data showed that the level of cleavage product formed by the M320V mutant enzyme was 2.2-fold that of the wild-type ETOP in the absence of Mg2+. Similar to wild-type YTOP, the presence of 2 mM or higher MgCl2 promoted religation by ETOP, so there was a decrease in the cleavage products formed with both wild-type and M320V ETOP upon addition of MgCl2. Religation of cleavage product in the presence of MgCl2 and high salt was complete within 10 s at 37°C, so the religation reaction was carried out on ice at 0°C. At this temperature, religation by wild-type ETOP was not complete at 10 s. Religation rate by the ETOP-M320V enzyme was found to be slightly greater (∼20%) at 10 s.
Figure 3.
The M320V mutation enhanced DNA cleavage by ETOP without affecting the relaxation and religation activities significantly. (A) Similar relaxation activity for wild-type ETOP and ETOP-M320V assayed in the presence of 6 mM MgCl2 at 37°C for 30 min. S: supercoiled plasmid DNA. (B) Comparison of cleavage of oligonucleotide substrate by wild-type ETOP and ETOP-M320V enzymes. The indicated amount of enzyme was incubated with 0.5 pmole of substrate for 30 min at 37°C in the absence of any MgCl2. (C) ETOP-M320V does not require magnesium for DNA cleavage. 400 ng of enzyme was incubated with 0.5 pmole of substrate at 37°C for 10 min. (D) DNA religation by ETOP-M320V mutant enzyme. The olignonucleotide substrate was incubated with 400 ng of enzyme for 10 min at 37°C in the absence of added MgCl2 before being placed on ice and the addition of 5 mM MgCl2 along with 1 M NaCl to promote DNA religation of the cleavage product and subsequent dissociation of the enzyme from the religated DNA. The reactions were allowed to proceed at 0°C for the indicated length of time before being stopped for analysis of the remaining cleavage product by gel electrophoresis.
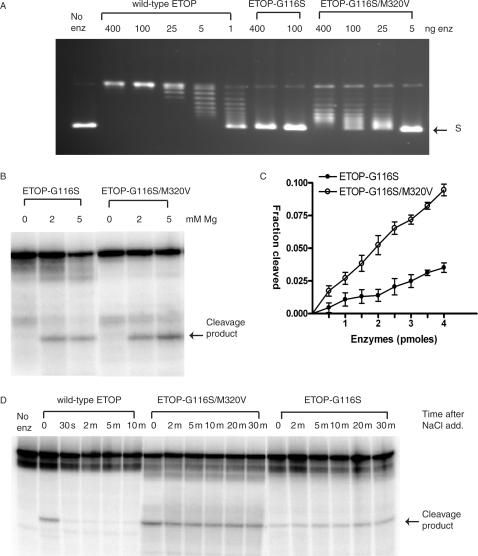
However, any small enhancement of DNA religation rate from the Met to Val substitution was overcome by the presence of the more dominant Gly to Ser mutation in the double mutants. The relaxation and religation activities of the double mutant ETOP-G116S/M320V enzyme were strongly inhibited (Figure 4A and D), as expected from the known effect of the G116S mutation on ETOP from previous study (12). DNA cleavage activity was Mg2+ dependent as in the case of the ETOP-G116S mutant (Figure 4B), and the level of DNA cleavage product formed by the ETOP-G116S/M320V mutant was increased over the ETOP-G116S mutant by ∼3-fold (Figure 4C). The increase in DNA cleavage provided a basis for the enhancement of cell killing in vivo when the Met to Val mutation in subdomain III was combined with the subdomain I TOPRIM Gly to Ser mutation.
Figure 4.
Effect of addition of the M320V mutation to the ETOP-G116S mutant enzyme activities. (A) Restoration of a low level of relaxation activity to the ETOP-G116S mutant enzyme by the M320V mutation. The indicated amount of enzyme was incubated with supercoiled plasmid DNA in buffer containing 6 mM MgCl2 at 37°C for 30 min. S: supercoiled plasmid DNA. (B) Dependence of magnesium for DNA cleavage. 400 ng of enzyme was incubated with 0.5 pmole of substrate for 30 min at 37°C. (C) Quantitation of oligonucleotide (5 pmole) cleavage by ETOP-G116S (filled circles) and ETOP-G116S/M320V (open circles) enzymes in the presence of 5 mM MgCl2. The data plotted represents the average and standard deviation from three different experiments. (D) DNA religation inhibition for the ETOP-G116 and ETOP-G116S/M320V mutants. The oligonucleotide substrate was incubated with 400 ng of enzyme in the presence of 5 mM MgCl2 at 37°C for 10 min at 37°C before the addition of 1 M NaCl and further incubation at 37°C for the indicated length of time.
The addition of the M320V mutation did not alter the Mg2+ or non-covalent DNA binding affinity of the ETOP-G116S enzyme
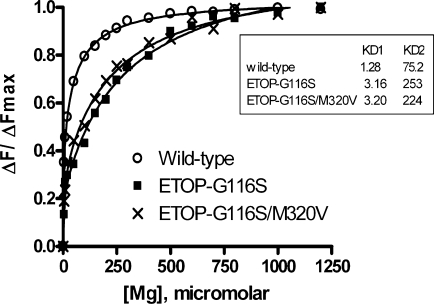
The acidic residues of the TOPRIM motif of ETOP have been shown to be involved in binding of two Mg2+ ions (20). The complete loss of relaxation and religation activities of the ETOP-G116S mutant enzyme is at least in part due to reduced Mg2+-binding affinity (12). The addition of the M320V mutation to the ETOP-G116S enzyme restored around 4–5% of the relaxation activity and 25% of the religation activity (Figure 4A and D). To determine if the addition of the M320V mutation altered the Mg2+-binding affinity of ETOP-G116S to account for this partial restoration of activities, change in intrinsic protein fluorescence from tryptophan residues upon the addition of Mg2+ was monitored. KD1 and KD2 values for binding of two Mg2+ ions were obtained by curve fitting of the experimental data. The results (Figure 5) showed that the double mutant ETOP-G116S/M320V had very similar Mg2+-binding affinity when compared with ETOP-G116S. As in the case of the ETOP-G116S mutant (12), the relaxation activity of the ETOP-G116S/M320V mutant could not be enhanced by increasing the Mg2+ concentration of the relaxation reaction buffer from 6 mM to 20 mM (data not shown).
Figure 5.
Magnesium binding measured by intrinsic tryptophan fluorescence of wild-type and mutant ETOP enzymes. Decrease of tryptophan fluorescence signal at 334 nm (excitation at 295 nm) with increasing concentration of MgCl2 was monitored. Fraction of maximal decrease of fluorescence signal was determined and curve fitted for binding of two Mg2+/enzyme molecule with the GraphPad Prism program. The KDs shown in the insert represent the average from two sets of data for ETOP-G116S/M320V and three sets of data for wild-type ETOP and ETOP-G116S.
Gel shift assay with the labeled oligonucleotide substrate used for the DNA cleavage assay was carried out to determine if the addition of the M320V mutation to the ETOP-G116S mutant enhanced the initial non-covalent interaction with DNA as a possible explanation for the higher amount of DNA cleavage product observed for the double mutant. The results (Supplementary Figure 1) showed that the ETOP-G116S and ETOP-G116S/M320V mutant enzymes had similar non-covalent binding affinity for the oligonucleotide DNA substrate used in the cleavage reaction. Therefore the biochemical effect of the M320V mutation on ETOP activity is likely to be directly on the step of DNA cleavage during the catalytic cycle.
DISCUSSIONS
In this study, it was determined that the Ser substitution at the strictly conserved Gly residue of the topoisomerase I TOPRIM motif was unique in its consequence of inhibiting DNA religation while retaining DNA cleavage activity, thus accounting for an effect on enzyme activity and cell viability similar to that expected from a topoisomerase poison. Among all amino acids, glycine has the smallest and most flexible side chain. The size of the side chain increases in the order of Gly to Ala to Ser. When the TOPRIM Gly was substituted with Ala, Mg2+-binding affinity was likely to have also been affected, as DNA cleavage became Mg2+ dependent as in the case of the Gly to Ser substitution. However, the Ala substitution might not have altered the positioning of the DNA 3′-hydroxyl group formed after DNA cleavage as much as the Ser substitution to have an inhibitory effect on DNA religation. With all the other 17 amino acid substitutions tested here, the steric effect from replacing the strictly conserved Gly side chain with a much larger group probably accounted for the complete loss of DNA cleavage activity, even in the presence of up to 5 mM Mg2+. The strict requirement of Gly for its steric properties probably accounts for its high degree of conservation following the DXD residues in the TOPRIM domain of DNA topoisomerases (26,27). The results from these experiments illustrate how the two events of DNA cleavage and DNA religation in the catalytic cycle, can be differentially affected by molecular perturbations even though the DNA religation is the reverse of DNA cleavage step when considered only for overall chemical changes. Small molecules could potentially bind adjacent to the Gly residue of the topoisomerase IA TOPRIM motif in subdomain I and exert effect selectively on the DNA religation step while allowing the enzyme to cleave DNA and form a covalent complex, similar to the effect of the Ser substitution.
Single mutation of the Met residue adjacent to the active site Tyr to Val was found not to have SOS-inducing or cell killing phenotypes in vivo, but nevertheless resulted in increase in DNA cleavage activity in vitro. There have been no mutagenesis results on this residue until this study. The Met to Val is a relatively conservative substitution so it is not surprising that the substitution did not inhibit the relaxation activity. Nevertheless, substitutions between Met and Val in other proteins have been shown to have significant effects on protein folding or catalytic properties (28–30). This result demonstrated for the first time that perturbation proximal to the active site tyrosine in subdomain III of topoisomerase I could influence DNA cleavage efficiency.
When this second Met to Val mutation adjacent to the active site tyrosine was added to the YTOP-G122S or ETOP-G116S TOPRIM mutants, DNA cleavage activity was enhanced while DNA rejoining was still inhibited effectively by the steric effect of the Gly to Ser substitution in subdomain I, resulting in a higher level of bacterial cell killing when the double mutant enzyme was induced by arabinose in vivo. This biochemical study showed that a perturbation in subdomain I of type IA topoisomerase that inhibits DNA religation can be combined with a second perturbation in subdomain III that enhances DNA cleavage in the process of ‘poisoning’ the topoisomerase enzyme. It might be possible to achieve the goal of creating a type IA topoisomerase poison by combining two small molecule fragments that interact separately with the TOPRIM motif and the region adjacent to the active site tyrosine (Figure 6). Bacterial type IA topoisomerases could potentially be a useful new target for discovery of novel antibacterial compounds to combat the serious public health problem of rapidly increasing instances of bacterial pathogens resistant to the antibacterial therapy currently in use (31–34).
Figure 6.
Regions of E. coli topoisomerase I structure important for the control of the DNA cleavage-religation equilibrium. The active site nucleophile Tyr-319 and adjacent Met-320 residues in subdomain III are shown along with the TOPRIM residues Asp-111, Asp-113 and Gly-116 in subdomain I in the structure (Protein Data Bank number 1ECL) of the 67 kDa N-terminal fragment of E. coli topoisomerase I (41).
High through-put screening of compound libraries is one approach that is being utilized in the attempt of identifying small molecules that can act as bactericidal poisons and increase the accumulation of covalent complex formed by bacterial type IA topoisomerases and cleaved DNA (14). The identification of various regions of the enzyme structure and associated molecular perturbations that can influence the DNA cleavage-religation equilibrium of topoisomerase I could facilitate a second alternative approach of drug discovery through molecular modeling (35–37). For such molecular modeling efforts, it would be extremely useful to have a crystal structure of the covalent complex formed between a type IA topoisomerase and cleaved DNA available. Structure of the covalent complex would also complement the currently available 3-dimensional structures of the non-covalent complexes in different conformational states formed between type IA topoisomerases and DNA (38–40) to provide key information on the catalytic mechanism and protein conformational changes that take place during the catalytic cycle. The mutant enzyme molecules that have been identified in our ongoing study as those that could form stabilized covalent intermediate with cleaved DNA might be more amenable to efforts of obtaining a crystal structure of the covalent complex between type IA topoisomerase and cleaved DNA substrate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We gratefully acknowledge Shikha Shukla and Somshuvra Mukhopadhyay, who were involved in early aspects of this work. Funding was provided by National Institutes of Health (GM54226, AI06933 to Y.T.) Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 3.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 5.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 6.Chen AY, Liu LF. DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 1994;34:191–218. doi: 10.1146/annurev.pa.34.040194.001203. [DOI] [PubMed] [Google Scholar]
- 7.Nitiss JL. DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr. Opin. Investig. Drugs. 2002;3:1512–1516. [PubMed] [Google Scholar]
- 8.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 9.Giles GI, Sharma RP. Topoisomerase enzymes as therapeutic targets for cancer chemotherapy. Med. Chem. 2005;1:383–394. doi: 10.2174/1573406054368738. [DOI] [PubMed] [Google Scholar]
- 10.Drlica K, Malik M. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 2003;3:249–282. doi: 10.2174/1568026033452537. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Q, Pongpech P, DiGate RJ. Type I topoisomerase activity is required for proper chromosomal segregation in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:9766–9771. doi: 10.1073/pnas.171579898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, Tse-Dinh YC. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J. Biol. Chem. 2005;280:38489–38495. doi: 10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng B, Liu I, Tse-Dinh YC. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 2007;59:640–645. doi: 10.1093/jac/dkl556. [DOI] [PubMed] [Google Scholar]
- 14.Tse-Dinh YC. Exploring DNA topoisomerases as targets of novel therapeutic agents in the treatment of infectious diseases. Infect. Disord. Drug Targets. 2007;7:3–9. doi: 10.2174/187152607780090748. [DOI] [PubMed] [Google Scholar]
- 15.Gupta M, Zhu CX, Tse-Dinh YC. Mutations of vaccinia virus DNA topoisomerase I that stabilize the cleavage complex. J. Biol. Chem. 1994;269:573–578. [PubMed] [Google Scholar]
- 16.Heitman J, Model P. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene. 1991;103:1–9. doi: 10.1016/0378-1119(91)90383-m. [DOI] [PubMed] [Google Scholar]
- 17.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 2001;147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 18.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl Acad. Sci. USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu CX, Tse-Dinh YC. Overexpression and purification of bacterial DNA topoisomerase I. Methods Mol. Biol. 1999;94:145–151. doi: 10.1385/1-59259-259-7:145. [DOI] [PubMed] [Google Scholar]
- 20.Zhu CX, Tse-Dinh YC. The acidic triad conserved in type IA DNA topoisomerases is required for binding of mg(II) and subsequent conformational change. J. Biol. Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 21.Kirkegaard K, Pflugfelder G, Wang JC. The cleavage of DNA by type-I DNA topoisomerases. Cold Spring Harb. Symp. Quant. Biol. 1984;49:411–419. doi: 10.1101/sqb.1984.049.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Cheng B, Feng J, Mulay V, Gadgil S, Tse-Dinh YC. Site-directed mutagenesis of residues involved in G strand DNA binding by Escherichia coli DNA topoisomerase I. J. Biol. Chem. 2004;279:39207–39213. doi: 10.1074/jbc.M405891200. [DOI] [PubMed] [Google Scholar]
- 23.Zhu CX, Roche CJ, Tse-Dinh YC. Effect of mg(II) binding on the structure and activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1997;272:16206–16210. doi: 10.1074/jbc.272.26.16206. [DOI] [PubMed] [Google Scholar]
- 24.Depew RE, Liu LF, Wang JC. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J. Biol. Chem. 1978;253:511–518. [PubMed] [Google Scholar]
- 25.Liu LF, Wang JC. Interaction between DNA and Escherichia coli DNA topoisomerase I. formation of complexes between the protein and superhelical and nonsuperhelical duplex DNAs. J. Biol. Chem. 1979;254:11082–11088. [PubMed] [Google Scholar]
- 26.Caron PR. Compendium of DNA topoisomerase sequences. Methods Mol. Biol. 1999;94:279–316. doi: 10.1385/1-59259-259-7:279. [DOI] [PubMed] [Google Scholar]
- 27.Aravind L, Leipe DD, Koonin EV. Toprim–a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 29.Wohlers TM, Fridovich-Keil JL. Studies of the V94M-substituted human UDPgalactose-4-epimerase enzyme associated with generalized epimerase-deficiency galactosaemia. J. Inherit. Metab. Dis. 2000;23:713–729. doi: 10.1023/a:1005682913784. [DOI] [PubMed] [Google Scholar]
- 30.Tahiri-Alaoui A, Gill AC, Disterer P, James W. Methionine 129 variant of human prion protein oligomerizes more rapidly than the valine 129 variant: implications for disease susceptibility to creutzfeldt-jakob disease. J. Biol. Chem. 2004;279:31390–31397. doi: 10.1074/jbc.M401754200. [DOI] [PubMed] [Google Scholar]
- 31.Rice LB. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006;43(Suppl. 2):S100–S105. doi: 10.1086/504487. [DOI] [PubMed] [Google Scholar]
- 32.Alekshun MN, Levy SB. Commensals upon us. Biochem. Pharmacol. 2006;71:893–900. doi: 10.1016/j.bcp.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 33.Dorman SE, Chaisson RE. From magic bullets back to the magic mountain: the rise of extensively drug-resistant tuberculosis. Nat. Med. 2007;13:295–298. doi: 10.1038/nm0307-295. [DOI] [PubMed] [Google Scholar]
- 34.Furuya EY, Lowy FD. Antimicrobial-resistant bacteria in the community setting. Nat. Rev. Microbiol. 2006;4:36–45. doi: 10.1038/nrmicro1325. [DOI] [PubMed] [Google Scholar]
- 35.Jain AN. Virtual screening in lead discovery and optimization. Curr. Opin. Drug Discov. Devel. 2004;7:396–403. [PubMed] [Google Scholar]
- 36.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 37.Stoermer MJ. Current status of virtual screening as analysed by target class. Med. Chem. 2006;2:89–112. doi: 10.2174/157340606775197750. [DOI] [PubMed] [Google Scholar]
- 38.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 39.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J. Mol. Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.