Abstract
The analysis of chromatin fine structure and transcription factor occupancy of differentially expressed genes by in vivo footprinting and ligation-mediated-PCR (LMPCR) is a powerful tool to understand the impact of chromatin on gene expression. However, as with all PCR-based techniques, the accuracy of the experiments has often been reduced by sequence similarities and the presence of GC-rich or repeat sequences, and some sequences are completely refractory to analysis. Here we describe a novel method, pyrophosphorolysis activated polymerization LMPCR or PAP-LMPCR, which is capable of generating accurate and reproducible footprints specific for individual alleles and can read through sequences previously not accessible for analysis. In addition, we have adapted this technique for automation, thus enabling the simultaneous and rapid analysis of chromatin structure at many different genes.
INTRODUCTION
It has become increasingly clear that the establishment of a correct chromatin fine structure is crucial for the coordinated regulation of the ∼20 000 or more genes in the mammalian genome. The treatment of living cells with different in vivo footprinting agents, such as dimethylsulphate (DMS), nucleases or UV-light followed by ligation-mediated PCR (LMPCR) has been an important tool for determining in vivo DNA accessibility, transcription factor occupancy and chromatin fine structure. All these agents induce lesions into DNA with a frequency modulated by transcription factor binding, chromatin compaction and nucleosome positioning. In essence, LMPCR is a method for detecting single-strand breaks or other lesions that terminate primer extension. Most DNA lesions or adducts formed by the treatment of living cells can be detected by LMPCR (1). The method generally consists of five steps: (i) primer extension using a gene-specific primer; (ii) addition of a linker to each blunt end generated in step (i); (iii) exponential PCR amplification using a second, gene-specific primer and a linker-specific primer; (iv) labelling by linear PCR using a single, 32P or fluorescently labelled third gene-specific primer and (v) separation and visualization of the fragments using sequencing gels, either flat or capillary (2). In some cases, the linker-primer can be used for labelling (2). The method is sensitive, requiring only 0.5–1.0 μg of DNA, and has even been partially automated (2,3). However, there are some genes and DNA sequences that are difficult to analyse with current methods; these include most parentally imprinted genes and other genes that are monoallelically expressed. For example, most of the 1000 or more X-linked genes in female cells have one allele in the active chromatin state and the other in the inactive state. It would be advantageous to be able to separately analyse these alternate chromatin states.
Another limitation of current LMPCR technology is that some sequences have proven difficult to analyse; these include dinucleotide repeats such as TG/CA repeats, triplet repeats, some CpG islands and very GC-rich regions. Many of these difficult-to-footprint sequences have been shown to influence chromatin structure and gene expression. For instance, it has been shown that (TG/CA)n repeats ≥12 downregulates transcription and that this effect increases with length (4), while previously TG/CA repeats have been shown to up- or downregulate transcription dependent on exact length (5). These variations are possibly due to a change in DNA conformation from B to Z that affects the movement of the polymerase. However TG/CA repeats have been shown to bind nuclear factors with strongest affinity at ≥(GT)16 (6) and these maybe responsible for the transcriptional changes. Triplet repeat expansions are associated with diseases such as myotonic dystrophy and Friedrich's ataxia. These triplet repeats have been shown to stimulate position-effect-variegation (PEV) (7) through heterochromatin protein 1 (HP1). The assembly of nucleosomes is also affected by triplet repeats, the propensity to form nucleosomes can either be increased or decreased dependent on the makeup of the triplet repeat (8). It is therefore of intense interest to develop a method that enables analysis of chromatin fine structure and transcription factor association of such sequences at high resolution.
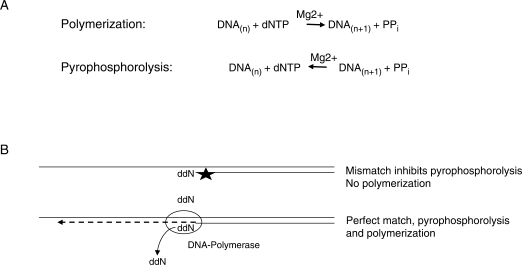
To this end, we sought to develop a LMPCR procedure that more robustly distinguishes single nucleotide polymorphisms (SNPs), shows improved analysis of difficult DNA sequences and is able to distinguish genes with differentially expressed alleles. To increase specificity we turned to pyrophosphorolysis activated polymerization (PAP) (9,10), which is a PCR-like amplification that utilizes 3′-blocked primers that are activated by pyrophosphorolysis while annealed to the complementary DNA strand in the presence of pyrophosphate. The general principle of PAP-LMPCR is depicted in Figure 1. During DNA synthesis, the incorporation of NTPs into the growing chain releases pyrophosphate, a high-energy compound. Since DNA polymerization is a reversible reaction so long as the pyrophosphate is not degraded to phosphate, high concentrations of pyrophosphate drive a pyrophosphorolysis reaction which removes nucleotides. Thus in the presence of pyrophosphate some DNA polymerases can remove a blocking nucleotide such as acycloNMP or ddNMP from the 3′ end of a primer (11,12). The use of a blocked primer increases specificity because removal of the blocked nucleotide by pyrophosphorolysis only occurs if the primer is perfectly annealed; any mismatches at or near the 3′ end of the primer prevents pyrophosphorolysis from occurring and hence elongation cannot take place (12). Once the 3′ blocked nucleotide is removed, efficient polymerization takes place because even under PAP conditions the rate of the forward, polymerization reaction is faster than the reverse reaction. We find that the use of 3′ dideoxy-blocked primers and PAP conditions permits allele-specific analysis of the CpG-rich, X-linked PGK promoter and improves analysis of difficult regions such as the promoter of the mouse colony stimulating factor 1 receptor gene (csf1r or c-fms), which contains a TC/GA repeat. In addition, we have adapted this procedure to be performed in an automated fashion, permitting the simultaneous analysis of multiple sequences.
Figure 1.
Pyrophosphorolysis in combination with a blocked primer reduces the production of non-specific products. (A) Polymerization is reversible under high PPi and favourable pH conditions. (B) Pyrophosphorolysis is inhibited by mismatches between the primer and DNA in at least the last 12 3′ nucleotides, leading to greater specificity (12).
MATERIALS AND METHODS
Allele-specific in vivo footprinting
Steps prior to the labelling step were done by our standard, published procedures (13) with modifications as indicated below. Primer 1 was 5′ biotin tagged and magnetic beads were used to pull down the extension product. Vent (exo-) polymerase was used for the first primer extension step. The washed beads were used for ligation with the LP25 linker and then PCR was done for 22 cycles using Primer 2, LP25, Qiagen Taq and 1× Qiagen PCR buffer and 1× Qiagen Q solution. Before labelling, the PCR primers were eliminated by treatment with Exonuclease 1 (3). The primer set used for the mouse Pgk-1 promoter is: Primer 1 (F1), 5′-biotin-CCG GAG ATG AGG AAG AGG AGA AC [Temperature (Tm) 58°C]; Primer 2 (F2), 5′-CAG CGC GGC AGA CGT GCG CTT TTG (Tm 66°C); Linker-Primer LP25, 5′-GCG GTG ACC CGG GAG ATC TGA ATT C (Tm 65.5°C). Primers used for labelling (F3a and F3b) were synthesized by LI-COR, Inc. 5′-labelled with infrared dyes, either IRD700 or IRD 800. We used these primers to make allele-specific, dideoxy-terminated primers using the procedure given below. The sequence of F3a (specific for Pgk-1 a allele) is 5′-GAA GCG TGC AGA ATG CCG GGC CTC G-3′ (Tm 68.5°C), and F3b (specific for Pgk-1 b allele) is 5′-GAA GCG TGC AGA ATG CCG GGC CTC C-3′ (Tm 68.5°C). The bold, 3′ nucleotide is the SNP position.
Preparation of 3′ blocked primers for PAP labelling and gel analysis
The appropriate 3′-terminal dideoxynucleotide was added with terminal transferase, according to (12). The mixture contained, in a total volume of 30 µl, 100 mM potassium cacodylate, pH 7.2, 2.0 mM CoCl2, 0.2 mM DTT, 400 µM oligonucleotide, 2 mM 2′,3′-ddNTP (the molar ratio of the 3′-OH terminus to ddNTP was 1:24) (Boehringer Mannheim), 100 U terminal transferase (Gibco BRL). The reaction was incubated at 37°C for 4 h and then stopped by adding EDTA to a 5 mM final concentration. After desalting using a Centri-spin column (Princeton Separations), the PAP-primer was purified by preparative 7 M urea–20% polyacrylamide gel electrophoresis in TBE buffer (90 mM Tris–borate, 1 mM EDTA, pH 8.3).
For PAP labelling, a master mix was prepared consisting of: 50 mM Tris–HCl (pH 7.8); 16 mM (NH4)2SO4; 3.5 mM MgCl2; 100 µM each dNTP with 7deaza dGTP replacing dGTP; 90 µM pyrophospate; 0.1 µM each of two allele-specific, 3′ dideoxy-terminated primers, one labelled at the 5′ end with Li-Cor dye IRD 700 and the other with Li-Cor dye IRD 800; 1× Q solution (Qiagen, Inc.), 5 U/reaction Klen Taq (Ab Peptides, Inc.), 2 U/reaction Qiagen Taq and distilled H2O to a total volume of 20 ul/reaction. Ten microlitres of the PCR amplification reaction (in Qiagen buffer pH 8.7) was added to 20 µl of labelling master mix, giving a total reaction volume of 30 µl. Final buffer composition is: 36.3 mM Tris–HCl; 14 mM (NH4)2SO4; 16.5 mM KCl; 3 mM MgSO4; 60 µM pyrophosphate; 100 µM dNTPs. Importantly, the final pH should be 8.0. Incubation for repeated, linear primer extension is: one cycle of 95°C for 2 min, 60°C for 2 min, 68°C for 1.5 min and 72°C for 2.5 min; and 15 cycles of 94°C for 45 s, 60°C for 2 min, 68°C for 1.5 min and 72°C for 2.5 min.
A LI-COR sequencing system was used as previously described (3). Two microlitres of 5× loading buffer was added to 8 µl of labelled product and heated at 95°C for 2 min prior to loading the gel. Fluorescence emission data at 700 and 800 nm were collected simultaneously but displayed as separate lanes.
PAP-LMPCR analysis of the csf1r promoter
DMS treatment of cells and DNA preparation have been described (14). DNase1 treatment of permeabilized cells and preparation of DNA was performed as described (15). Primers: For PAP-LMPCR the biotinylated extension primer contained a blocked dideoxy end, 5′Bio/TAAGTCTCTCAAACTCCATCATCTddC3′ (Integrated DNA Technologies Inc), the amplification primer was 5′TCTCCCTTCAGGATCAGTTTGAGCCT, LP25: generic linker primer was 5′GCGGTGACCCGGGAGATCTGAATTC3′
PAP-primer extension reactions were carried out in 1× reaction buffer [20 mM Tris pH 8, 10 mM KCl, 10 mM (NH4)SO4, 3 mM MgCl2, 0.1% Triton, 60 µM Na pyrophosphate], 0.3 M Sulpholan (Sigma), 2 mM tetramethylammonium oxalate (TMA ox) (gift from Sachem Inc.), 1 pM biotinylated dideoxyterminated primer, 250 µM dNTPs, 2 U Therminator™ DNA polymerase (NEB) and 1 µg DNA. dH2O was added to give a volume of 5 µl and the reaction mix was overlaid with 10 µl mineral oil. PCR conditions were 95°C for 20 min, annealing 65°C for 20 min, 60°C for 2 min, 68°C for 5 min and 76°C for 20 min for one cycle.
Primer extension product capture was carried out by adding 150 µg M280 streptavidin coated Dynabeads (Invitrogen) per sample. Beforehand, beads were washed twice in B&W buffer (10 mM Tris–HCl pH 7.5, 1 mM EDTA, 2.0 M NaCl), re-suspended in 5 µl B&W buffer and added to the primer extension reaction. The primer extension/bead mix was rotated at room temperature for 1–2 h. The beads were then separated and washed three times with 1 × ligation buffer (33 mM Tris–HCl pH 7.8, 66 mM K acetate, 10 mM Mg acetate). For ligation, the beads were re-suspended in 15 µl of ligation mix [1.5 µl 10 × ligation buffer, 3 µl 50% PEG6000, 60 pM LP25-LP21 linker, 1 mM ATP, 5 U T4 ligase (Epicentre Biotechnologies) dH2O to 15 µl] and rotated overnight at room temperature. A more detailed description of the procedure can be found in Supplementary Methods.
Denaturation
80 μl of TE (10 mM Tris–HCl pH 7.5, 1 mM EDTA) was added to each reaction and the beads separated on a magnet. The supernatant was removed and the beads washed 2 × with 100 µl TE and 1 × 100 µl 1 mM Tris–HCl pH 7.5, then re-suspended in 10 µl 1 mM Tris–HCl pH 7.5 before heating to 95°C for 15 min.
Amplification
5 μl 10× Pfu buffer (Stratagene), 0.3 M sulpholan, 2 mM TMA ox, (alternatively 1.4 M Betaine can be used instead of the sulpholan and TMA ox) 250 µM dNTPs, 10 pM LP25 primer, 10 pM specific 2nd primer, 2.5 U Pfu turbo (Stratagene), dH2O to 40 µl then added to 10 µl bead solution. PCR program: 95°C for 5 min (95°C for 45 s, 56°C for 3 min, 72°C for 5 min) 25 cycles, 72°C for 10 min.
Automation of PAP-LMPCR
All reagents used in the automated procedure were the same as in the manual method described above. The automated procedure uses a Biomek 2000 and the running program was basically as described in Ref. (2) with one minor alteration. Because the ligation reaction was very inefficient in the presence of the pyrophosphorolysis-inducing buffer of the primer extension reaction, the primer extension product capture and ligation reaction were switched around. Following the primer extension reaction, 150 µg M280 streptavidin-coated Dynabeads, washed twice in B&W buffer and re-suspended in 5 µl of B&W buffer, were added to the PCR reaction. The binding reaction was incubated at 25°C for 60 min with mixing by pipetting every 20 min. The plate was then transferred to the magnet where 80 µl of 1× ligation buffer were added to the samples to aid the separation of the beads, after 3 min separation all the supernatant was removed. The beads were then washed 2× in 100 µl 1× ligation buffer before being re-suspended in 15 µl of the ligation mix, (as described above). A more detailed description of the Biomek 2000 settings is available upon request. Labelling of the reaction products and their analysis on a CEQ capillary sequencer were exactly as described in Ref. (2).
RESULTS AND DISCUSSION
PAP-LMPCR results in improved allele specificity
To investigate the effect of PAP conditions on allele specificity, we examined the mouse X-linked PGK promoter, which has a known G/C SNP at position –97 relative to the translation start point (Allele a is C and Allele b is G). Cis to the a allele there is also a 5-bp micro deletion at nucleotide position –318, which is useful to confirm separate chromatin analysis of the active and inactive PGK promoters. We prepared 3′-dideoxy blocked primers for the G/C SNP in order to compare their performance in LMPCR with normal, unblocked primers. Figure 2 illustrates the results obtained when PAP primers and PAP conditions are used only for the labelling reaction. The primer specific for the a allele was 5′-labelled with IRD700, a near-infrared dye absorbing at 700 nm, while the primer specific for the b allele was labelled with IRD800, which absorbs at 800 nm. These primers were mixed and used in competition with each other for labelling C57B6 DNA, which has the Pgk-1 b allele only. The primer-extended DNA molecules labelled with the two dyes were run in the same gel lane of a LI-COR sequencing gel system, with each dye being detected separately. Figure 2 shows G ladders obtained using DNA cleaved at every G position by Maxam–Gilbert chemistry. When unblocked primers were used, only small differences were seen between matched and mismatched. This means that the IRD700-labelled primer, specific for a, was priming on the b allele template. However, almost no signal was seen for the IRD700-labelled dideoxy-blocked primers under PAP conditions. Other data (data not shown) confirmed similar increased specificity when A, T and C cleaved DNA was used, and Figure 3 shows that the IRD700-labelled primer functioned well on a perfectly matched template. pH was found to be important, with pH 8.0 being the best compromise between signal strength and specificity. It should be noted that this PAP-labelling procedure is not dependent on special equipment. 32P-labelled, 3′ dideoxy-blocked primers and standard sequencing gels can be used.
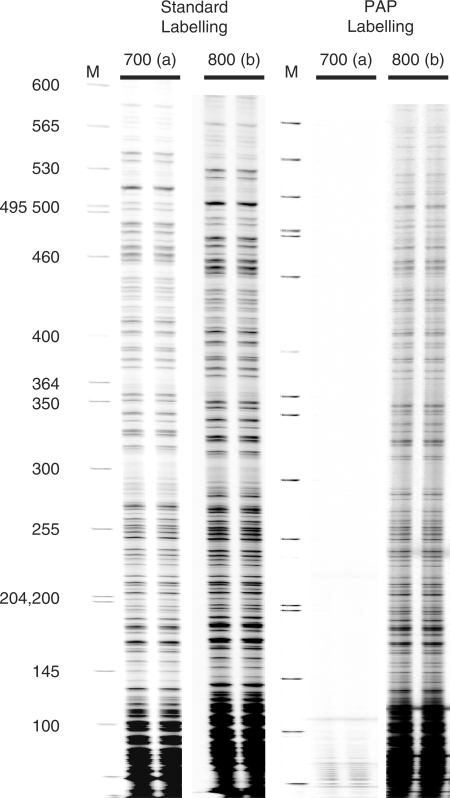
Figure 2.
PAP labelling reduces non-specific products. LMPCR G ladders showing that labelling done using 3′ dideoxy-blocked primers and PAP conditions (PAP labelling) provides excellent specificity for single nucleotide differences. Standard LMPCR with normal, unblocked primers is shown on the left. LMPCR with PAP labelling is shown on the right. F-set third primers (F3, see Materials and Methods section) labelled with either IRD700 (specific for the Pgk-1 a allele) or IRD800 (specific for the b allele) were used together in the labelling reaction. The DNA used was Maxam–Gilbert-treated C57B6 DNA (Pgk-1 b allele only); therefore, the IRD700 dye-labelled primers are mismatched at the 3′ end and give no extension product. To verify reproducibility, each reaction was repeated from start to finish. Lanes marked M are size markers.
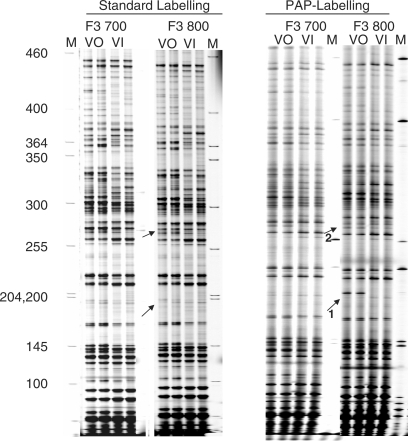
Figure 3.
PAP labelling allows allele-specific in vivo footprinting. Allele-specific UV photofootprinting, comparing standard labelling and PAP labelling. BMSL2 cells, which are heterozygous (a/b) for a G/C SNP, were UV treated prior to DNA extraction (lanes VO) or in vitro after purification (lanes VI). PAP conditions and dideoxy-blocked primers were used only for the labelling reaction. Arrow 1 marks the position of a CAAT box site known to be hyper-reactive with UV on the active allele (b allele). Arrow 2 marks the position of a 5-bp deletion in the Pgk-1 a allele; above this position the IRD700 (a allele) and IRD800 (b allele) patterns are shifted.
Figure 3 shows a UV photo footprinting experiment comparing dideoxy-blocked primers and PAP-labelling conditions with normal, non-blocked primers. For lanes marked VO, cells heterozygous for the a/b polymorphism were treated with UV before preparing DNA. For lanes marked VI, the UV treatment was done on naked DNA. A band at position –246 relative to the translation start point (arrow 1, fragment size 198), indicating strongly enhanced UV reactivity, was seen for the in vivo-treated sample only for the b allele if PAP conditions were used (arrow 1). In these cells, the active X carried the b allele, and from previous work with male cells, which have only an active X, the footprint was as expected; the active promoter was clearly distinguished at this position from the inactive promoter, which showed no UV photo footprint. Also consistent with clean, separate detection, was a shift in the ladder at position –318 relative to the translation start (arrow 2), where a 5-bp deletion is known to be cis to the a allele. This shift was not seen if unblocked primers were used; in this case, the ladder patterns for an allele and b alleles were very similar. Analysis of each DNA was done in duplicate and run in adjacent lanes; it is apparent that reproducibility and read length is excellent for both standard LMPCR and LMPCR with PAP labelling. However, PAP labelling gives greater specificity.
PAP-LMPCR is suitable for the analysis of repeat sequences
The next experiments were aimed at addressing the following issues. First, we wanted to optimize PAP-LMPCR for the analysis of difficult sequences, such as nucleotide repeat sequences. In addition, we previously published an automated LMPCR procedure (2) and we tested whether PAP-LMPCR could be adapted for automation. Last, but not least we wanted to see whether the improved specificity of PAP-LMPCR would allow us to label all products with a linker-specific third generic primer, thus significantly reducing costs.
The csf1r locus in the mouse contains both an A repeat and a (TG/CA)23 segment (Figure 4A). Previous attempts to footprint this region have largely failed (data not shown). While the polymerase can pass through the poly(A) stretch it tends to stutter resulting in indistinct bands on the other side. In addition, we have failed to get LMPCR to pass through the TG/CA repeat from the other side. Hence, this region was chosen to test out the ability of PAP-LMPCR to pass through difficult sequences.
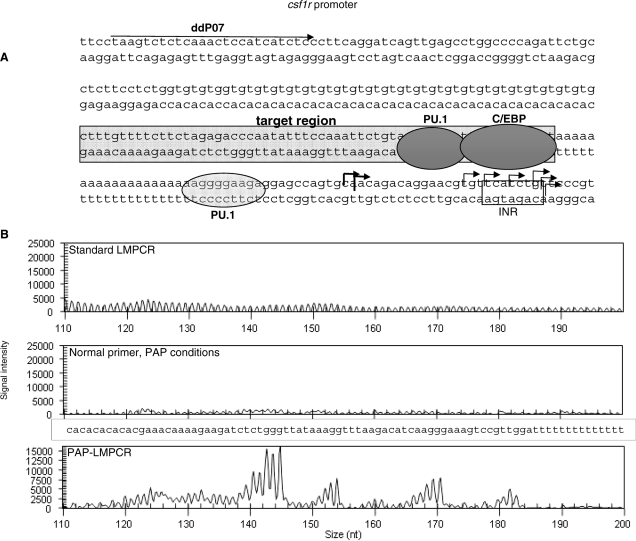
Figure 4.
PAP-LMPCR of the csf1r promoter allows reading through TG/CA-repeats. (A) Schematic diagram of the csf1r promoter showing the footprinting target area and known transcription factor binding sites. The position of the first primer is marked with an arrow labelled ddP07; transcription initiation region is labelled INR and the right-angled arrows mark the known transcription start sites. (B) Comparison of footprinting reactions of the csf1r promoter using DMS-treated DNA labelled with fluorescent LP25 primer and run on a CEQ8000 capillary sequencer. Only the footprinting reaction using the full pyrophosphorolysis conditions generated a G ladder.
A crucial step defining the specificity of in vivo footprinting is the first primer extension reaction and it is this step that is disturbed by repeat sequences. As shown above, PAP-LMPCR worked well when the blocked primer was used in the labelling step, however it had never been applied to primer extension reactions before and the reaction conditions therefore needed to be optimized. The standard LMPCR method uses Vent Exo, a polymerase commonly used in primer extension reactions, which has higher fidelity than Taq but lacks exonuclease activity. However, we found Vent Exo to be inefficient at removing the blocking nucleotide under PAP conditions, hence an alternative polymerase was sought. Therminator™ has similar properties to Vent but has the added ability to incorporate modified nucleotides such as dideoxynucleotides and acyclonucleotides and was found here to be able to remove such nucleotides from a blocked primer efficiently. During the optimization of the PAP-LMPCR for the csf1r promoter, several different chemical PCR enhancers were tried, as our previous standard enhancer (5% DMSO) did not perform satisfactorily in the PAP reaction. The low molecular weight sulphone, sulpholan, (tetramethylene sulphone) (16) used at 0.3 M was found to give good results and that these were further improved by the addition of 2 mM tetramethylammonium oxalate (17). When used alone, these enhancers were still not sufficient to reliably push the reaction through the CA/GT repeat, but when combined with the use of a dideoxyterminated primer under pyrophosphate conditions, Therminator™ was able to read through the repeat as illustrated in Figure 4B. This figure shows a LMPCR experiment in which DMS-treated naked DNA was used in either standard LMPCR or under PAP conditions with or without a blocked primer. DMS mainly methylates guanosines, which are then specifically cleaved using piperidine. The reaction products were labelled with a generic linker primer and were analysed on a capillary sequencer as previously described in (2). The experiment clearly shows that under standard LMPCR conditions, or using PAP conditions but a standard, unblocked primer, no readable G reaction was achieved. In contrast, once a dideoxyterminated primer was used that can only extend if accurately annealed, a readable G reaction was observed.
In vivo footprinting of the csf1r promoter by PAP-LMPCR using a robotic workstation
We next tested the performance of PAP-LMPCR with respect to the analysis of chromatin fine structure in living cells and the adaptation of the PAP-LMPCR procedure to a robotic workstation. Figure 5 shows a DMS in vivo footprinting experiment analysing the csf1r promoter in the RAW264 macrophage cell line, where the csf1r gene is active, as compared to the NIH3T3 line, where it is not. Macrophage-specific alterations in DMS reactivity indicative of transcription factor binding were seen in the same area as on the other strand (18) where we have previously been able to detect the binding of the transcription factors C/EBPβ and PU.1.
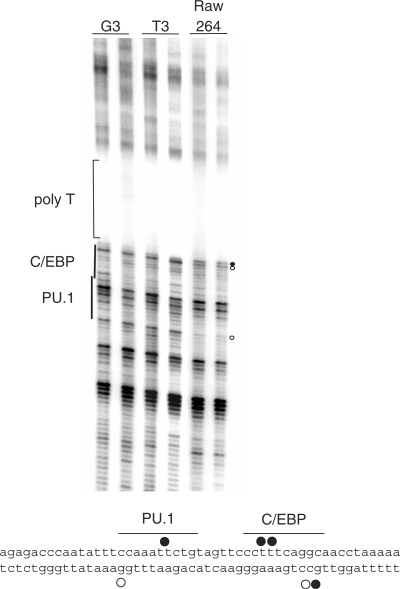
Figure 5.
In vivo DMS footprinting of the csf1r promoter using PAP-LMPCR. Radiolabelled PAP-LMPCR products from genomic DNA purified from DMS-treated NIH3T3 fibroblast cells (csf1r non-expressing) or RAW264 macrophage cells (csf1r expressing) as well as DMS-treated genomic DNA as indicated were run on a 6% sequencing gel. The experiment shows clear footprints within the target region in RAW264 cells closely associated with the C/EBP- and PU.1-binding sites. Samples are in duplicates, demonstrating the reproducibility of the reaction. Footprints are marked with either closed circles for guanines showing DMS hyper-reactivity or open circles indicating hypo-reactivity. The lower panel shows the position of footprints previously demonstrated on the upper strand (18); the footprints on the lower strand are from the experiment described here.
DMS footprinting is a useful tool for analysing transcription factor occupancy of DNA but is limited by the occurrence of Gs in the interacting DNA sequence. DNaseI accessibility studies give a wider view of the chromatin structure, indicating the level of compaction, nucleosome positioning and the presence of transcription factors. Figure 6A shows overlaid CEQ capillary sequencer traces of a DNaseI analysis of the csf1r promoter in RAW264 and 3T3 cells after PAP-LMPCR was performed on a Biomek 2000 robotic workstation. Care was taken to ensure equal DNaseI digestion of samples. Digestion was monitored by performing an LM-PCR looking at rDNA genes, which are expressed in each cell type (19). The sample from DNaseI-treated cells is shown in black overlaying the naked DNA control in grey. The analysis uncovered significant differences in chromatin fine structure of csf1r between the two cell types. The RAW264 trace showed a larger number of high peaks relative to the naked DNA control than the NIH3T3 trace, indicating increased DNaseI accessibility indicative for a more open chromatin structure. In addition, clusters of peaks specific for macrophages were seen in the vicinity of the known transcription start representing increased DNase1 accessibility in these areas. A further group of peaks between –190 and –205 bp, indicates what appeared to be a binding site for an unknown transcription factor (marked by a question mark). As might be expected, the region around the transcription start site (–115 bp) also appeared to be more accessible in RAW264 cells. In this case, we saw no major difference in the CA repeat between the two cell lines, with both giving a pattern similar to the naked DNA, suggesting that the DNA was not bound by proteins. Figure 6B shows the same samples labelled with radioactive primers and run on a standard sequencing gel, confirming these results.
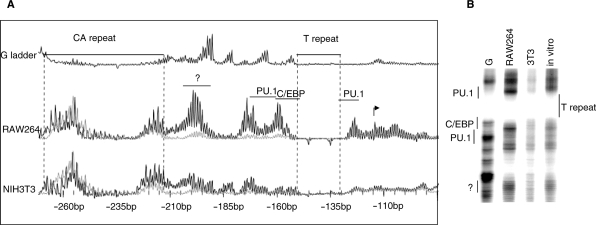
Figure 6.
Automated in vivo DNaseI footprinting of the csf1r promoter using PAP-LMPCR. Panel A shows DNase1 accessibility of the csf1r promoter using PAP-LMPCR. The samples were labelled with a fluorescent LP25 primer and run on a CEQ8000. The sample (black line) has been overlaid on to the naked DNA control (grey line). Note the increased DNase1 accessibility of the C/EBP and PU.1 sites, and over the transcription start site in the active RAW264 cell line as compared to the inactive NIH3T3 line. The binding of an unknown protein is further indicated by increased accessibility between –190 and –205 bp in RAW264 cells. (B) The samples from (A) were radiolabelled and run on a sequencing gel for comparison; the reduced accessibility of the region as a whole in the csf1r non-expressing NIH3T3 line is clearly visible.
Taken together, our experiments demonstrate the feasibility of using PAP-LMPCR for the analysis of previously unreadable sequences, which should significantly reduce the areas of the genome that are inaccessible for footprinting studies. Using DMS-treated DNA, we have shown reproducible footprints in a previously inaccessible area adding further evidence for the binding of PU.1 and C/EBP family members to this region of the csf1r promoter. This is further supported by the DNase1 accessibility study. The overlaying of traces for naked DNA and the DNase1 in vitro treated cell DNA, obtained from the capillary sequencer gives a very clear picture of alterations in chromatin structure occurring as a result of transcription factor binding—in this case PU.1 and C/EBP—on this stretch of DNA.
Another interesting result from our study is that the repeat sequences have a very low DNaseI accessibility and this is the same in csf1r expressing RAW264) and csf1r non-expressing (NIH3T3) cells. This suggests that the CA/GT repeat does not bind sequence-specific DNA-binding proteins involved in regulation of csf1r since this interaction would cause changes in conformation. This may be as expected since in the csf1r promoter this repeat is only found in mouse and rat and not in a variety of other species such as human and rabbit, indicating that its insertion is a relatively recent event and the sequence has not acquired any particular function
LMPCR is a long procedure with repeated washing steps that we have previously automated. PAP-LMPCR lends itself readily to automation with only a few program changes from the conventional LMPCR method, thus removing the need for the investigator to be present as each stage comes to an end, hence shortening and streamlining the procedure. The increased specificity and robustness of PAP-LMPCR allows the use of a common, fluorescent-labelled linker (LP25 primer) for all reactions (Figures 4 and 6), thus greatly reducing costs. These advantages of PAP-LMPCR over conventional LMPCR should make it a useful addition in the ongoing efforts to analyse and understand the structure of the epigenome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hiromi Tagoh, Leeds for her generous help and advice. This research was funded by the Biotechnology and Biological Sciences Research Council (BBSRC), the Leukaemia Research Fund and a contract from the City of Hope Medical Centre. Funding to pay the Open Access publication charges for this article was provided by the BBSRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pfeifer GP. Measuring the formation and repair of DNA damage by ligation-mediated PCR. Methods Mol. Biol. 2006;314:201–214. doi: 10.1385/1-59259-973-7:201. [DOI] [PubMed] [Google Scholar]
- 2.Ingram R, Tagoh H, Riggs AD, Bonifer C. Rapid, solid-phase based automated analysis of chromatin structure and transcription factor occupancy in living eukaryotic cells. Nucleic Acids Res. 2005;33:e1. doi: 10.1093/nar/gni001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai SM, Chen HH, Chang C, Riggs AD, Flanagan SD. Ligation-mediated PCR for quantitative in vivo footprinting. Nat. Biotechnol. 2000;18:1108–1111. doi: 10.1038/80323. [DOI] [PubMed] [Google Scholar]
- 4.Sharma VK, Kumar N, Brahmachari SK, Ramachandran S. Abundance of dinucleotide repeats and gene expression are inversely correlated: a role for gene function in addition to intron length. Physiol. Genomics. 2007;31:96–103. doi: 10.1152/physiolgenomics.00183.2006. [DOI] [PubMed] [Google Scholar]
- 5.Gebhardt F, Zanker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J. Biol. Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 6.Gao PS, Heller NM, Walker W, Chen CH, Moller M, Plunkett B, Roberts MH, Schleimer RP, Hopkin JM, Huang SK. Variation in dinucleotide (GT) repeat sequence in the first exon of the STAT6 gene is associated with atopic asthma and differentially regulates the promoter activity in vitro. J. Med. Genet. 2004;41:535–539. doi: 10.1136/jmg.2003.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 8.Mulvihill DJ, Nichol Edamura K, Hagerman KA, Pearson CE, Wang YH. Effect of CAT or AGG interruptions and CpG methylation on nucleosome assembly upon trinucleotide repeats on spinocerebellar ataxia, type 1 and fragile X syndrome. J. Biol. Chem. 2005;280:4498–4503. doi: 10.1074/jbc.M413239200. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Sommer SS. Pyrophosphorolysis-activated polymerization (PAP): application to allele-specific amplification. Biotechniques. 2000;29:1072–1076. doi: 10.2144/00295rr03. 1078, 1080 passim. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Sommer SS. PAP: detection of ultra rare mutations depends on P* oligonucleotides: “sleeping beauties” awakened by the kiss of pyrophosphorolysis. Hum. Mutat. 2004;23:426–436. doi: 10.1002/humu.20036. [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Sommer SS. Pyrophosphorolysis by type II DNA polymerases: implications for pyrophosphorolysis-activated polymerization. Anal. Biochem. 2004;324:22–28. doi: 10.1016/j.ab.2003.08.037. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Sommer SS. Pyrophosphorolysis-activatable oligonucleotides may facilitate detection of rare alleles, mutation scanning and analysis of chromatin structures. Nucleic Acids Res. 2002;30:598–604. doi: 10.1093/nar/30.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfeifer GP, Dammann R. Measuring the formation and repair of UV photoproducts by ligation-mediated PCR. Methods Mol. Biol. 1999;113:213–226. doi: 10.1385/1-59259-675-4:213. [DOI] [PubMed] [Google Scholar]
- 14.Kontaraki J, Chen HH, Riggs A, Bonifer C. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 2000;14:2106–2122. [PMC free article] [PubMed] [Google Scholar]
- 15.Lefevre P, Lacroix C, Tagoh H, Hoogenkamp M, Melnik S, Ingram R, Bonifer C. Differentiation-dependent alterations in histone methylation and chromatin architecture at the inducible chicken lysozyme gene. J. Biol. Chem. 2005;280:27552–27560. doi: 10.1074/jbc.M502422200. [DOI] [PubMed] [Google Scholar]
- 16.Chakrabarti R, Schutt CE. The enhancement of PCR amplification by low molecular-weight sulfones. Gene. 2001;274:293–298. doi: 10.1016/s0378-1119(01)00621-7. [DOI] [PubMed] [Google Scholar]
- 17.Kovarova M, Draber P. New specificity and yield enhancer of polymerase chain reactions. Nucleic Acids Res. 2000;28:E70. doi: 10.1093/nar/28.13.e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tagoh H, Himes R, Clarke D, Leenen PJ, Riggs AD, Hume D, Bonifer C. Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev. 2002;16:1721–1737. doi: 10.1101/gad.222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoogenkamp M, Krysinska H, Ingram R, Huang G, Barlow R, Clarke D, Ebralidze A, Zhang P, Tagoh H, Cockerill PN, et al. The Pu.1 locus is differentially regulated at the level of chromatin structure and non-coding transcription by alternate mechanism at distinct developmental stages of hematopoiesis. Mol. Cell Biol. 2007;27:7425–7438. doi: 10.1128/MCB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.