Abstract
The chromatin immunoprecipitation (ChIP) assay is a major tool in the study of genomic processes in vivo. This and other methods are revealing that control of gene expression, cell division and DNA repair involves multiple proteins and great number of their modifications. ChIP assay is traditionally done in test tubes limiting the ability to study signaling of the complex genomic events. To increase the throughput and to simplify the assay we have developed a microplate-based ChIP (Matrix ChIP) method, where all steps from immunoprecipitation to DNA purification are done in microplate wells without sample transfers. This platform has several important advantages over the tube-based assay including very simple sample handling, high throughput, improved sensitivity and reproducibility, and potential for automation. 96 ChIP measurements including PCR can be done by one researcher in one day. We illustrate the power of Matrix ChIP by parallel profiling 80 different chromatin and transcription time-course events along an inducible gene including transient recruitment of kinases.
INTRODUCTION
Chromatin-dependent processes respond to extracellular signals, and control gene transcription, cell division and DNA repair (1–3). The dynamic chromatin structure is at the center of regulating these nuclear events. ChIP has proven to be a powerful tool to investigate chromatin structure (1,4,5). Along with other techniques (6,7), ChIP assays are revealing complexity of chromatin, transcriptional, RNA processing and DNA replication events (1,6,8–13). These processes are regulated by multiple histone modifications mediated by kinases, methyltransferases, acetyltransferases, ubiquitintransferases and others (14–19). Given that there are hundreds of different transcription and RNA processing factors, the list of regulatory modifications of DNA-bound and accessory factors is far greater than that documented for histones alone. The ability to follow a multitude of these modification events simultaneously would enhance our understanding of nuclear processes.
The traditional ChIP method has notable limitations, in that it takes several days to complete, requires bead-based immunoprecipitation, phenol-chloroform or spin column-based DNA extractions and involves multiple tube transfers (4). Such DNA extractions and precipitations, are not only time consuming and tedious but also introduce steps for potential sample loss, cross-contamination and variability. This becomes a bigger problem in experiments involving multiple chromatin samples. We have previously developed the Fast ChIP method that improved the standard ChIP protocol by simplifying the DNA purification step (20–23). Although Fast ChIP increased the efficiency and reproducibility of the procedure, it is done in test tubes and requires bead-based precipitation and elution steps limiting the number of samples that one researcher can handle and making it unsuitable for automation.
This article describes a novel ChIP strategy, Matrix ChIP, that utilizes surface-immobilized antibodies in a 96-well plate, where the entire procedure from chromatin precipitation to PCR-ready DNA purification is done on the same plate without sample transfers. The high-throughput potential of the Matrix ChIP is illustrated by simultaneous profiling of recruitment of three kinases, Erk1/2 (24), PKCδ (25) and Fyn (26), and several chromatin and transcriptional events along the PMA inducible egr-1 locus in mesangial cells (27). These data demonstrate that the pattern of Erk1/2, PKCδ and Fyn recruitment to the inducible egr-1 gene resemble that of RNA Polymerase II. We suggest that these kinases phosphorylate their targets in situ to regulate chromatin dynamics and transcription (28–32).
MATERIALS AND METHODS
96-well microplates
Reactin-Bind™ Protein A (Pierce cat. no. 15132) and Polystyrene High Binding Capacity 96-well microplates (Corning cat. no. 9018) were used.
Reagents
Chelex 100 (Bio-Rad, cat. no. 142-1253), Protein A (Sigma, cat. no. P7837), Proteinase K (Invitrogen, cat. no. 25530-015), Protein A–Sepharose (Amersham, cat. no. 17-5280-01), Formaldehyde (J.T. Baker, cat. no. 2106-02), BSA (Sigma cat. No. A9647), beta-Glycerol phosphate diSodium salt hydrate (Sigma cat. no. G-6251), Sodium molybdate dihydrate (Sigma cat. no. S-6646), Sodium fluoride (Sigma cat. no. S-1504), Sodium orthovanadate (Sigma cat. no. S-6508), p-nitrophenyl phosphate diTris salt (Calbiochem cat. no. 487655), PMSF (Sigma, cat. no. P-7626), Leupeptin (Sigma, cat. no. L-2884), SYBR Green PCR Master Mix (Quantace, 2xSensiMix, cat. no. QT6T3) and Salmon sperm DNA [Sigma, cat. no. D1626] were the reagents used.
Equipment
ChIP: Misonix Sonicator 3000 with micro tip (Misonix, cat. no. S3000); Ultrasonic bath (Branson, cat. no. B3510-MT CPN-952-316); Heat blocks (Analog Heat Block VWR Scientific 13259032, and Isotemp 125, Fisher Scientific); Eppendorf Repeater Pipette model 4780.
PCR: Quantitative PCR ABI 7900HT system, ABI Biotechnology; MixMate (Eppendorf).
Buffers
PBS: 137 mM NaCl, 10 mM Sodium phosphate, 2.7 mM KCl, pH 7.4; TE: 10 mM Tris, 1 mM EDTA, pH 7.5; Immunoprecipitation (IP) buffer: 150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, NP-40 (0.5% vol/vol), Triton X-100 (1.0% vol/vol); Blocking buffer: 5% BSA, 100 μg/ml sheared salmon sperm DNA in IP buffer; Elution buffer: 25 mM Tris base, 1 mM EDTA, (pH 9.8), 200 μg/ml proteinase K. A stock solution of 500 ml of Tris base-EDTA buffer is prepared at a time and stored at room temperature for at least four months. A 20 mg/ml proteinase K stock solution in water is stored at –20°C.
Cells
Rat mesangial cells were grown in 150 mm plastic cell culture dishes in RPMI 1640 media supplemented with 10% FBS, 2 mM glutamine, penicillin (100 units/ml), streptomycin (0.01%) and humidified with 5/95% CO2/air gas mixture (33).
Preparation of sheared chromatin and tube-based Fast ChIP assay
Preparation was done as described previously using IP buffer containing protease and phosphatase inhibitors (20,34). Sheared chromatin of 1 ml was prepared from 1–2 × 107 mesangial cells and stored in 50 μl aliquots (–70°C) to avoid repeated thawing/freezing.
Matrix ChIP assay using polystyrene plates coated with protein A
All washes were done using needle connected to vacuum pump and Eppendorf Repeater Pipette. Polystyrene plates washed once with 200 μl PBS/well were incubated overnight with 0.2 μg Protein A in 100 μl PBS/well. After a wash (200 μl PBS/well) well walls were blocked with 200 μl blocking buffer (15–60 min, RT). Wells were cleared and then incubated with antibodies in 100 μl blocking buffer/well (60 min, RT). Wells were cleared and chromatin samples (2.0–5.0 μl chromatin prep/100 μl blocking buffer) were added to wells (100 μl/well) and plates were floated in ultrasonic water bath (60 min, 4°C) to accelerate protein–antibody binding (20,34,35). Well walls were washed three times with 200 μl IP buffer and once with 200 μl TE buffer. Wells were incubated with 100 μl elution buffer (15 min at 55°C, followed by 15 min at 95°C). DNA samples were stored (–20°C) in the same Matrix ChIP plates for repeated use.
Real-time PCR
The reaction mixture contained 2.5 μl 2X SYBR Green PCR master mix (SensiMix, Quantace), 2.3 μl DNA template and 0.2 μl primers (10 μM) in 5 μl final volume in 384-Well Optical Reaction Plate (Applied Biosystems). Amplification (three steps, 40 cycles), data acquisition and analysis were carried out using the 7900HT Real Time PCR system and SDS Enterprise Database (Applied Biosystems). All PCR reactions were run in triplicates.
Calculations
ChIP DNA data were expressed as a fraction of input DNA calculated using the following method. PCR calibration curves were generated for each primer pair from a dilution series of total cellular DNA. The PCR-primer efficiency curve were fit to the ln (Standard DNA dilution) versus CT points measured for the DNA samples using an R-squared best fit. DNA concentration values for each ChIP and input DNA samples were calculated from their respective average PCR CT values using the formula
| 1 |
where b and m are the curve fit parameters from the primer calibration curve that is generated for each PCR experiment. Dilution is the cumulative dilution of ChIP DNA compared to input DNA sample. Final results are expressed as
| 2 |
where DNA concentrations were computed from Equation (1), DNAsample, ChIP DNA sample; DNAmock, IgG mock IP control and DNAinput; input DNA used in ChIP.
RESULTS AND DISCUSSION
Fast ChIP is done in test tubes with antibody immobilized to protein A-agarose beads that require centrifugation and use of Chelex-100 resin to purify the DNA (20). Our goal was to develop a simple microplate-based ChIP method that would not only increase the throughput but would also be suitable for automation. The adaptation of the Fast ChIP assay to a 96-well microplate format required several key modifications: (i) Purification of PCR-ready DNA without Chelex-100 resin. (ii) Immobilization of antibodies to well walls. (iii) Minimizing non-specific adsorption to the well surface.
DNA purification
Isolation of PCR-ready DNA from immunoprecipitated chromatin requires not only elution of the DNA from the protein A agarose beads but also reversal of the cross-links between DNA and proteins. In the Fast ChIP assay we introduced Chelex-100 resin to extract DNA (20). For the plate-based ChIP, we chose to develop a simpler method for the isolation of PCR-ready DNA from the surface-bound antibody by using a buffer that reverses cross-links and facilitates DNA extraction.
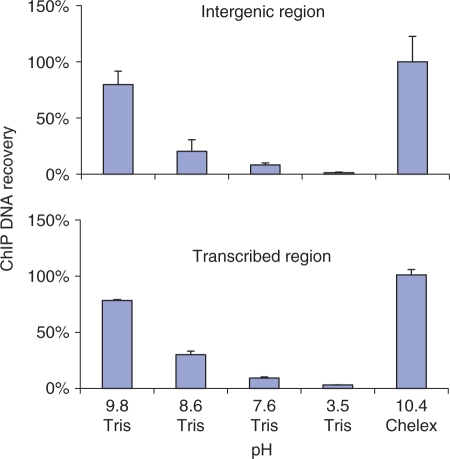
The Chelex-100 resin is a styrene-divinylbenzene copolymer containing paired iminodiacetate groups which chelate polyvalent metal ions. The Chelex suspension has pH ∼10. We reasoned that a high pH buffer and EDTA could be substituted for the Chelex beads. The test-tube format with chromatin-loaded Protein A beads was used to test buffers with a range of pH values. The Chelex-based Fast ChIP protocol was used as a positive control (20). Briefly, test tubes with chromatin-loaded Protein A beads (anti-H3K4m3 antibody) suspended in elution buffer were first incubated with proteinase K at 55°C for 15 min and then at 95°C for another 10 min. After centrifugation of the tubes, supernatant was collected and used in real-time PCR to compare the DNA recoveries to the Chelex-based Fast ChIP protocol. The results demonstrated that 25 mM Tris base, 1 mM EDTA (pH 9.8) performs comparably to Chelex (Figure 1), Therefore, we used this buffer in the following experiments.
Figure 1.
Extraction of PCR-ready DNA from immunoprecipitated chromatin with Tris-base/EDTA buffer. All steps were done in 1.5 ml tubes. Sheared chromatin from MC (0.5 ml) was incubated with anti-H3K4m3 antibody in an ultrasonic water bath (15 min, 4°C). After centrifugation (10 min at 17 000g) the supernatant was transferred to fresh tubes containing Protein A agarose beads. The slurry was rotated for 45 min (4°C) and then the beads were washed with IP buffer (20,34). Equal aliquots of beads were first incubated with proteinase K (200 μg/ml) in either 25 mM Tris, 1 mM EDTA (pH 9.8, 8.6, 7.6 or 3.5 titrated with HCl) or 10% Chelex/H2O for 15 min at 55°C and then at 95°C for 15 min. After centrifugation the supernatant was collected and analyzed by qPCR using primers to either 5′-flanking intergenic (-5kb to egr-1 transcription start site) or transcribed region (exon 1 of egr-1). PCR results are shown as percent of the ChIP DNA extracted with Chelex (mean ± SD, n=3).
Antibody immobilization
The 96-well microplate format requires the protein capturing antibodies to be attached to the surface of the microplate well wall. Antibody-coated microplates have been widely used in multi-well enzyme-sensitive immunosorbent assays (ELISA) (36) and in standard co-immunoprecipitations (37). For immobilizing antibodies, the surface needs to be modified to maintain the antibodies in an active state and in the correct orientation. It is known that specific orientation of antibodies increases the binding capacity of target molecules up to 10-fold compared to surfaces with random-oriented antibodies (38). Several antibody immobilization methods have been introduced to achieve this goal (38–42). Surface coating with Protein A, G or A/G mixture is one way to increase antibody-binding capacity of a surface (43,44).
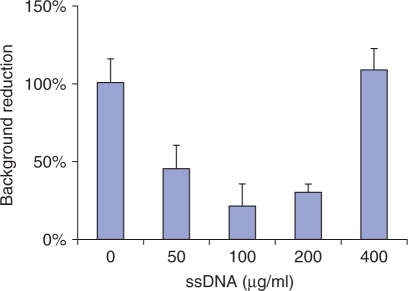
To test this strategy, we utilized commercially available polystyrene Protein A coated 96-well microplates (Pierce). The Fast ChIP done in tubes in parallel was used as a control. Results of these experiments done with H3K4m3 antibody demonstrated that the performance of well-attached antibodies was comparable to Fast ChIP. However, the unspecific background signal (Mock IP done with non-immune IgGs) was higher in Matrix ChIP. We found that using blocking buffer that contains BSA and sheared salmon sperm DNA reduced the non-specific binding to polystyrene well walls (Figure 2). The reason for higher background with high concentration of salmon sperm DNA is not clear, it is possible that excess of DNA causes aggregation of chromatin to the well walls.
Figure 2.
Reducing non-specific chromatin binding to well walls with sheared salmon DNA (ssDNA). Polystyrene well walls were pre-blocked with 5%BSA and either 0, 50, 100, 200, 400 μg/ml of ssDNA. After washing with IP/TE buffers DNA was eluted from the walls and assayed in real-time PCR using primers to egr-1 exon1. PCR results are shown as percent of chromatin DNA bound to walls without blocking buffer (mean ± SD, n = 3).
Although pre-coated Protein A microplates are commercially available they are costly. We found that passive adsorption of Protein A to the Corning Polystyrene High Bind Microplate performed well in the ChIP assay. Thus, Protein A-coated ChIP microplates can be easily fabricated in the laboratory (‘Methods’ section).
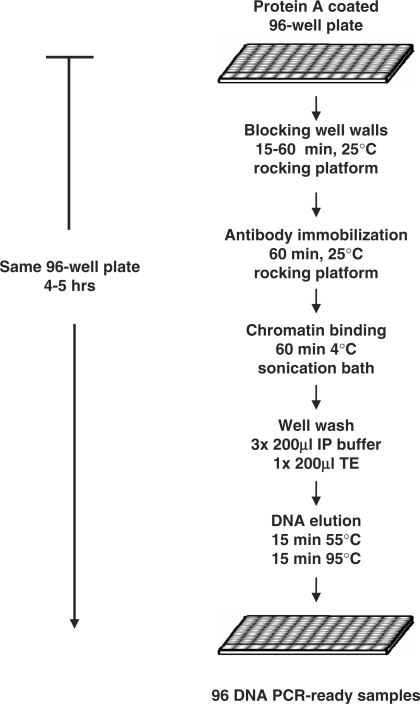
The Matrix ChIP protocol for Protein A coated polystyrene plates is summarized in Figure 3. The entire ChIP procedure is done in the same microplate well. The PCR-ready ChIP samples are stored in the same plates (−20°C) for repeated use. Starting with sheared chromatin, 96 PCR-ready ChIP DNA samples can be generated in 4–5 h.
Figure 3.
Diagram of the Matrix ChIP protocol. Detailed protocol is described in ‘Methods’ Section. Starting with sheared chromatin and Protein A coated plates 96 ChIPs can be completed in 4–5 h.
Verification of Matrix ChIP assay
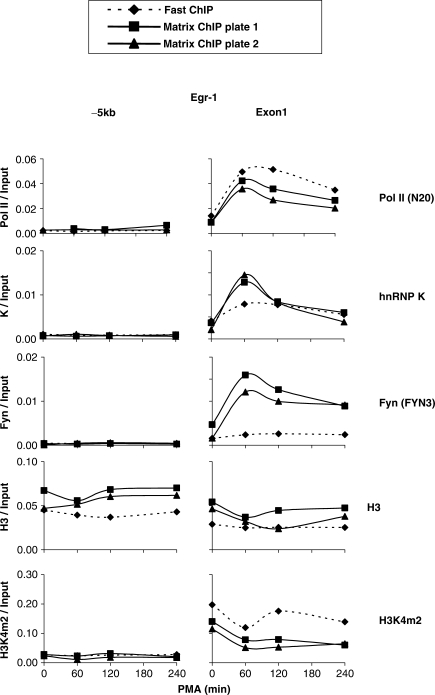
To validate the Matrix ChIP method, we profiled several inducible changes along the phorbol 12-myristate 13-acetate (PMA)-inducible egr-1 (27) locus in rat mesangial cells using the Matrix ChIP method and compared it to the tube-based Fast ChIP method (20,34). We used both the commercially available (Pierce) and in-lab made Protein A-coated microplates. The following affinity-purified antibodies were used: to RNA polymerase II, Pol II (45), histone H3 (15) hnRNP K protein (46) and the tyrosine kinase Fyn (26) as well as whole antiserum to histone H3 dimethylated at lysine 4, H3K4m2 (Figure 4). The detailed information about antibodies that were used in this article is included in Table S1. The list includes mouse monoclonal and rabbit polyclonal that either affinity, or Protein A- and Protein G-purified antibodies as well as whole antisera.
Figure 4.
Validation of Matrix ChIP assay. Sheared chromatin was prepared from PMA-treated rat mesangial cells. Matrix ChIP method using either in-lab (Plate 1) or commercial (Pierce) (Plate 2) Protein A-coated polystyrene plates was compared to tube-based Fast ChIP assay (20). The following rabbit polyclonal antibodies were used; anti-Pol II (N20), anti-hnRNP K, anti-Fyn (FYN3), anti-histone H3 and whole antiserum, anti-H3K4m2 (Table S1). Real-time PCR was done using primers to either egr-1 exon 1 (+232 bp from transcription start site, TSS) or 5′ flanking site (-5344bp from TSS), -5kb. Non-immune IgG fraction was used as a mock IP control. ChIP results are calculated as fraction of input DNA (see ‘Methods’ Section).
The Pol II, H3, H3K4m2 and hnRNP K (46) densities at the egr-1 gene measured with the Matrix and Fast ChIP assays were overall similar. While transient binding of Fyn to the egr-1 gene was observed with Matrix ChIP, the level of recruitment detected with Fast ChIP was much lower. The reproducibility of the results obtained with the Matrix ChIP method is illustrated by similar kinetics of changes of Pol II, hnRNP K and Fyn at the egr-1 locus observed in two independent experiments (Figure 4). Also, this comparison shows that the in-lab coated and the commercially available plates performed equally well.
Matrix ChIP enables tracking of kinase binding in parallel with many chromatin and transcriptional events along the transcribed gene locus
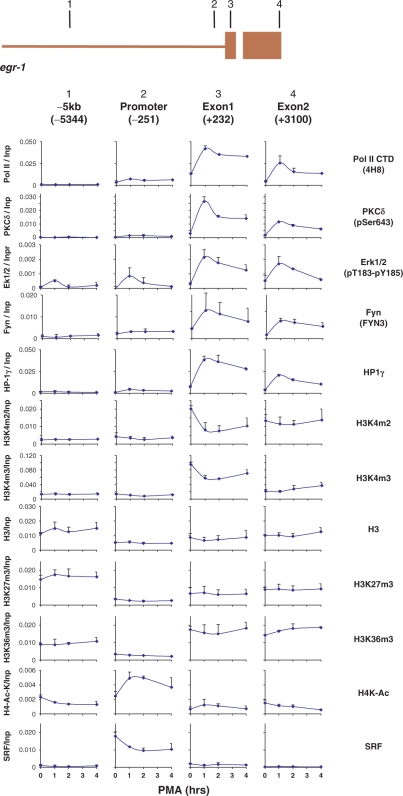
Extracellular stimuli activate signaling cascades that result in changes of gene expression, cell division and other processes. Protein kinase-mediated phosphorylation plays a major role in signal transmission (47). In many cases the steps required for signaling have been defined. Compartmentalization of protein kinases has been recognized as an important mode to control events at discrete intracellular locations (48–52). However, it was not until very recently that inducible kinases were discovered to be directly recruited to genomic sites (28–32). Most of these studies were done in yeast and looked at a single kinase at a time. MAPK, PKC and tyrosine kinase cascades are the major pathways that signal inducible genomic events in response to mitogens and other stimuli (31,32). We used a panel of MAPK, PKC and tyrosine anti-kinase antibodies in Matrix ChIP to identify which kinases target the mitogen-inducible egr-1 locus. Among 18 anti-kinase antibodies tested, this search identified strong recruitment of active phospho Erk1/2 and PKCδ as well as Fyn. We used Matrix ChIP and 20 different antibodies to correlate recruitment of these kinases with chromatin and transcription events (Figures 5 and S1). Pol II density is highest at the transcription start sites (17, 53, 54). Parallel profiling revealed that the kinetics of Erk1/2, PKCδ and Fyn binding are similar to each other and resemble the spatiotemporal patterns of Pol II, hnRNP K (46) and HP1γ (55) recruitment. Higher spatiotemporal resolution is needed to determine if these profiles are identical. HnRNP K acts as a docking platform at sites of nucleic-acids-directed processes (46,56,57). HnRNP K may bind pre-mRNA and/or Pol II. Since hnRNP K interacts with Erk1/2, PKCδ and Fyn (46) it may serve to recruit and/or regulate activity of these enzymes along the egr-1 locus. Several chromatin factors have recently been shown to be associated with transcription including HP1γ (55). It has been suggested that HP1 proteins may provide a dynamic platform for recruitment of regulatory proteins (55,58). Thus, HP-1γ may serve to transmit kinase signals to chromatin and/or to recruit these enzymes. It is also possible that recruited kinases are activated in situ and transmit signals downstream by phosphorylating components of chromatin components and transcriptional machinery.
Figure 5.
Time-course pattern of Erk 1/2, PKCδ and Fyn recruitment profiled in parallel with several chromatin and transcriptional events along the PMA-induced egr-1 gene. Matrix ChIP analysis of sheared chromatin from PMA-treated cells was done using in-lab coated Protein A polystyrene plates and 20 antibodies to different proteins. A non-immune IgG fraction was used as a mock IP control. The results of following ChIPs are shown. Top-to-bottom (all antibodies and amounts used are listed in Table S1), Pol II, RNA polymerase II (4H8), PKCδ (anti-phosphoSer643), Erk1/2 (anti-phosphoThr183-phosphoTyr185), Fyn, HP-1γ, H3K4m2, H3K4m3, H3, H3K27m3, H3K36m3, H4-Ac-K5,8,12,16 and Serum Responsive Factor, SRF. Real-time PCR was done using primers to the regions shown in the diagram of the egr-1 gene (the two exons are shown as boxes). The ChIP results corrected for background (IgG mock IP) are expressed as fraction of input DNA. The graphs represent mean ± SD of two experiments. The full data set (20 different antibodies) is shown in Figure S1.
The constitutive and inducible profiles of chromatin factors and histone marks observed at the egr-1 promoter and the transcribed region (Figures 5 and S1) are in good agreement with the general patterns seen along transcribed genes (17,53,54). For example, lower nucleosome density in regions flanking transcription start sites is a common feature of genes (14,53). We found that total histone H3 levels and several of its marks were lowest at the promoter of transcribed egr-1 gene (Figure 5). Acetylation of lysines eliminates their positive charge which, when it occurs on certain histone tails, has been shown to have a negative effect on the higher order structure of chromatin, essentially making it more open (15). In agreement with previous reports we found inducible changes in histone H3 and H4 acetylation (59). Di- and trimethylation of histone H3 lysine 4, H3K4m2/m3, are associated with actively transcribed genes (60–62). Consistent with previous reports, H3K4m2 and m3 marks were enriched in the egr-1 transcribed region and there was a decrease with induction. As there was no change in total H3 levels this decrease could reflect demethylation or histone exchange (2,63). Trimethylation of histone H3 lysine 36, H3K36m3, is a mark associated with transcription elongation (15,64). H3K36m3 levels were lowest at the promoter and highest in the transcribed region, this pattern of H3K36 methylation, is a common feature of transcribed genes (18,54). Trimethylation of lysine 27 of histone H3, H3K27m3, is a marker of silent genes (60). In agreement with previous studies on H3K27 methylation (54,65,66), the level of H3K27m3 was significantly higher within the 5′ flanking region (Figure S1).
Serum responsive factor (SRF) binds several elements within the egr-1 promoter (67–69). Matrix ChIP revealed SRF presence in the promoter region in uninduced cells and following induction its levels decreased (Figure 5). The significance of the decrease remains to be determined.
The coupled chromatin, transcription and RNA processing and other genome-directed events have emerged as some of the most intensively studied fields of biology today (15,16,18,70). Matrix ChIP introduces the much needed high-throughput technology to effectively use the large collection of antibody probes. With these advances, the Matrix ChIP platform enables us for the first time to follow hundreds of antigens including the less abundant but critical factors such as kinases (Figures 5 and S1). Thus, the Matrix ChIP platform greatly increases the potential to study the complexity of the inducible and dynamic chromatin, transcription and RNA-processing events. Studies of this type also promise to facilitate discovery of epigenetic biomarkers of diseases.
In summary, we have developed a microplate-based ChIP technology that has several important advantages over the test-tube-based assay. (i) The entire procedure is done on the same plate without sample transfers, which simplifies the procedure, increases sample to sample measurement consistency, reduces the time of the assay and reduces sample loss. (ii) Increased throughput allows for efficient sample processing so that one can run 96 or more ChIPs followed by PCR analysis in one day. (iii) Microplate Matrix ChIP could be automated for studies to follow complex chromatin and transcriptional events or high volume screening applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by NIH R37-DK45978 and RO1-GM45134 (K.B.) and NIH R01-GM074511 and P41-EB002027 (D.G.C.). Funding to pay the Open Access publication charges for the article was provided by NIH DK54978. This paper is dedicated to the memory of Helen Minski who passed away after a long and courageous battle with cancer.
Conflict of interest statement. None declared.
REFERENCES
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, III, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Thiriet C, Hayes JJ. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol. Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 5.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 6.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 7.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 8.Spector DL. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 9.Lorincz MC, Schubeler D. RNA polymerase II: just stopping by. Cell. 2007;130:16–18. doi: 10.1016/j.cell.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 10.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 11.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 13.Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 14.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 20.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res. 2006;34:e2. doi: 10.1093/nar/gnj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L'Honore A, Rana V, Arsic N, Franckhauser C, Lamb NJ, Fernandez A. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol. Biol. Cell. 2007;18:1992–2001. doi: 10.1091/mbc.E06-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007;21:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seger R, Krebs EG. The MAPK signaling cascade. Faseb J. 1995;9:726–735. [PubMed] [Google Scholar]
- 25.Gschwendt M. Protein kinase Cd. Eur. J. Biochem. 1999;259:555–564. doi: 10.1046/j.1432-1327.1999.00120.x. [DOI] [PubMed] [Google Scholar]
- 26.Resh MD. Fyn, a Src family tyrosine kinase. Int. J. Biochem. Cell Biol. 1998;30:1159–1162. doi: 10.1016/s1357-2725(98)00089-2. [DOI] [PubMed] [Google Scholar]
- 27.Ostrowski J, Kawata Y, Schullery DS, Denisenko ON, Bomsztyk K. Transient recruitment of the hnRNP K protein to inducibly transcribed gene loci. Nucleic Acids Res. 2003;31:3954–3962. doi: 10.1093/nar/gkg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow CW, Davis RJ. Proteins kinases: chromatin-associated enzymes? Cell. 2006;127:887–890. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 31.Edmunds JW, Mahadevan LC. Cell signaling. Protein kinases seek close encounters with active genes. Science. 2006;313:449–451. doi: 10.1126/science.1131158. [DOI] [PubMed] [Google Scholar]
- 32.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, O'Neill BC, Suzuki Y, Denisenko ON, Bomsztyk K. Activation of a nuclear DNA-binding protein recognized by a transcriptional element, bcn-1, from the laminin B2 chain gene promoter. J. Biol. Chem. 1996;271:18981–18988. doi: 10.1074/jbc.271.31.18981. [DOI] [PubMed] [Google Scholar]
- 34.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 35.Chen R, Weng L, Sizto NC, Osorio B, Hsu CJ, Rodgers R, Litman DJ. Ultrasound-accelerated immunoassay, as exemplified by enzyme immunoassay of choriogonadotropin. Clin. Chem. 1984;30:1446–1451. [PubMed] [Google Scholar]
- 36.Mendoza LG, McQuary P, Mongan A, Gangadharan R, Brignac S, Eggers M. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA) Biotechniques. 1999;27:778–788. doi: 10.2144/99274rr01. [DOI] [PubMed] [Google Scholar]
- 37.Desai S, Dworecki BR. Coated microwell plate-based affinity purification of antigens. Anal. Biochem. 2004;328:162–165. doi: 10.1016/j.ab.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Peluso P, Wilson DS, Do D, Tran H, Venkatasubbaiah M, Quincy D, Heidecker B, Poindexter K, Tolani N, Phelan M, et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 39.Briand E, Salmain M, Compere C, Pradier CM. Immobilization of Protein A on SAMs for the elaboration of immunosensors. Colloids Surf B Biointerfaces. 2006;53:215–224. doi: 10.1016/j.colsurfb.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Dai J, Bao Z, Sun L, Hong SU, Baker GL, Bruening ML. High-capacity binding of proteins by poly(acrylic acid) brushes and their derivatives. Langmuir. 2006;22:4274–4281. doi: 10.1021/la0600550. [DOI] [PubMed] [Google Scholar]
- 41.Hollmann O, Gutberlet T, Czeslik C. Structure and protein binding capacity of a planar PAA brush. Langmuir. 2007;23:1347–1353. doi: 10.1021/la061881b. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka G, Funabashi H, Mie M, Kobatake E. Fabrication of an antibody microwell array with self-adhering antibody binding protein. Anal. Biochem. 2006;350:298–303. doi: 10.1016/j.ab.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 43.Danczyk R, Krieder B, North A, Webster T, HogenEsch H, Rundell A. Comparison of antibody functionality using different immobilization methods. Biotechnol. Bioeng. 2003;84:215–223. doi: 10.1002/bit.10760. [DOI] [PubMed] [Google Scholar]
- 44.Anderson GP, Jacoby MA, Ligler FS, King KD. Effectiveness of protein A for antibody immobilization for a fiber optic biosensor. Biosens. Bioelectron. 1997;12:329–336. doi: 10.1016/s0956-5663(96)00074-7. [DOI] [PubMed] [Google Scholar]
- 45.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 46.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 47.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 48.Denis V, Cyert MS. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot. Cell. 2005;4:36–45. doi: 10.1128/EC.4.1.36-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timms JF, Swanson KD, Marie-Cardine A, Raab M, Rudd CE, Schraven B, Neel BG. SHPS-1 is a scaffold for assembling distinct adhesion-regulated multi-protein complexes in macrophages. Curr. Biol. 1999;9:927–930. doi: 10.1016/s0960-9822(99)80401-1. [DOI] [PubMed] [Google Scholar]
- 50.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Rongish BJ, Kinsey WH. Transient nuclear localization of Fyn kinase during development in zebrafish. Anat. Rec. 2000;260:115–123. doi: 10.1002/1097-0185(20001001)260:2<115::AID-AR10>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 52.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 53.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 54.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Ostrowski J, Schullery DS, Denisenko ON, Higaki Y, Watts J, Aebersold R, Stempka L, Gschwendt M, Bomsztyk K. Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem. 2000;275:3619–3628. doi: 10.1074/jbc.275.5.3619. [DOI] [PubMed] [Google Scholar]
- 57.Adolph D, Flach N, Mueller K, Ostareck DH, Ostareck-Lederer A. Deciphering the crosstalk between hnRNP K and c-Src: the c-Src activation domain in hnRNP K is distinct from a second interaction site. Mol. Cell Biol. 2006;27:1758–1770. doi: 10.1128/MCB.02014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 59.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 60.Miao F, Natarajan R. Mapping global histone methylation patterns in the coding regions of human genes. Mol. Cell Biol. 2005;25:4650–4661. doi: 10.1128/MCB.25.11.4650-4661.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morillon A, Karabetsou N, Nair A, Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 62.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 63.Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shilatifard A. Transcriptional elongation control by RNA polymerase II: a new frontier. Biochim. Biophys. Acta. 2004;1677:79–86. doi: 10.1016/j.bbaexp.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 65.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 66.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 67.Wu SQ, Minami T, Donovan DJ, Aird WC. The proximal serum response element in the Egr-1 promoter mediates response to thrombin in primary human endothelial cells. Blood. 2002;100:4454–4461. doi: 10.1182/blood-2002-02-0415. [DOI] [PubMed] [Google Scholar]
- 68.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol. Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper SJ, Trinklein ND, Nguyen L, Myers RM. Serum response factor binding sites differ in three human cell types. Genome Res. 2007;17:136–144. doi: 10.1101/gr.5875007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.