Abstract
RNA chaperone proteins are essential partners of RNA in living organisms and viruses. They are thought to assist in the correct folding and structural rearrangements of RNA molecules by resolving misfolded RNA species in an ATP-independent manner. RNA chaperoning is probably an entropy-driven process, mediated by the coupled binding and folding of intrinsically disordered protein regions and the kinetically trapped RNA. Previously, we have shown that the core protein of hepatitis C virus (HCV) is a potent RNA chaperone that can drive profound structural modifications of HCV RNA in vitro. We now examined the RNA chaperone activity and the disordered nature of core proteins from different Flaviviridae genera, namely that of HCV, GBV-B (GB virus B), WNV (West Nile virus) and BVDV (bovine viral diarrhoea virus). Despite low-sequence similarities, all four proteins demonstrated general nucleic acid annealing and RNA chaperone activities. Furthermore, heat resistance of core proteins, as well as far-UV circular dichroism spectroscopy suggested that a well-defined 3D protein structure is not necessary for core-induced RNA structural rearrangements. These data provide evidence that RNA chaperoning—possibly mediated by intrinsically disordered protein segments—is conserved in Flaviviridae core proteins. Thus, besides nucleocapsid formation, core proteins may function in RNA structural rearrangements taking place during virus replication.
INTRODUCTION
Members of the positive-strand RNA virus family Flaviviridae are small, enveloped viruses containing a single-stranded RNA genome. Flaviviridae is classified in three genera (Flavivirus, Pestivirus and Hepacivirus), with important human and/or animal pathogens present in each genus, having considerable global health and socio-economic impacts. Hepatitis C virus (HCV; a hepacivirus) is estimated to infect more than 120 million persons worldwide, corresponding to a prevalence of 2.2–2.5% in the human population (1). Chronic HCV infection may entail serious progressive liver disease, including liver cirrhosis and hepatocellular carcinoma (HCC), and is the most important indication for liver transplantation (2). Major arthropod-borne pathogens of the flaviviruses include West Nile virus (WNV), yellow fever virus (YFV), dengue virus (DEN) and tick-borne encephalitis virus (TBEV); while pestiviruses, including bovine viral diarrhoea virus (BVDV) and classical swine fever virus (CSFV), are animal pathogens infecting livestock and wild ruminants. Viruses from the three genera share fundamental features in their genome organization, replication strategy and virion morphology, but at the same time they exhibit an impressive collection of genus- and virus-specific adaptations (3).
The genome of Flaviviridae encodes a single, long polyprotein which is co- and post-translationally processed by cellular and viral proteases to yield the mature structural and non-structural proteins of the virus (3). The core (capsid) protein, located at the N-terminal region of the polyprotein, is a small, highly basic RNA-binding protein that presumably encapsidates and coats the genomic RNA in the viral particle. Besides their highly basic character, core proteins from the three Flaviviridae genera do not exhibit significant sequence similarities or any apparent common features in domain organization. Hepacivirus core proteins (Figure 1) are generated in their mature form via successive cleavages of the polyprotein by the endoplasmic reticulum-associated signal peptidase (SP) and signal peptide peptidase (SPP) enzymes (4–7). The mature proteins consist of two domains: the N-terminal domain 1 (D1) which constitutes the RNA-binding region, and the C-terminal hydrophobic domain (D2) which targets HCV and GBV-B core proteins to intracellular lipid droplets (5,8,9). Flavivirus capsid proteins are released from the viral polyprotein by the action of the virus-encoded NS2B–NS3 serine protease complex (10–12). In contrast to hepaci- and flaviviruses, where the core protein is located at the exact N-terminus of the polyprotein, in pestiviruses core is preceded by the N-terminal protease (Npro). The N-terminus of mature core is generated by an autocatalytic cleavage of Npro, while the C-terminus is processed successively by SP and SPP, similarly to hepacivirus core processing (13–16). At present, pestivirus core proteins are biochemically and structurally poorly characterized.
Figure 1.
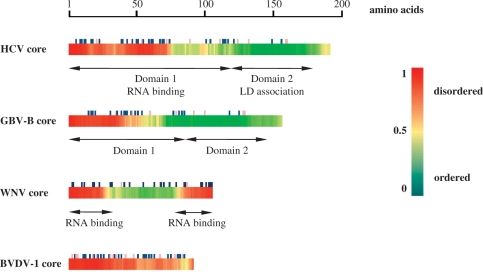
Disorder prediction in Flaviviridae core proteins. Disordered regions in HCV, GBV-B, WNV and BVDV core proteins (accession numbers D89872; AF179612; AF481864; and AF220247, respectively) were predicted using the DisProt VL3-H predictor (http://www.ist.temple.edu/disprot/predictor.php). An amino acid with a disorder score above 0.5 is considered to be in a disordered environment, while below 0.5 to be ordered. The predicted disorder is illustrated by a colour scale, with highly flexible segments in red and well-folded domains in green. Basic and acidic amino acid residues are indicated by dark blue and mauve symbols, respectively.
RNA chaperones are abundant proteins in all living organisms and some viruses. They are thought to function by providing assistance to the correct folding of RNA molecules by preventing their misfolding or by resolving misfolded RNA species (17). At every stage of cellular RNA metabolism, including transcription, RNA transport, translation and storage, RNA molecules are associated with a distinct subset of chaperone molecules, which protect them and help in their folding process (18–20). Examples of cellular RNA chaperone proteins include the major messenger ribonucleoprotein particle (mRNP)-associated protein YB-1, the fragile X mental retardation protein (FMRP), and several hnRNP proteins (18). In viruses, human immunodeficiency virus type 1 (HIV-1) nucleocapsid protein represents a canonical example of a multifunctional RNA chaperone (21–23).
Importantly, RNA chaperones are able to promote profound structural rearrangements in RNA without ATP consumption. Based on the high proportion of intrinsically unstructured regions in RNA chaperones, Tompa and Csermely (24) recently suggested an elegant model for the ATP-independent mechanism of chaperone function. According to the ‘entropy exchange model’, highly flexible protein regions would undergo disorder-to-order transition upon binding to RNA, concomitantly melting the RNA structure through an entropy exchange process. The de-stabilized RNA then could search the conformational space again, eventually reaching its most stable conformation upon cyclic protein binding and release (24).
We previously reported that the core protein of HCV possesses RNA chaperone activities in vitro (25,26). In this study, we examined whether this activity is conserved in the Flaviviridae family. We found that the core proteins of the hepacivirus GBV-B, the pestivirus BVDV and the flavivirus WNV all exhibit RNA chaperone activities in vitro. In addition, since core proteins from the three genera show marked differences in their sequence and domain organization, their shared features may give insights into the underlying common characteristics of RNA chaperones. Thus, using the Flaviviridae core proteins as a model system, we examined whether highly flexible, intrinsically unstructured protein regions play an important role in mediating RNA chaperoning, as predicted by the entropy exchange model. By a combination of far-UV circular dichroism (CD) spectroscopy, heat denaturation and in vitro chaperone assays, we show that short, highly basic and flexible protein segments are a hallmark of active RNA chaperone domains in Flaviviridae core proteins.
MATERIALS AND METHODS
Proteins and peptides
Recombinant HCV genotype 1b core (corresponding to amino acids 2–169 and 2–117; accession number D89872) and GBV-B core (amino acids 2–155; accession number AF179612) proteins were expressed in Escherichia coli and purified as previously described (25,27–28). HCV core proteolytic fragments (amino acids 2–54 and 90–159) were generated by endoproteinase Glu-C cleavage of HCV core(2–169) and purified by HPLC as previously reported (25). WNV core (amino acids 2–105; accession number AF481864) and BVDV core (amino acids 2–91; accession number AF220247) were cloned, expressed and purified as previously reported for HCV core (27) except that the proteins were purified from the E. coli soluble fraction under non-denaturating conditions. All the purified proteins were stored in a buffer containing 20 mM sodium phosphate (pH 7.4) and 5 mM 2-mercaptoethanol and were >95% pure as revealed by SDS–PAGE (Supplementary Figure 1).
WNV core peptides were synthesized on an ABI 433 apparatus with Fmoc-OH/DCC/Hobt chemistry. The peptides were cleaved by a TFA solution with classical scavengers and precipitated in diethyl ether. The precipitate was centrifuged and the pellet was solubilized in water and lyophilized. The crude peptides were dissolved in water and purified on Vydac column (C18, 5 µm, 250 × 10 mm) with an appropriate gradient of B (70% acetonitrile, 0.09% TFA solution in water). Purified peptides were characterized by electrospray mass spectrum (SCIEX API 165) at 2567 UMA for peptide WNV 1–24 and 3041 UMA for peptide WNV 80–105, and by HPLC apparatus HP 1100 on analytical column Vydac (C18, 5 µm, 250 × 4.6 mm) in a gradient of 30 min from 10 to 90% of B.
Oligodeoxynucleotide (ODN) labelling
Oligonucleotides corresponding to the HIV-1 TAR sequence (MAL strain) in the sense and anti-sense orientation were purchased from Eurogentec (Belgium). Tar(+): 5′ G G T C T C T C T T G T T A G A C C A G G T C G A G C C C G G G A G C T C T C T G G C T A G C A A G G A A C C C; Tar(–): 5′ G G G T T C C T T G C T A G C C A G A G A G C T C C C G G G C T C G A C C T G G T C T A A C A A G A G A G A C C. Tar(−) was 32P-labelled with 50 µCi of γ32P-ATP using T4 polynucleotide kinase (Invitrogen), and subsequently purified by 10% PAGE, 7 M urea in 0.5× TBE.
In vitro RNA synthesis
In order to obtain template DNAs, plasmids pR3 and pS14 were digested by PstI and treated with Klenow polymerase (Invitrogen) to remove 3′ overhangs. In vitro transcription was carried out using T7 RNA polymerase, according to the manufacturer's instructions (Promega). RNAs were labelled by incorporation of α32P-UMP during in vitro transcription. RNAs were purified on a 8% denaturing polyacrylamide gel containing 7 M urea in 50 mM Tris–borate, pH 8.3, 1 mM EDTA (0.5× TBE) and recovered by elution in 0.3 M sodium acetate–0.1% SDS for 4 h at 37°C, followed by ethanol precipitation.
DNA annealing
Fifteen femtomoles of Tar(+) and equal amounts of 32P-labelled Tar(−) ODNs were incubated with or without protein in 10 µl of annealing buffer (20 mM Tris–HCl, pH 7.0, 30 mM NaCl, 0.1 mM MgCl2, 10 µM ZnCl2 and 5 mM DTT). Standard reactions were performed at 37°C for 5 min, except for the heat-annealed positive control which was incubated at 62°C for 30 min. Protein (peptide)-to-nucleotide molar ratios are indicated in the figure legends. In order to assess the resistance of RNA chaperone activity to heat denaturation, proteins were boiled for 5 min and chilled on ice prior to incubation with the ODNs. Annealing reactions were stopped with 5 µl of 20% glycerol, 20 mM EDTA, pH 8.0, 0.2% SDS, 0.25% bromophenol blue and 0.4 mg/ml calf liver tRNA (29). DNAs were resolved by 8% native PAGE in 50 mM Tris–borate, pH 8.3, 1 mM EDTA (0.5× TBE). Double-stranded versus single-stranded DNA ratios were determined by autoradiography and PhosporImager quantification.
Hammerhead ribozyme cleavage
R3 ribozyme and S14 substrate RNAs were independently heated for 1 min at 90°C in water. Reaction buffer was added to yield final concentrations of 5 mM MgCl2, 100 mM NaCl, 20 mM Tris–HCl, pH 7.5 and RNAs were slowly cooled down to 37°C. Following a further 5 min incubation at 20°C, 5 nmol R3 and 30 nmol S14 were combined in a final volume of 10 µl, and proteins were added at a final protein-to-nucleotide ratio as indicated in the figure legends. Standard reactions were incubated for 25 min at 37°C and stopped by adding 20 µl of stop solution (0.5% SDS, 25 mM EDTA). RNAs were phenol–chloroform extracted, followed by ethanol precipitation and resuspension of the pellet in 10 µl of loading buffer (45% formamide, 0.5× TBE, and 0.1% bromophenol blue). RNAs were resolved on an 8% denaturing polyacrylamide gel containing 7 M urea in 50 mM Tris–borate, pH 8.3 and 1 mM EDTA (0.5× TBE). Product-to-substrate ratios were determined by autoradiography and PhosporImager quantification.
CD spectroscopy
CD spectra were recorded on a Chirascan (Applied Photophysics) spectrophotometer. Routinely, measurements were done at 20°C in a 0.02 cm path-length quartz cuvette (Hellma) with protein concentration of 20 µM in 20 mM sodium phosphate buffer (pH 7.4) containing 5 mM 2-mercaptoethanol. Spectra were recorded in the 180–260 nm wavelength range with 0.1 nm increments and 2 s integration times. Protein secondary structure content was determined by using the k2d method of spectral deconvolution at the Dichroweb web facility (http://www.cryst.bbk.ac.uk/cdweb/html/home.html). Denaturation and renaturation were recorded at 222 nm from 20°C to 95°C with a denaturation speed of 1°C/min and with measurement each 0.5°C.
RESULTS
Prediction of disordered regions in Flaviviridae core proteins
RNA chaperone proteins do not share a consensus RNA-binding domain or motif that would make possible their identification from amino acid sequences or structural information alone. However, mostly disordered regions with a highly basic character are probably a hallmark of RNA chaperones (20,24) and, together with clues from protein function, can be considered as an indication for RNA chaperone activities. Indeed, these flexible regions may undergo disorder-to-order transition upon binding to a misfolded RNA structure, and help in its folding process by an entropy exchange mechanism, as proposed by Tompa and Csermely (24). As a proof-of-concept, successful identification of an active RNA chaperone domain in the Gypsy retrotransposon Gag protein—based on disorder prediction and charge distribution—has been reported (30).
We used the DisProt VL3-H neural network predictor, developed by Dunker et al. [http://www.ist.temple.edu/disprot/predictor.php (31)] to assess intrinsic disorder in core proteins from the three Flaviviridae genera (Figure 1). DisProt predictors can identify relatively long unstructured regions with a reasonable accuracy from sequence information alone. The prediction gave a good overall agreement with data available on the structural and domain organization of core proteins, and identified the known RNA-binding domains as highly flexible regions within the proteins (Figure 1). In order to increase the confidence of the prediction, the same sequences were also submitted to the IUPred [http://iupred.enzim.hu/index.html (32)] and FoldIndex [http://bip.weizmann.ac.il/fldbin/findex (33)] servers, which use different parameters for disorder prediction, based on the estimated pair-wise energy content or the ratio of the hydrophobicity and net charge of a sequence, respectively (34,35). Both IUPred and FoldIndex gave similar disorder-order profiles to that of the VL3-H predictor shown on Figure 1 (data not shown).
HCV and GBV-B core proteins are believed to share a common domain organization (9,36), with an N-terminal, highly basic RNA-binding domain (domain 1 or D1) and a C-terminal, fairly hydrophobic domain (D2), which mediates lipid droplet association of the proteins (7,9). As shown in Figure 1, the two domains are clearly separated by their disorder profile and their basic amino acid content, where the RNA-binding region is highly flexible and rich in basic residues. In its unbound state, HCV core protein has been shown to lack considerable structure [(36) and Figure 5]. Interaction with intracellular membranes was proposed to induce the formation of a helix-loop-helix structure in D2, which is thought to be essential for the concomitant folding of the full-length (FL) protein into an α helix-rich conformation (36,37). Importantly, the isolated N-terminal domain of HCV core was shown to mediate RNA-binding (5), RNA chaperoning (25,26) and in vitro particle assembly (38,39), indicating that D2-mediated folding is not essential for these processes.
Figure 5.
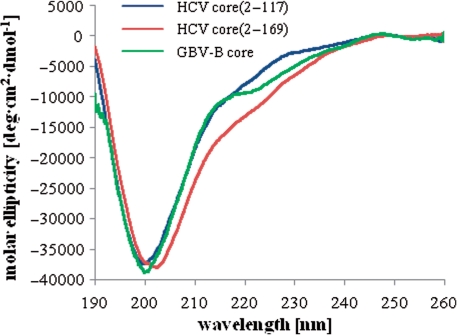
Far-UV CD analysis of HCV and GBV-B core proteins. CD spectra were recorded at 20°C, in 20 mM sodium phosphate buffer (pH 7.4) containing 5 mM 2-mercaptoethanol.
The crystal structure of WNV core protein and the NMR structure of the related DEN core have recently been reported (40,41). With the exception of amino acid residues 1–20 and a short C-terminal tail, which appear to be highly flexible (40–42), these flavivirus core proteins adopt a compact dimeric fold consisting of four α-helices per monomer, with distinct putative RNA binding and membrane interaction surfaces, proposed based on the asymmetric spatial charge distribution of the dimer (41). According to this model, RNA binding is mediated by the most C-terminal α-helix of the protein. In addition, specific in vitro association of the isolated N- and C-terminal regions (32 and 26 amino acids, respectively) with viral RNA fragments has been reported (43).
While hepaci- and flavivirus core proteins contain additional domains besides the markedly basic RNA-binding region(s), BVDV core protein seems to lack distinct functional domains, and shows a uniform charge distribution along its length. Interestingly, BVDV core is predicted to be completely disordered (Figure 1), indicating that it may function as an intrinsically unstructured protein.
Flaviviridae core proteins possess DNA-annealing activity
Full-length Flaviviridae core proteins were expressed in E. coli with a C-terminal (His)6-tag to facilitate their purification (see Materials and Methods section). The ability of purified proteins to stably bind RNA and DNA was verified by means of mobility shift assays. All four proteins bound both to RNA and DNA without a strict sequence specificity, and they caused complete retention of the nucleic acids at the top of the gel (indicative of the formation of large nucleoprotein complexes) at a protein-to-nucleotide molar ratio of ∼1:20 (data not shown).
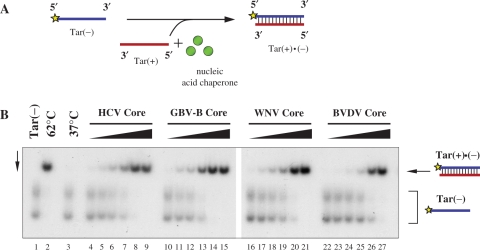
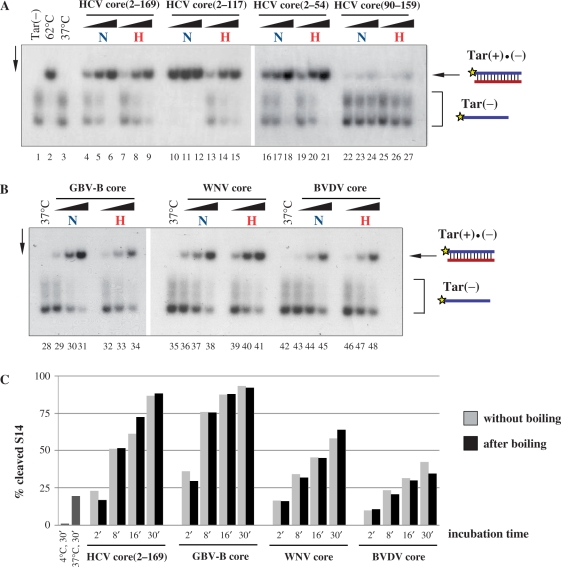
In order to assess the putative nucleic acid chaperone activity of Flaviviridae core proteins, their capacity to enhance the annealing of complementary ODNs was examined. The 56-mer Tar(−) ODN can form a stable hairpin structure, which impedes its hybridization with the complementary Tar(+) ODN. Strand annealing can occur only at high temperatures or in the presence of a protein with nucleic acid chaperone activity [Figure 2A, (18)]. The helix-destabilizing activity associated with nucleic acid chaperones is essential for efficient hybridization, since molecular crowding or charge neutralization caused by basic peptides or single-stranded nucleic acid-binding proteins without chaperone activity were shown not to be sufficient for duplex formation (25). 32P-labelled Tar(−) and Tar(+) were incubated with increasing amounts of proteins at 37°C for 5 min. After dissociation of the DNA–protein complexes, duplex formation was analysed by native polyacrylamide gel electrophoresis. In the absence of protein, no significant annealing was detected at 37°C (Figure 2A, lane 3), while complete hybridization was achieved upon 30 min incubation at 62°C (lane 2). Co-incubation of the complementary ODNs with any of the Flaviviridae core proteins (at increasing protein-to-nucleotide molar ratios) strongly enhanced annealing at 37°C (lanes 4–27), in all cases leading to complete complex formation at a protein-to-nucleotide molar ratio of 1:5. Annealing of Tar(−) and Tar(+) ODNs was found to be extremely rapid with all core proteins tested, reaching almost maximal duplex formation at very early time points, after 10–30 s incubation with the proteins (data not shown), indicating that core protein chaperoning probably involves only a limited number of binding-and-release cycles.
Figure 2.
Flaviviridae core proteins exhibit DNA annealing activity. (A) Schematic representation of the DNA annealing assay. Radioactively labelled Tar(−) and the complementary Tar(+) ODNs are incubated in the absence or presence of the putative nucleic acid chaperone. Efficient hybridization of the oligonucleotides takes place only at elevated temperatures or in the presence of a protein with nucleic acid chaperone activity. (B) Annealing of complementary oligonucleotides is promoted by core proteins. 32P-labelled Tar(−) ODN was incubated together with Tar(+) ODN in the presence of increasing amounts of the proteins, as indicated at the top of the figure. Protein-to-nucleotide molar ratios were 1:160, 1:80, 1: 40, 1:20, 1:10 and 1:5 for each protein tested (corresponding to 1, 2, 4, 8, 16 and 32 nM protein concentrations, respectively). Lane 1: labelled Tar(−) ODN alone; lane 2: Tar(−)/Tar(+) complex formed by heat annealing at 62°C, without protein; lane 3: Tar(−)/Tar(+) complex formation at 37°C in the absence of protein; lanes 4–9, 10–15, 16–21 and 22–27: complex formation at 37°C in the presence of increasing concentrations of HCV, GBV-B, WNV and BVDV core proteins, respectively. Tar(−) migrates as two distinct bands due to its extensive secondary structures.
Overall, these results show that Flaviviridae core proteins, despite their highly divergent sequences and markedly different domain organization, all facilitate nucleic acid annealing.
Enhancement of hammerhead ribozyme cleavage by Flaviviridae core proteins
RNA chaperone proteins can destabilize existing interactions within and between RNA molecules, thus allowing formation of new contacts (44). Due to this mechanism, RNA chaperones can facilitate both the annealing of RNA strands and the unwinding of pre-formed helices. A canonical in vitro assay to examine both facets of RNA chaperoning is the hammerhead ribozyme cleavage assay [(21,45–46), Figure 3A]. In the absence of protein cofactors, hammerhead ribozyme-mediated cleavage is relatively slow, limited either by the slow rate of substrate-ribozyme complex formation, especially at subsaturating substrate concentrations (step 1 on Figure 3A), or by the slow release of the cleavage products (step 3). RNA chaperones, such as hnRNP A1, the FMRP and HIV-1 NCp7 were shown to accelerate both annealing and product release, thus allowing fast recycling of the ribozyme (21,45–47).
Figure 3.
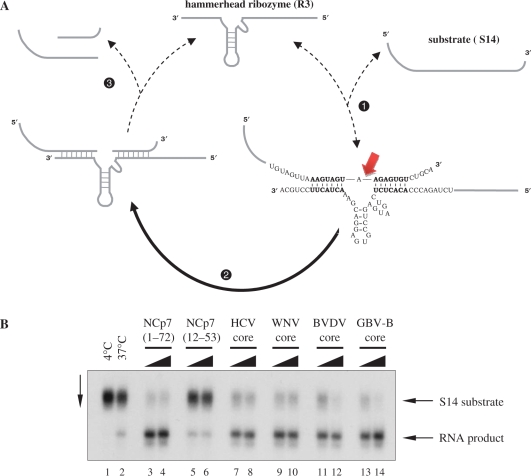
Enhancement of hammerhead ribozyme cleavage by Flaviviridae core proteins. (A) Schematic representation of the hammerhead ribozyme RNA cleavage reaction. Efficient cleavage of the 32P-labelled RNA substrate (S14) by the R3 hammerhead ribozyme necessitates hybridization of the substrate to the complementary region in the ribozyme sequence (step 1), and release of the cleaved products (step 3), allowing the cyclic reuse of the ribozyme. In the absence of a nucleic acid chaperone, steps 1 and 3 are slow, while both annealing and ribozyme turnover are greatly accelerated by proteins with nucleic acid chaperone activity. S14 RNA substrate cleavage is indicated by the arrow. (B) Hammerhead ribozyme-mediated cleavage is enhanced by Flaviviridae core proteins. R3 ribozyme and S14 substrate RNA were incubated and analysed as described in Materials and Methods section. Lanes 1 and 2: substrate cleavage in the absence of protein at 4°C and 37°C; lanes 3 and 4, 5 and 6, 7 and 8, 9 and 10, 11 and 12 and 13 and 14: substrate cleavage at 37°C in the presence of proteins (as indicated at the top of the figure), at 1:18 and 1:9 protein-to-nucleotide molar ratios (corresponding to 16.6 and 33.3 nM protein concentrations), respectively.
R3 hammerhead ribozyme and 32P-labelled S14 substrate (with a 14-nt long region base-pairing with the ribozyme) were incubated at 37°C for 25 min, together with Flaviviridae core proteins. Following incubation, proteins were removed by phenol–chloroform extraction and RNA products were analysed by PAGE under denaturing conditions. In the absence of protein, cleavage of the substrate occurred slowly, yielding ∼15% of product in 25 min at 37°C (Figure 3B, lane 2). In contrast, all Flaviviridae core proteins caused a clear activation of ribozyme-directed cleavage of the S14 RNA substrate (compare lanes 7–14 with lane 2). The nucleocapsid protein of HIV-1 (NCp7, aa 1–72) was included as a well-characterized RNA chaperone and, as expected, it greatly facilitated the cleavage reaction (lanes 3–4). Conversely, a deletion mutant of NCp7 (aa 12–53) was inactive in this assay (lanes 5 and 6), despite its highly basic nature, confirming that bona fide RNA chaperone activity is necessary for the enhancement of ribozyme-mediated cleavage.
The chaperoning activity of Flaviviridae core proteins is resistant to heat denaturation
As a first approach to assess whether RNA chaperone activity is really mediated by unstructured proteic regions (24), we examined the heat resistance of Flaviviridae core chaperone function. In contrast to well-folded proteins that usually undergo irreversible denaturation upon heat treatment, intrinsically unstructured proteins (IUPs) are known to be mostly heat resistant (48,49), a property that has been exploited for their purification (50) and large-scale identification (51,52).
FL HCV core protein and its deletion mutants, as well as FL GBV-B, WNV and BVDV core proteins were boiled for 5 min, followed by immediate quenching on ice. Chaperone activity without and after boiling was analysed by the capacity of proteins to promote strand annealing of the complementary Tar(−) and Tar(+) ODNs (Figure 2A). FL HCV and GBV-B core proteins retained most of their chaperone activity after boiling for 5 min (∼75%, based on PhosporImager quantification, see lanes 4–6 versus 7–9 in Figure 4A and lanes 29–31 versus 32–34 in Figure 4B). Surprisingly, both WNV core, a protein with a well-defined 3D structure (40), and BVDV core, which was predicted to be completely unstructured (Figure 1), retained their full activity upon boiling (compare lanes 36–38 and 39–41, and 43–45 and 46–48 in Figure 4B). Kinetic analysis of the heat resistance of WNV core protein chaperone activity indicated that it remains fully active up to 10 min of boiling, but further incubation at 100°C led to rapid loss of strand annealing activity (data not shown).
Figure 4.
Heat resistance of Flaviviridae core protein chaperoning activity. (A) and (B) 32P-labelled Tar(−) ODN was incubated together with Tar(+) ODN in the presence of increasing amounts of the proteins, as indicated at the top of the figure. Proteins were either kept on ice (labelled with ‘N’ for native) or boiled (labelled with ‘H’ for heat) for 5 min before mixing with the ODNs. Protein-to-nucleotide molar ratios were 1:40, 1:20 and 1:10 for each protein tested (corresponding to 4, 8 and 16 nM protein concentrations, respectively). Lane 1: labelled Tar(−) ODN alone; lane 2: Tar(−)/Tar(+) complex formed by heat annealing at 62°C, without protein; lanes 3, 28, 35 and 42: Tar(−)/Tar(+) complex formed at 37°C in the absence of protein; lanes 4–6, 10–12, 16–18, 22–24, 29–31, 36–38 and 43–45: complex formation at 37°C in the presence of increasing concentrations of proteins without prior boiling; lanes 7–9, 13–15, 19–21, 25–27, 32–34, 39–41 and 46–48: complex formation at 37°C in the presence of increasing concentrations of proteins after boiling. (C) Kinetics of hammerhead ribozyme cleavage in the presence of Flaviviridae core proteins. R3 ribozyme and 32P-labelled S14 substrate were incubated with core proteins at 1:20 protein-to-nucleotide molar ratio (corresponding to 15 nM protein concentrations) at 37°C. Reactions were stopped at different time points, as indicated in the figure. Proteins were either kept on ice (grey bars) or boiled for 5 min (black bars) before incubation with the RNAs. RNAs were resolved on an 8% denaturing polyacrylamide gel and the percentages of the cleaved S14 substrate were determined by autoradiography and PhosporImager quantification. As a control, R3 and S14 were co-incubated without protein either at 4°C or at 37°C (dark grey bars). Results of a representative experiment are shown.
C-terminally truncated HCV core proteins [core(2–117), corresponding to the D1 domain of the protein, and core(2–54)] exhibited potent annealing activities (Figure 4A, lanes 10–12 and 16–18), in agreement with our earlier findings (25,26). Duplex formation was slightly reduced with heat treated core(2–117) but was not affected with core(2–54) (lanes 13–15 and 19–21, respectively).
Since Tar(−)/Tar(+) complex formation reaches a maximum level extremely rapidly in the presence of Flaviviridae cores, it was possible that a potential decrease in RNA chaperone activity upon boiling of the proteins would be masked by the relatively long incubation time in these assays. In order to compare the kinetics of chaperoning associated with core proteins before and after boiling, we took advantage of the relatively slow reaction rates of the ribozyme cleavage reaction. R3 hammerhead ribozyme and 32P-labelled S14 substrate were incubated together with Flaviviridae core proteins with or without prior boiling. Reactions were stopped after 2, 8, 16 or 30 min of incubation, and the ratio of cleaved versus uncleaved substrate RNA was determined by autoradiography following denaturing gel electrophoresis (Figure 4C). After 30 min incubation, ∼20% of the substrate RNA was cleaved in the absence of protein. As expected, all core proteins induced a considerable increase in the cleavage rates, with hepacivirus core proteins demonstrating a higher activity compared to WNV and BVDV cores (Figure 4C). Importantly, boiling of the proteins for 5 min before incubation with the RNAs did not have an effect either on the kinetics or on the end-point of the reaction, indicating that heating does not lead to a decrease in the RNA chaperone activity of Flaviviridae core proteins.
Overall, the potent strand annealing activity and facilitation of ribozyme cleavage retained after boiling of the proteins provide convincing evidence that heat resistance is a general feature of Flaviviridae cores.
Investigating the secondary structure content of Flaviviridae core proteins
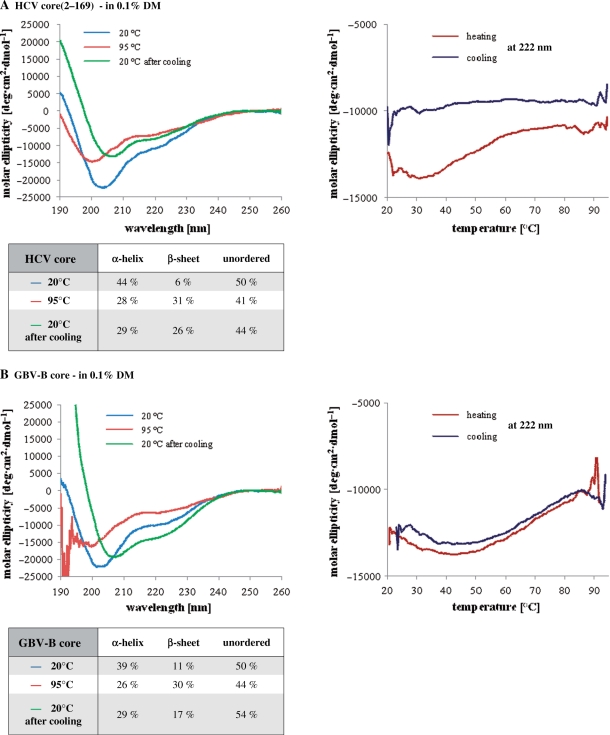
In order to get further insights into the structural requirements for RNA chaperoning, we carried out far-UV CD measurements on FL HCV, GBV-B, WNV and BVDV core proteins, and on HCV core(2–117). The secondary structure of HCV core has been well characterized before (36,53), and was included in these studies only for a direct comparison with other Flaviviridae core proteins (Figure 5). The isolated N-terminal domain of HCV core [core(2–117)], as well as the FL HCV and GBV-B core proteins are mostly unstructured in solution, showing an ellipticity minimum at ∼198 nm, characteristically of random-coil like peptides [(36) and Figure 5]. Indeed, estimation of the secondary structure content by CD deconvolution indicated <10% α-helical structure for hepacivirus core proteins [(36) and data not shown]. As previously reported, FL HCV core [core(2–169)] requires the presence of mild, non-ionic detergents [such as 0.1% n-dodecyl β-d-maltoside (DM)] for efficient solubilization, presumably mimicking the effect of lipid droplet association. Under these conditions HCV core adopts a mostly α-helical conformation [(36), and Figure 6A].
Figure 6.
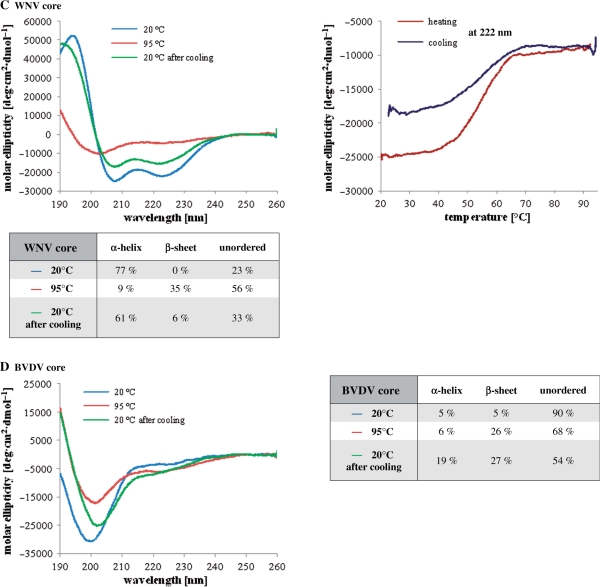
Far-UV CD analysis of core proteins. Far-UV CD spectra of HCV (A), GBV-B (B), WNV (C) and BVDV (D) core proteins. Measurements were done in 20 mM sodium phosphate buffer (pH 7.4) containing 5 mM 2-mercaptoethanol. For HCV and GBV-B core proteins, the buffer also contained 0.1% n-dodecyl β-d-maltoside (DM). Spectra were recorded at 20°C, at 95°C after slow heating of the protein, and again at 20°C after slow cooling. Melting curves were recorded at 222 nm, during the heating and cooling processes. Secondary structure content was calculated by the k2d method of spectral deconvolution (http://www.cryst.bbk.ac.uk/cdweb/html/home.html).
In order to examine the effect of temperature on the conformation of core proteins, far-UV CD spectra were recorded at 20°C, followed by slow (1°C/min) heating of the samples up to 95°C, with constant monitoring of thermal unfolding at 222 nm (54,55). A second CD spectrum was recorded at 95°C, and conformational changes associated with the slow (1°C/min) cooling of the samples were followed again at 222 nm. Upon heating of HCV core(2–169) in 0.1% DM, the protein underwent irreversible denaturation, as indicated by the markedly different spectra recorded at 222 nm upon heating and cooling (Figure 6A). The denaturation was characterized by a decrease in α-helix content (from 44% to 28%), a decrease in the content of unordered structures (from 50% to 41%), and by an increase in β-sheet content (from 6% to 31%). After renaturation, the content in secondary structures remained nearly the same as it was at 95°C (Figure 6A).
Similarly to HCV core, adding 0.1% DM to GBV-B core protein led to the formation of partly α-helical structure (Figure 6B). Ellipticity changes upon heating and cooling indicated that GBV-B core denaturation was mostly reversible in the presence of DM (Figure 6B). The denaturation was characterized by a decrease in α-helix content (from 39% to 26%), a decrease in the content of unordered structures (from 50% to 44%), and by an increase in β-sheet content (from 11% to 30%). After renaturation, the content of β-sheets and of unordered structures was nearly the same as it was before heating, while the content of α-helices remained the same as it was at 95°C (Figure 6B).
In agreement with the available stuctural data on flavivirus capsid proteins (40–42), WNV core exhibited the characteristics of an α-helical protein, with local molar ellipticity minima at 208 and 222 nm (Figure 6C). Estimating the secondary structure content of WNV core by CD deconvolution gave ∼77% α-helix, in accordance with the published crystal structure of the protein (40). Ellipticity changes at 222 nm upon heating of the protein showed a classical denaturation curve (Figure 6C). Surprisingly, denaturation of WNV core was mostly reversible, as shown by the renaturation curve and the spectrum recorded at 20°C after cooling of the sample (Figure 6C). In separate experiments, we obtained full renaturation of WNV core protein after heating at 95°C (data not shown). The reason for this discrepancy is currently unknown. The reversibility of the WNV core protein denaturation was confirmed by the determination of similar secondary structure content prior to heating and after cooling of the protein (Figure 6C).
BVDV core protein, in agreement with the disorder prediction (Figure 1), was found to be completely unstructured at 20°C, as evidenced by the pronounced minimum in the CD spectrum observed at ∼200 nm (Figure 6D).
The chaperoning activity of the C-terminal RNA-binding region of WNV core
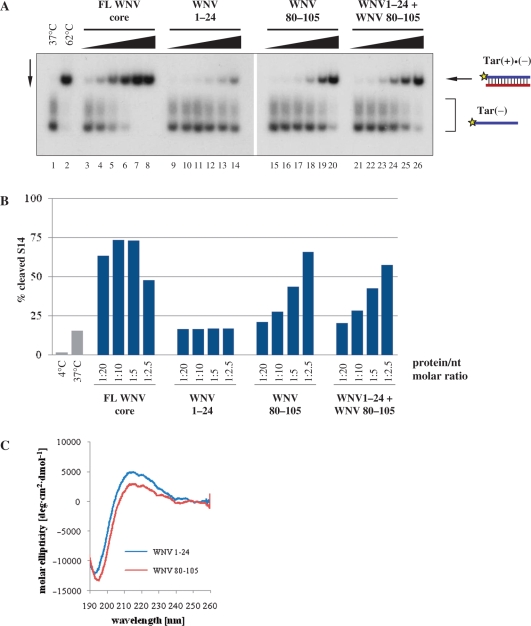
The isolated N- and C-terminal regions of the WNV core protein were found to independently bind RNA (43). Using the strand-annealing assay (Figure 2A), we examined the chaperoning activity of the N-terminal (WNV 1–24) and C-terminal (WNV 80–105) core peptides. The two peptides are similar in length and basic amino acid content (7 basic residues for WNV 1–24 versus 8 for WNV 80–105). As shown in Figure 7A, WNV 1–24 exhibited only a low level of strand-annealing activity, while WNV 80–105 caused efficient DNA duplex formation. Incubation of the ODNs with WNV 1–24 and WNV 80–105 together did not result in a considerable increase in annealing compared to WNV 80–105 alone, indicating that the two peptides do not act in a cooperative manner (Figure 7A).
Figure 7.
Strand-annealing activity of WNV core peptides. (A) 32P-labelled Tar(−) ODN was incubated together with Tar(+) ODN in the presence of increasing amounts of the proteins, as indicated at the top of the figure. Protein-to-nucleotide molar ratios were 1:160, 1:80, 1:40, 1:20, 1:10 and 1:5 for each protein tested (corresponding to 1, 2, 4, 8, 16 and 32 nM protein concentrations, respectively). Lane 1: Tar(−)/Tar(+) complex formed at 37°C in the absence of protein; lane 2: Tar(−)/Tar(+) complex formed by heat annealing at 62°C, without protein; lanes 3–8, 9–14, 15–20 and 21–26: complex formation at 37°C in the presence of increasing concentrations of proteins. (B) Enhancement of hammerhead ribozyme cleavage by WNV core peptides. R3 ribozyme and 32P-labelled S14 RNA substrate were incubated at 37°C for 25 min in the presence of increasing amounts of the proteins, as indicated in the figure. Protein-to-nucleotide molar ratios were 1:20, 1:10, 1:5 and 1:2.5 for each protein tested (corresponding to 15, 30, 60 and 120 nM protein concentrations, respectively). RNAs were resolved on an 8% denaturing polyacrylamide gel and the percentage of the cleaved S14 substrate was determined by autoradiography and PhosporImager quantification. As a control, R3 and S14 were co-incubated without protein either at 4°C or at 37°C (grey bars). Results of a representative experiment are shown. (C) Far-UV CD spectra of WNV core peptides. CD spectra were recorded at 20°C, in 20 mM sodium phosphate buffer (pH 7.4) containing 5 mM 2-mercaptoethanol.
The RNA chaperone activity of the WNV (80–105) peptide was further confirmed with the ribozyme cleavage assay, which requires both the strand annealing and helix unwinding activities of a chaperone. In agreement with the results of the strand-annealing assay, WNV (80–105) peptide caused a clear activation of the ribozyme-directed S14 RNA cleavage, while WNV peptide (1–24) did not accelerate the cleavage reaction at all (Figure 7B). Interestingly, at the highest protein-to-nucleotide molar ratio used in this experiment (one protein molecule per 2.5 nt), the FL WNV core protein demonstrated sub-optimal cleavage enhancement, emphasizing that RNA chaperoning occurs in a relatively narrow ‘window of activity’ (18,20). While a low protein–RNA ratio probably favours high affinity interactions (selection of RNA substrates) over chaperoning, a high occupancy of RNA molecules by the chaperone could ‘freeze’ RNA structure, hindering conformational rearrangements (18,20).
CD spectroscopy of WNV peptides (1–24) and (80–105) revealed that they are both completely unfolded (Figure 7C; deconvolution data not shown), suggesting that RNA chaperone activity of WNV core protein does not require a well-defined structure.
DISCUSSION
RNA chaperone activity of Flaviviridae core proteins and possible functional implications
The genomic RNA (gRNA) of non-segmented positive sense RNA viruses plays complex, temporally and spatially regulated roles throughout the virus life cycle. It serves both as a template for minus-strand RNA or DNA synthesis and as an mRNA directing the translation of viral proteins, and lastly, it is specifically packaged into newly made progeny virions. To accomplish these functions, the gRNA relies at least in part on short specific cis-acting RNA elements (CREs), which regulate viral translation, replication with possible recombination events and virion assembly in infected cells (22–23,56–57). Thus, the gRNA and its CREs most probably undergo complex structural rearrangements, assisted by a virus-encoded protein with nucleic acid chaperone activities. The best-characterized example of a viral nucleic acid chaperone is the small nucleocapsid protein (NCp7) of human immunodeficiency virus type 1 (HIV-1). NCp7 mediates several RNA–RNA and RNA–DNA interactions and gRNA rearrangements that are required at multiple stages of the virus replication cycle, such as initiation and completion of viral DNA synthesis, virus assembly, genome dimerization and packaging (22,23,56). In addition, NCp7 most likely contributes to the genetic variability of HIV-1 by promoting gRNA dimerization, thus enhancing the frequency of copy-choice recombination (58).
The core protein of HCV exhibits striking similarities to HIV-1 NCp7, as evidenced by its potent in vitro nucleic acid chaperone activities, facilitating RNA–RNA interactions and structural rearrangements (25,26). Further strengthening the analogy with retroviruses, the genomic RNA of HCV contains a short palindromic CRE mediating dimerization of the gRNA 3′ untranslated region (UTR) upon core protein binding in vitro (25,26). Albeit the physiological relevance of this interaction is still not clear, it is tempting to speculate that RNA structural rearrangements induced by HCV core chaperoning—including genomic RNA dimerization—may constitute regulatory switch(es) between translation/replication or replication/packaging of the viral RNA.
The closest relative of HCV is GB virus-B, a hepatotropic virus with unknown natural host range that was shown to be infectious in New World monkeys (59). Even though the similarity between HCV and GBV-B is only ∼25–30% at the protein level (60), the two viruses show striking resemblance in virtually every aspect of their replication strategy and structural features, including the tripartite organization of the 3′ UTR, IRES structure and translation mechanism, and lipid droplet association of the core protein (3). By means of classical in vitro RNA chaperone assays (e.g. strand annealing, strand exchange and ribozyme assays; Figures 2 and 3, and data not shown), we showed that nucleic acid chaperone activity is also conserved between the two hepacivirus core proteins, and that GBV-B core also efficiently facilitates the formation of the most stable nucleic acid structure. In addition, similarly to HCV, GBV-B core protein binding induced dimerization of the 3′ UTR of the GBV-B gRNA (R.I.-N. and J.-L.D., unpublished data), suggesting that core-mediated dimerization is a common feature in the hepacivirus genus.
Despite a lack of significant similarity with hepacivirus cores in amino acid sequence or domain organization (Figure 1), core proteins of the flavivirus WNV and the pestivirus BVDV both demonstrated potent RNA chaperone activities in vitro (Figures 2 and 3). Currently, we are investigating potential CREs in the WNV and BVDV genomic RNAs possibly regulated by core chaperoning. Preliminary results show that the 5′ and 3′ conserved sequence (CS) elements of the WNV genome—involved in long-range RNA–RNA interaction, leading to genome cyclization necessary for viral replication (61–64)—require the strand annealing activity provided by RNA chaperones for efficient interaction (R.I-.N. and J.-L.D., unpublished data).
Intrinsic disorder and RNA chaperone activity
The mechanism of action of RNA chaperone proteins is still not well understood. According to the recently proposed entropy exchange model, intrinsically unstructured, highly flexible protein regions would play an essential role in facilitating RNA structural rearrangements in an ATP-independent manner, probably by providing the energy necessary for partially melting the misfolded RNA structure through an entropy exchange process, coupled with the cyclic RNA binding and release of the protein (24). Experimental evidence for the role of unstructured domains in RNA chaperone function is still scattered and incomplete (20,24). Based on bioinformatic analysis of a dataset consisting of 27 RNA chaperone proteins, Tompa and Csermely (24) found that the frequency of long, continuous disordered fragments in RNA chaperones is considerably higher than in any other protein class examined so far. However, in most cases the protein region responsible for the RNA chaperone function is not precisely mapped, preventing the straightforward assessments of the correlation between disorder and chaperoning.
In spite of significant differences in their sequence and domain organization, Flaviviridae core proteins all possess RNA chaperone activities, thus providing an ideal model system to study the common (sequence and structural) requirements for chaperone function. In order to examine whether intrinsic disorder really plays an important role in chaperone activity, we analysed the secondary structure content of Flaviviridae core proteins by far-UV CD spectroscopy both at room temperature, and during thermal denaturation and renaturation. In agreement with in silico predictions (Figure 1), the four proteins were found to contain various amounts of secondary structure. Hepacivirus (HCV and GBV-B) core proteins are mostly unstructured in solution (Figure 5), and they only gain an α-helical conformation in membrane-mimetic environments, provided in our experiments by 0.1% dodecyl maltoside (Figure 6A and B). This property is due to the presence of hydrophobic domain D2 (Figure 1) involved in lipid droplet association (9,37). Nevertheless, even in these conditions, approximately half of the protein remained highly flexible, as shown by the estimation of secondary structure content for both HCV and GBV-B core (Figures 6A and B). In sharp constrast, WNV core was found to be mostly structured by CD spectroscopy (Figure 6C), in agreement with the published crystal structure of the protein (40). Despite these structural differences between hepacivirus and flavivirus core proteins, relatively short unstructured peptides were shown to be responsible for the chaperone activity of both HCV and WNV cores [(26), and Figures 4 and 7]. Surprisingly, BVDV core protein was found to completely lack a well-defined structure, as evidenced by its CD spectrum at 20°C and 95°C (Figure 6D). Further supporting the hypothesis that a well-defined structure is not required for RNA chaperoning, Flaviviridae core proteins retained most of their chaperone activity after heat ‘denaturation’, a characteristic feature of intrinsically unstructured proteins (Figure 4).
Overall, these experiments show that even closely related RNA chaperones may utilize different strategies and structural features to carry out the same function, and that RNA chaperone activity of Flaviviridae core proteins is mediated by disordered, highly charged protein segments, lending experimental support to the entropy exchange hypothesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
R.I-N. is the recipient of an ANRS PhD fellowship. We are grateful to Roland Montserret (IBCP CNRS, Lyon) for help and advice on CD spectroscopy measurements and to François Penin (IBCP CNRS, Lyon) for his support and discussions. We thank Philippe Desprès (Institut Pasteur, Paris), Jens Bukh (NIH, Bethesda, USA) and Till Rümenapf (Justus-Liebig-Universität, Giesen, Germany) for their kind gift of plasmids. This work was funded by Agence nationale de recherches sur le sida (to J.-L.D. and J.-P.L.); CNRS and Université Lyon I (to J.-P.L.). Funding to pay the Open Access publication charges for this article was provided by French ANRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Brown RS. Hepatitis C and liver transplantation. Nature. 2005;436:973–978. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1101–1152. [Google Scholar]
- 4.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl Acad. Sci. USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 1994;68:3631–3641. doi: 10.1128/jvi.68.6.3631-3641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 7.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hope RG, McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 2000;81:1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 9.Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 2002;277:4261–4270. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- 10.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl Acad. Sci. USA. 1993;90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amberg SM, Nestorowicz A, McCourt DW, Rice CM. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 1994;68:3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamshchikov VF, Compans RW. Processing of the intracellular form of the West Nile virus capsid protein by the viral NS2B-NS3 protease: an in vitro study. J. Virol. 1994;68:5765–5771. doi: 10.1128/jvi.68.9.5765-5771.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rümenapf T, Unger G, Strauss JH, Thiel HJ. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 1993;67:3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stark R, Meyers G, Rümenapf T, Thiel HJ. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J. Virol. 1993;67:7088–7095. doi: 10.1128/jvi.67.12.7088-7095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rümenapf T, Stark R, Heimann M, Thiel HJ. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J. Virol. 1998;72:2544–2547. doi: 10.1128/jvi.72.3.2544-2547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heimann M, Roman-Sosa G, Martoglio B, Thiel HJ, Rümenapf T. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J. Virol. 2006;80:1915–1921. doi: 10.1128/JVI.80.4.1915-1921.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 18.Cristofari G, Darlix JL. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- 19.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 20.Ivanyi-Nagy R, Davidovic L, Khandjian EW, Darlix JL. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62:1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertrand EL, Rossi JJ. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J. Mol. Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 23.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 24.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 25.Cristofari G, Ivanyi-Nagy R, Gabus C, Boulant S, Lavergne JP, Penin F, Darlix JL. The hepatitis C virus Core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32:2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fossé P, Darlix JL. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boulant S, Becchi M, Penin F, Lavergne JP. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 2003;278:45785–45792. doi: 10.1074/jbc.M307174200. [DOI] [PubMed] [Google Scholar]
- 28.Boni S, Lavergne JP, Boulant S, Cahour A. Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA. J. Biol. Chem. 2005;280:17737–17748. doi: 10.1074/jbc.M501826200. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabus C, Ivanyi-Nagy R, Depollier J, Bucheton A, Pelisson A, Darlix JL. Characterization of a nucleocapsid-like region and of two distinct primer tRNALys,2 binding sites in the endogenous retrovirus Gypsy. Nucleic Acids Res. 2006;34:5764–5777. doi: 10.1093/nar/gkl722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53(Suppl. 6):566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 32.Dosztányi Z, Csizmók V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 33.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 34.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 35.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne JP. Hepatitis C virus core protein is a dimeric alpha-helical protein exhibiting membrane protein features. J. Virol. 2005;79:11353–11365. doi: 10.1128/JVI.79.17.11353-11365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 2006;281:22236–22247. doi: 10.1074/jbc.M601031200. [DOI] [PubMed] [Google Scholar]
- 38.Klein KC, Polyak SJ, Lingappa JR. Unique features of hepatitis C virus capsid formation revealed by de novo cell-free assembly. J. Virol. 2004;78:9257–9269. doi: 10.1128/JVI.78.17.9257-9269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majeau N, Gagne V, Boivin A, Bolduc M, Majeau JA, Ouellet D, Leclerc D. The N-terminal half of the core protein of hepatitis C virus is sufficient for nucleocapsid formation. J. Gen. Virol. 2004;85:971–981. doi: 10.1099/vir.0.79775-0. [DOI] [PubMed] [Google Scholar]
- 40.Dokland T, Walsh M, Mackenzie JM, Khromykh AA, Ee KH, Wang S. West Nile virus core protein; tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl Acad. Sci. USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 2003;77:7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khromykh AA, Westaway EG. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 1996;141:685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldsich C, Grossberger R, Schroeder R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 2002;16:2300–2312. doi: 10.1101/gad.231302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- 46.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 49.Receveur-Bréchot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 50.Kalthoff C. A novel strategy for the purification of recombinantly expressed unstructured protein domains. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2003;786:247–254. doi: 10.1016/s1570-0232(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 51.Csizmók V, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol. Cell. Proteomics. 2006;5:265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Csizmók V, Dosztányi Z, Simon I, Tompa P. Towards proteomic approaches for the identification of structural disorder. Curr. Protein Pept. Sci. 2007;8:173–179. doi: 10.2174/138920307780363479. [DOI] [PubMed] [Google Scholar]
- 53.Kunkel M, Watowich SJ. Biophysical characterization of hepatitis C virus core protein: implications for interactions within the virus and host. FEBS Lett. 2004;557:174–180. doi: 10.1016/s0014-5793(03)01486-8. [DOI] [PubMed] [Google Scholar]
- 54.Kim TD, Ryu HJ, Cho HI, Yang CH, Kim J. Thermal behavior of proteins: heat-resistant proteins and their heat-induced secondary structural changes. Biochemistry. 2000;39:14839–14846. doi: 10.1021/bi001441y. [DOI] [PubMed] [Google Scholar]
- 55.Uversky VN. What does it mean to be natively unfolded? Eur. J. Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 56.Darlix JL, Garrido JL, Morellet N, Mély Y, Rocquigny H. Properties, functions, and drug targeting of the multifunctional nucleocapsid protein of the human immunodeficiency virus. Adv. Pharmacol. 2007;55:299–346. doi: 10.1016/S1054-3589(07)55009-X. [DOI] [PubMed] [Google Scholar]
- 57.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat. Rev. Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 58.Negroni M, Buc H. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl Acad. Sci. USA. 2000;97:6385–6390. doi: 10.1073/pnas.120520497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bukh J, Apgar CL, Govindarajan S, Purcell RH. Host range studies of GB virus-B hepatitis agent, the closest relative of hepatitis C virus, in New World monkeys and chimpanzees. J. Med. Virol. 2001;65:694–697. doi: 10.1002/jmv.2092. [DOI] [PubMed] [Google Scholar]
- 60.Muerhoff AS, Leary TP, Simons JN, Pilot-Matias TJ, Dawson GJ, Erker JC, Chalmers ML, Schlauder GG, Desai SM, et al. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001;75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo MK, Tilgner M, Bernard KA, Shi PY. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 2003;77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA–RNA interactions circularize the dengue virus genome. J. Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006;20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.