Abstract
Much of the research on insulators in Drosophila has been done with transgenic constructs using the white gene (mini-white) as reporter. Hereby we report that the sequence between the white and CG32795 genes in Drosophila melanogaster contains an insulator of a novel kind. Its functional core is within a 368 bp segment almost contiguous to the white 3′UTR, hence we name it as Wari (white-abutting resident insulator). Though Wari contains no binding sites for known insulator proteins and does not require Su(Hw) or Mod(mdg4) for its activity, it can equally well interact with another copy of Wari and with unrelated Su(Hw)-dependent insulators, gypsy or 1A2. In its natural downstream position, Wari reinforces enhancer blocking by any of the three insulators placed between the enhancer and the promoter; again, Wari–Wari, Wari–gypsy or 1A2–Wari pairing results in mutual neutralization (insulator bypass) when they precede the promoter. The distressing issue is that this element hides in all mini-white constructs employed worldwide to study various insulators and other regulatory elements as well as long-range genomic interactions, and its versatile effects could have seriously influenced the results and conclusions of many works.
INTRODUCTION
The two definitive properties of insulator elements are (i) the ability to block stimulation of a downstream gene promoter by an upstream enhancer (supposed to restrict ‘cross-talk’ in complex genetic loci) and (ii) the ability to put up a barrier between active and suppressive chromatin (1–7). We more or less understand now what molecular mechanisms may be involved in the chromatin barrier function (1,3,4,7). In contrast, no one really knows how a single insulator can block the enhancer–promoter communication. Perhaps for this reason the widespread models simply shun this question and regard insulators just as ‘clothes pegs’ that tether the chromatin fibre to the nuclear matrix/scaffold/envelope, or as ‘snap halves’ that bind with each other to close a chromatin loop; in either case, this is supposed to result in partitioning of the genome into ‘independent transcription units’. Only the enhancer-blocking function is considered in this work.
Most of the progress in this challenging field of research has been achieved in the transgenic approach, examining the effects of insulator(s) in a construct comprising enhancer(s) and reporter gene(s) that is inserted into the genome. With the standing problem of genomic position effects, it is still more important to manipulate a well-defined ‘autonomous unit’ with predictable/controllable interactions at least within the construct. Again, the general methodological requirement that data obtained in different laboratories must be comparable and reproducible should have been enforced by the availability of standard tools such as expression vectors with convenient reporters.
One such instance is the white gene, required for eye pigmentation in Drosophila and regulated by its eye-specific enhancer (8). The changes in gene expression are phenotypically obvious (brick red eyes in wild type, paling through shades of red and yellow with decreasing stimulation by the enhancer, down to white eyes when the gene is inactive) and easily assessed. The mini-white gene, which is an abridged white with most of the first intron deleted (9), is one of the most popular reporters in transgenic studies, which include testing the insulator properties of various sequences (10–18).
Here we expose a serious pitfall in the use of these ‘standard’ constructs: the mini-white insert proves to contain a 3′-adjacent insulator of a novel kind, which can pair not only with its twin but also—not less efficiently—with unrelated insulators, reinforcing or nullifying their enhancer-blocking activity depending on the position relative to the gene promoter.
MATERIALS AND METHODS
Drosophila strains, transgenes, germ line transformation and genetic crosses
All flies were maintained at 25°C on the standard yeast medium. The mutant alleles and chromosomes used in this study and the balancer chromosomes are described elsewhere (19). Transgenes were obtained with standard cloning techniques (Supplementary Data).
The construct, together with a P element containing defective inverted repeats (P25.7wc) that was used as a transposase source (20), was injected into y ac w1118 preblastoderm embryos as described (21,22). The resulting flies were crossed with y ac w1118 flies, and the transgenic progeny were identified by their eye and/or cuticle colour. The chromosome localization of various transgene inserts was determined by crossing the transformants with the y ac w1118 balancer stock carrying dominant markers: In(2RL),CyO for chromosome 2 and In(3LR)TM3,Sb for chromosome 3. The transformed fly lines were tested for transposon integrity and copy number by Southern blot hybridization. Only single-copy transformants were taken into study.
The lines with DNA fragment excisions were obtained by crossing the flies bearing the transposons with the Flp (w1118; S2CyO, hsFLP, ISA/Sco; +) or Cre (y1, w1; Cyo, P[w+,cre]/Sco; +) recombinase-expressing lines or with the I-SceI endonuclease-expressing line (v P{v+; hsp70-I-SceI}) (23–25). The Cre recombinase induces 100% excisions in the next generation. High levels of Flp recombinase (almost 90% efficiency) and I-SceI endonuclease (90% efficiency) were produced by heat shock treatment (2 h daily) during the first 3 days after hatching. All excisions were confirmed by PCR analysis; for details, see ‘Supplementary Data’.
The su(Hw)v/su(Hw)2 and mod(mdg4)u1/mod(mdg4)u1 mutations were combined with transgenes as previously described (26).
The phenotypic scoring assay
To estimate the levels of yellow and white expression, we visually determined the degree of pigmentation in the abdominal cuticle and wing blades (yellow) and in the eyes (white) of 3- to 5-day-old males developing at 25°C, with reference to standard colour scales. In the five-grade scale of yellow (Supplementary Figure S1 for the abdominal stripes), grade 5 corresponds to wild type and grade 1 to total loss of yellow expression. Identical data were obtained for the wing and body pigmentation in all experiments. In the nine-grade scale of white (Supplementary Figure S2), brick red (R) eyes correspond to wild type and white (W) to total loss of white expression. Intermediate levels of eye pigmentation are brownish red (BrR), brown (Br), dark orange (dOr), orange (Or), dark yellow (dY), yellow (Y) and pale yellow (pY) in the order of decreasing gene expression.
Two experts separately inspected 30–50 flies from each of two independent crosses for every transgenic line. For all data considered, there was full agreement between crosses and between experts. Each line thus assessed contributed a unit to the corresponding bin of the scoring table. Hence, each numerical entry in the distributions shown in the figures under the scales is the number of fly lines with the specified pigmentation grade (corresponding to the gene expression level decreasing from left to right).
Additionally, the central tendency in the distribution was estimated as the arithmetic mean (for this purpose, the R–W grades of white were temporarily converted into numerical grades 9–1). The values thus obtained proved stable against truncation (tested up to 10%), the shift never exceeding 0.1 of the mean. These statistical estimates are shown in Figures 2 and 3 as the positions of shaded ‘cursors’ on the distribution frames relative to the scale above. In an alternative assessment, the medians of these distributions always were either one or both grades enclosing the means; therefore, they are not shown.
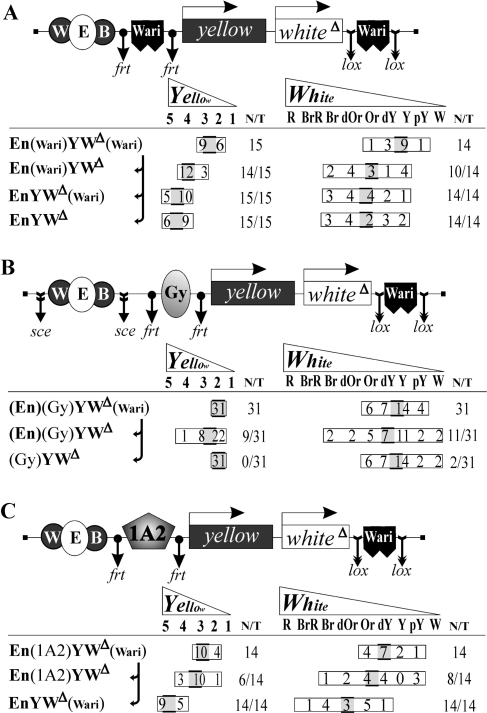
Figure 2.
Reinforcing influence of downstream Wari on the apparent activity of Wari (A), Gy (B) and 1A2 (C) insulators in standard enhancer blocking assays. Presentation and designations as in Figure 1B, except that all parental and excision-derived constructs are spelled out at the left of the expression data, with excisable elements parenthesized; sce in scheme B denotes the sites for I-SceI endonuclease; whiteΔ (WΔ) is the ‘purified’ mini-white from which the downstream insulator-containing sequence was removed; Wari is the novel white-abutting resident insulator, Gy is the Su(Hw)-dependent insulator from the gypsy retrotransposon, 1A2 is the endogenous Su(Hw)-dependent insulator found after the yellow gene. The shaded cursor at each distribution frame marks the ‘mean colour’ on the scale above; thus, cursor positions and shifts in different rows are directly comparable, but the cursors themselves are not associated with the numerals they may cover.
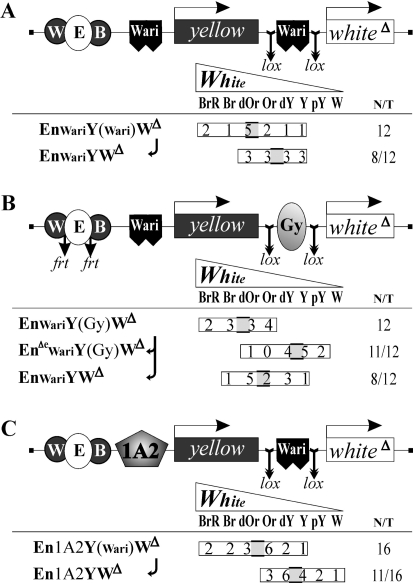
Figure 3.
Mutual neutralization of two insulators around yellow in the expression of white. A. by two Wari insulators. B. by Wari and Gy insulators. C. by Wari and 1A2 insulators. Presentation as in Figure 2; EnΔe in panel B denotes excision of the eye enhancer. The data for yellow expression (without any appreciable changes) are given in Supplementary Figure S3.
Assessment of changes in gene expression
The effects of insulator elements and their combinations and rearrangements on gene expression are deduced by comparing the phenotypic distributions of fly lines carrying the basic constructs and their derivatives produced by in vivo excision of a particular element (such elements were flanked with appropriate sites as shown in the construct schemes and parenthesized in construct names).
The data are presented as ‘tabular figures’ where the position of the frame enclosing the entire sample on the horizontal colour scale gives a rough idea of the expression range. Note that practically always the range is broader for white than for yellow, for the obvious reason that it is spread on a more detailed scale (nine standard grades versus five).
The study is made robust against the genomic position effects (including the possible influence of other nearby insulators) by the double assessment protocol whereby all essential conclusions are drawn from comparisons not between single transgenic lines but between groups of independent lines with single copies of the construct inserted at random in different places of the genome. Considering that interplay of insulators can both reinforce and neutralize their enhancer-blocking activity (14,15), the occasional effects of such extraneous elements are likely to be stochastically ‘levelled off’ besides being simply ‘diluted’ in group comparison. The processed data are presented as follows:
In the N/T column on the right of the framed sets of scoring data, the only or the last figure (T) is the total number of fly lines examined for the given construct, and the numerator N is the number of lines from this sample that acquire a new phenotype upon manipulation indicated by the change in the construct name on the left. A ratio N/T ≥0.5 is taken to be a reliable indication of the influence of the tested element on gene expression.
The distribution frames also carry ‘shaded cursors’ marking the central tendency determined as in the preceding subsection, so one can see the shifts in the averaged expression caused by removal of each element. Note that the cursors indicate the ‘mean colour’ on the scale above and should not be directly associated with the numerals they may be superimposed on.
The two kinds of assessment are to some extent complementary: e.g. if excision of an insulator is expected to increase gene expression but some opposite responses also occur within a group of fly lines (broadening of the distribution), the changes of interest will be overestimated by N/T but underestimated by the arithmetic mean, so one can always make a sober judgement.
In as much as insulator elements are thought to influence not the gene transcription as such but the stimulatory action of enhancers, it should be admitted that a more sophisticated analysis might include (apart from the conventional wild type and null extremes) a third reference point on the gene expression scale: a ‘ground level’ that can be established from the phenotypes of transgenic lines with the corresponding enhancer-less constructs. Thus in our experience the overwhelming majority of such flies exhibit wing and body pigmentation about grade 2 and eye pigmentation not exceeding orange; the same is observed for the enhancer-excised controls in this study [a single dOr ‘outlier’ (Figure 3B) out of 49 lines for white and none out of 37 for yellow]. A propos, these data confirm again that incidental activation of either transgene by resident genomic enhancers is a very rare event. Estimation of the central tendency from the aggregate data gives exactly grade 2 for yellow and exactly the 3rd grade (Y) for white.
Notwithstanding, for the purposes of the present study, we decided against any cut-offs or editing, and compared the whole samples; clearly, this most conservative assessment attenuates the relative changes in the enhancer action, i.e. the quantitative effects of insulators and their interactions, but on the other hand, augments the reliability of the qualitative conclusions drawn from such consideration.
Plasmid constructs, transient transfection and luciferase expression assay
See Supplementary Data
RESULTS AND DISCUSSION
An insulator resides immediately downstream of the white gene
In our previous experiments concerning the role of insulators (enhancer blockers) in gene expression control, from time to time we encountered some strange or equivocal data; retrospective analysis suggested that the mini-white used as a reporter gene might have itself carried insulator-like activity associated with its 3′ end. Indeed, the mini-white module in the pCaSpeR series routinely contains almost a 1000 bp of genomic DNA after the coding part (9).
To check this surmise, we isolated the corresponding stretch of pCaSpeR2 DNA totalling 825 bp to include the end of the white 3′UTR and the beginning of the P element that follows the reporter gene in the plasmid (Figure 1A), flanked it with recombinase sites for in vivo excision, and inserted it in a ‘standard’ white expression construct (pCaSpeR3) between the eye enhancer and the mini-white (scheme in Figure 1B-1).
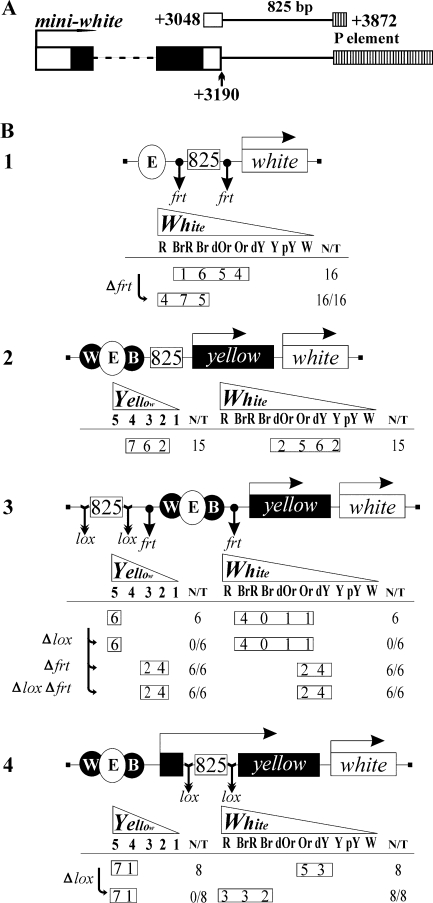
Figure 1.
Identification of an insulator at the 3′ end of the mini-white gene. (A) Demarcation of the pCaSpeR2 825 bp segment taken into study; positions given relative to the mini-white transcription start site. (B) Transgenic constructs used in the enhancer-blocking assay. The yellow and white genes are shown as rectangles with arrows indicating the direction of transcription. The Wing, Eye and Body enhancers are encircled and shaded as their target gene. The tested 825 bp sequence is boxed. Downward arrows indicate sites for Flp or Cre recombinases. Below the schemes are the expression data for each parental construct shown in the scheme and those derived from it by in vivo excision of the elements flanked by the specified sites. The horizontal colour scales are headed by tapered gene names, the reference images are shown in Supplementary Figures S1 and S2. For yellow, grade 5 pigmentation is that in wild type, grade 2 corresponds to complete blocking of wing and body enhancers and grade 1 is characteristic of completely lost expression. For white, the scale is from red (R) in wild type through brownish red (BrR), brown (Br), dark orange (dOr), orange (Or, maximal colour without eye enhancer), dark yellow (dY), yellow (Y) and pale yellow (pY) to white (W) in the absence of any expression. Each entry in the frame gives the number of transgenic lines with the corresponding pigmentation grade, while the frame itself shows the range; T is the total number of lines examined for each particular construct; for derivative constructs, N is the number of lines where the phenotype (i.e. expression level) changed as compared with the parental construct.
Eye pigmentation in such transgenic flies (ranging from orange to brown, 1st row of expression data in Figure 1B-1) was markedly weaker than usual (10–12), which meant that the action of the eye enhancer on the white promoter was partly blocked upon interposing the 825 bp duplicate; indeed, excision of this sequence largely restored the gene expression (red to brown eyes) (Figure 1B-1, 2nd row).
The same phenomenon was clearly observed with an analogous two-gene construct where white was preceded by yellow [another popular reporter gene, responsible for dark cuticular pigmentation (27,28)], the corresponding enhancers were grouped upstream (wing and body enhancers for yellow surrounding the eye enhancer for white, collectively designated W·E·B), and the 825 bp duplicate was placed in between (Figure 1B-2): the promoters of both genes enclosed by two copies of the supposed insulator were only weakly stimulated by their enhancers, proving that the enhancer-blocking effect of the tested sequence was not unique for the ‘aboriginal’ gene.
Next, we tested the position dependence of the enhancer-blocking activity. The 825 bp duplicate on the other side of the enhancers (Figure 1B-3) had no influence on the expression of either gene, be it with (cf. 1st and 2nd rows) or without enhancers (cf. 3rd and 4th rows). The duplicate inserted into the yellow intron (Figure 1B-4) allowed full stimulation of the (upstream) yellow promoter, as reported for other insulators and genes (10–12,29,30), but prevented stimulation of the (downstream) white promoter (1st row). Expectedly, removal of this insert (2nd row) did not change the expression of yellow but restored normal expression of white.
Thus, the 825 bp sequence from the 3′ end of white exhibits all the definitive features of an enhancer blocker: it can hinder the stimulatory action of enhancers on the promoters of different genes in a strictly position-dependent manner (as opposed to silencing) without irreversibly inactivating either element. It may be provisionally called white-abutting resident insulator (Wari, as an acronym).
Wari is not related to any known Drosophila insulator
Sequence analysis of the Wari-containing segment revealed no similarity with already known elements reported to have enhancer-blocking activity. Since insulators are generally held to exert their functions through specific associated proteins, we first of all looked for consensus binding sites, but TRANSFAC(R) Professional r10.2 found none for Su(Hw), dCTCF, GAGA or Zw5 (data not shown). This immediately set Wari apart from the most studied groups such as gypsy and other Su(Hw)-dependent insulators (31,32); Fab-6, Fab-8 and Mcp elements (33); or scs (34). As the binding consensuses are still a matter of debate, we nonetheless tried to check whether any of the major insulator-related proteins could be somehow involved in the Wari function.
In particular, the 825 bp DNA fragment was tested in electrophoretic mobility shift assays using in vitro-synthesized Su(Hw) or CTCF proteins. As positive controls, we used the 1A2 insulator, which has two sites for Su(Hw) (35,36), and Fab-8 with two sites for CTCF (37). However, no binding of these proteins to the Wari-containing sequence could be observed (data not shown).
In another approach, nine transgenic fly lines with two Waris around yellow and white were tested in the su(Hw)v/su(Hw)2 (38) or mod(mdg4)u1/mod(mdg4)u1 (39) background, i.e. in the absence of functional Su(Hw) or Mod(mdg4) proteins; these mutations did not influence the enhancer-blocking ability of the novel insulator (data not shown, being similar to those in Figure 1B-2 or Figure 2A).
It is yet to be discovered what Wari-binding protein(s) might mediate its function; thus far we can only say that Wari is clearly distinct from any type of insulator element known to date.
The resident insulator aggravates the effect of the same or unrelated enhancer blocker on the enclosed gene(s) in standard assays
It should be noted that in the first subsection (Figure 1) the enhancer-blocking properties of Wari were tested with constructs that originally contained this sequence within the mini-white module. Thus, the effect of the insulator placed between enhancers and genes could have been modulated by interaction between the two copies. There is ample, though often inconclusive, evidence for such functional interactions, concerning twin pairs as well as different insulator elements (14,15,17,40–43).
In the constructs tested further, the 825 bp Wari-containing stretch was removed from the 3′ end of conventional mini-white (this ‘purified’ module is denoted as WΔ) and, where specified, reinserted in the same (and/or other) position, frt- or lox-flanked to be excisable in vivo (which is denoted in parentheses) [WΔ(Wari)]. The procedure of assessing the changes in gene expression is detailed and substantiated in section ‘Methods’.
As demonstrated in Figure 2A, insertion of two Waris around yellow and white (i.e. reconstruction of the arrangement shown in Figure 1B-2) resulted in markedly attenuated expression of both genes (1st row: no flies with wing and body pigmentation exceeding grade 3 or eyes darker than orange). Removal of Wari from its ‘natural’ position largely restored the gene activities (2nd row); i.e. a single Wari between the enhancers and the promoters was only a modest blocker. Excision of the interposed duplicate admitted the same extent of gene stimulation as excision of both Waris (Figure 2A, 3rd and 4th rows), which means that the resident downstream insulator by itself does not perceptibly affect reporter gene expression. This is the most likely reason why it has remained hidden heretofore. These data once again confirm the positional dependence of Wari action, but more importantly, they strongly suggest functional interaction (pairing) between the two copies [as reported for other insulators (14,15,17,40–44)]; most plausibly, such pairing gives rise to a loop sequestering the two genes, which may indeed make their promoters less accessible to the enhancer ‘signals’.
The next obvious step was to test whether this hidden element could also modulate the effects of other, unrelated insulators. We made two analogous constructs with the gypsy insulator (Gy) or another Su(Hw)-dependent 1A2 insulator between the enhancers and the reporter genes, as required in a standard assay, and excisable Wari reinserted after white.
Figure 2 shows that the action of enhancers on yellow and white in such transgenic flies was completely blocked with Gy (panel B, compare 1st row with the 3rd where the enhancers had been excised from the construct) and largely blocked with 1A2 (panel C); this was quite in line with the literature data (11,35,36,42,44). However, the apparent blockage by 1A2 was appreciably weakened upon removal of the downstream Wari, and even with Gy quite a few Wari-excision lines showed increased pigmentation (cf. 1st and 2nd rows in each panel). Closer inspection of the expression data reveals that the less strong is the first (interposed) insulator (Gy > 1A2 > Wari), the greater is the ‘reinforcing contribution’ of Wari in its natural position, without discernible difference between the two enclosed genes. At the same time, one can see that removal of the downstream Wari may result in even lower white expression in occasional fly lines (e.g. two more pY lines in the 2nd row of panel C and even two W lines in panel C). Note that these changes occur in the very low expression range that is not associated with enhancer action, and hence can hardly be due to altered enhancer blocking. Most probably, in some genomic positions such ‘open’ constructs become more susceptible to the influence of neighbouring repressive chromatin. These observations suggest that Wari may also have the second, barrier function (see ‘Introduction’) and thus may be a full-fledged insulator element; work along this line is under way.
Since the constructs where Gy or 1A2 is combined with Wari (Figure 2B and C) behave very similarly to those with two interacting Waris (Figures 1B-2 and 2A), and there are no reasons to suppose that the same effects in analogous constructs are caused by basically different events, these results are the first evidence for pairing between insulators totally unrelated in nucleotide sequence and apparently having no common proteins. Of course the molecular mechanism(s) involved are yet to be elucidated, but this is so even for twin pairs of long-studied insulators.
‘Mutually neutralizing’ interaction of Wari with another Wari, Gy or 1A2 between the eye enhancer and the white promoter in white expression
Insulator pairing is still more vividly demonstrated with white expression in the other series of constructs where one of the insulators followed the enhancers and another was inserted between the two reporter genes (Figure 3).
With two identical or different insulators between the eye enhancer and the white promoter, the gene was expressed to the same or even higher level than in constructs without any interposed insulator (cf. 1st rows in all Figure 3 panels with, e.g. 3rd rows in Figure 2A and C). The 2nd row in Figure 3B proves that this high expression was due to promoter stimulation by the eye enhancer within the construct. Conversely, excision of the insulator right in front of the promoter (bottom rows in each Figure 3 panel) resulted in marked attenuation of white expression (consistent with the data for the corresponding arrangements in Figure 2). Note that yellow showed no response to these manipulations, remaining weakly or moderately insulated in accordance with the strength of the insulator preceding its promoter (Supplementary Figure S3).
This seemingly paradoxical behaviour of white is, however, another manifestation of the ‘insulator bypass’, ‘mutual neutralization’ or ‘cancellation’ phenomenon (14,15,17,40–43). It was first observed with tandemly placed identical insulators (14,40); later we demonstrated (42,45) that insulators such as Gy could interact with each other at considerable distances, over enhancers or promoters and coding sequences. Indeed, the only reasonable explanation of the data in Figure 3 is that here the insulators (even unrelated ones) pair around yellow to form a loop but no longer present any obstacle for stimulation of white; moreover, the distance between its enhancer and promoter becomes much shorter. That is, in one and the same construct, the same pair of elements acts as blockers for yellow but as facilitators for white.
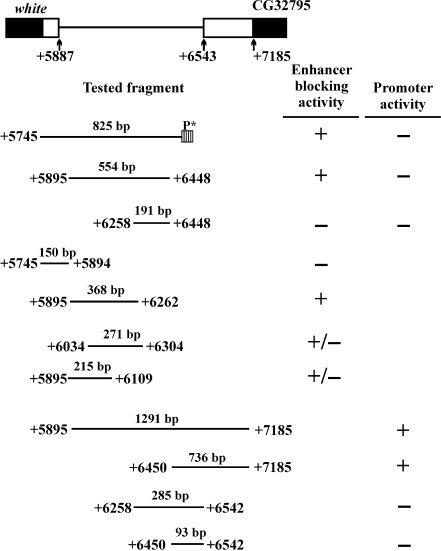
Mapping of the Wari core
Finally, we undertook an attempt to locate more precisely the novel insulator in the genomic sequence between the coding part of white and the next gene CG32795. Specifically, we wanted to check overlapping with the CG32795 promoter, overlapping with white 3′ UTR, and to isolate the Wari functional core. To this end, we obtained fragments of different lengths and positions (specified in Figure 4) and tested them for (i) enhancer-blocking activity in transgenic constructs analogous to the ‘standard’ shown in Figure 1B-2 and (ii) promoter activity in the luciferase reporter assay (Drosophila S2 cells, see ‘Supplementary Data’).
Figure 4.
Functional anatomy of the sequence between white and CG3295. All positions are given relative to the transcription start site of the unabridged white gene. Specified fragments were tested for enhancer-blocking activity as in Figure 1B2 and for promoter activity in the luciferase reporter assay. The ‘+/−’ marks indicate weak activity (mean expression shifts <1 grade) for yellow and only trace activity for white.
The full-sized 825 bp sequence ending shortly before the reported CG32795 regulatory region (see scheme in Figure 4) proved devoid of promoter activity (line 1), which indeed mapped to the most downstream part of the fragments tested (line 3 from bottom).
Removal of the 5′-terminal 150 bp did not affect the enhancer-blocking activity (line 2), and the fragment thus removed had none of its own (line 4), meaning that the insulator proper does not overlap with the white gene sequence. Moreover, the 3′-terminal quarter proved also inessential (meaning that the insulator is not even contiguous to the promoter region of the next gene), while the central 368 bp segment (45% of the initial length) retained full enhancer-blocking capacity (line 5). Another dissection in the middle part, however, reduced the activity to weak for yellow and insignificant for white (as in the two overlapping fragments of 271 and 215 bp). The 368 bp sequence encompassing the Wari core is given in Supplementary Figure S4.
Inferences and implications
Thus, we have found an insulator residing immediately downstream of the white gene in the Drosophila genome. This is a novel and somewhat surprising kind of enhancer blocker, as its functional requirements apparently do not include Su(Hw), CTCF, Zw5 or GAGA factor, though such zinc finger proteins heretofore appeared almost universal in insulator functions. Nonetheless, it interacts not only with another copy of itself in model transgenic constructs but equally well with the totally unrelated Su(Hw)-dependent insulators, markedly modulating their apparent enhancer-blocking activity; furthermore, this interaction implies physical pairing, as suggested by the bypass/neutralization phenomenon (Figure 3). All these findings make the obscure question of protein-mediated insulator function still more puzzling and still more challenging.
Anyway, it is clear that no responsible conclusions about the properties of any insulator element(s) can be drawn without considering the possible interactions with other insulators. Hence the general impact of our finding, stemming from the fact that this versatile insulator was inadvertently included in all transgenic constructs with white as reporter for testing the enhancer-blocking activity of various insulators (10–18,34,35,41–43), the anti-insulator ability of promoter-targeting sequences (46–49), the boundary activity of insulators and matrix attachment regions (50–53), or simply as selection marker (29–32,36,37,40,44,54,55). The cryptic downstream Wari could have aggravated the effects of single insulators, just as shown here for Gy and 1A2 (quantitative distortion of data); conversely, it could have disrupted their tandem pairing or simply masked their mutual neutralization in ‘insulator bypass’ assays (qualitative distortion).
In our previous work (45), pairing between Gy copies located in the same sites on homologous chromosomes facilitated the enhancer action in trans throughout the Drosophila genome. However, in some genomic positions the phenomenon was also observed in the absence of Gy; this ‘residual’ trans-activation could actually be due to pairing between Waris. Much the same applies to works concerning various kinds of long-distance genomic interactions presumably mediated by insulator elements (45,56–58).
Overall, the results and interpretations in a number of works (including ours) perhaps require re-evaluation, and care should be exercised in future studies regarding not only insulators proper but also enhancer–promoter communication and genomic control in general.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Marina Karakozova for expert technical assistance. This study was supported by the Russian Foundation for Basic Research (project no. 06-04-48360-a), the Molecular and Cellular Biology Program of the Russian Academy of Sciences, an International Research Scholar award from the Howard Hughes Medical Institute (to P.G.), a stipend from the Centre for Medical Studies, University of Oslo (to E.S.) and a stipend from the Russian Science Support Foundation (to O.M.). Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sun FL, Elgin SC. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 3.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol. Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Brasset E, Vaury C. Insulators are fundamental components of the eukaryotic genomes. Heredity. 2005;94:571–576. doi: 10.1038/sj.hdy.6800669. [DOI] [PubMed] [Google Scholar]
- 5.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 6.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 7.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 8.Qian S, Varjavand B, Pirrotta V. Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics. 1992;131:79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 10.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 13.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 15.Conte C, Dastugue B, Vaury C. Coupling of enhancer and insulator properties identified in two retrotransposons modulates their mutagenic impact on nearby genes. Mol. Cell. Biol. 2002;22:1767–1777. doi: 10.1128/MCB.22.6.1767-1777.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweinsberg S, Schedl P. Developmental modulation of Fab-7 boundary function. Development. 2004;131:4743–4749. doi: 10.1242/dev.01343. [DOI] [PubMed] [Google Scholar]
- 17.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer–promoter communication. Mol. Cell. Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brasset E, Bantignies F, Court F, Cheresiz S, Conte C, Vaury C. Idefix insulator activity can be modulated by nearby regulatory elements. Nucleic Acids Res. 2007;35:2661–2670. doi: 10.1093/nar/gkm140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsley DL, Zimm GG. The Genome of Drosophila Melanogaster. New York: Academic Press; 1992. [Google Scholar]
- 20.Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 21.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 22.Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 23.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 24.Siegal ML, Hartl DL. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 2000;136:487–495. doi: 10.1385/1-59259-065-9:487. [DOI] [PubMed] [Google Scholar]
- 25.Rodin S, Georgiev P. Handling three regulatory elements in one transgene: combined use of cre–lox, FLP–FRT, and I–Scel recombination systems. Biotechniques. 2005;39:871–876. doi: 10.2144/000112031. [DOI] [PubMed] [Google Scholar]
- 26.Georgiev P, Kozicina M. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy–induced mutations. Genetics. 1996;142:425–436. doi: 10.1093/genetics/142.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geyer PK, Corces VG. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 28.Martin M, Meng YB, Chia W. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 1989;218:118–126. doi: 10.1007/BF00330574. [DOI] [PubMed] [Google Scholar]
- 29.Cai H, Levine M. Modulation of enhancer−promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 30.Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos E, Ghosh D, Baxter E, Corces V. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–2349. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parnell T, Kuhn E, Gilmore B, Helou C, Wold M, Geyer P. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 2006;26:5983–5993. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organization of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golovnin A, Birukova I, Romanova O, Silicheva M, Parshikov A, Savitskaya E, Pirrotta V, Georgiev P. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003;130:3249–3258. doi: 10.1242/dev.00543. [DOI] [PubMed] [Google Scholar]
- 36.Parnell TJ, Viering MM, Skjesol A, Helou C, Kuhn EJ, Geyer PK. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl Acad. Sci. USA. 2003;100:13436–13441. doi: 10.1073/pnas.2333111100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison DA, Gdula DA, Coyne RS, Corces VG. A leucine zipper domain of the suppressor of hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 1993;7:1966–1978. doi: 10.1101/gad.7.10.1966. [DOI] [PubMed] [Google Scholar]
- 39.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 40.Cai H, Shen P. Effects on cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 41.Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc. Natl Acad. Sci. USA. 2004;101:14806–14811. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savitskaya E, Melnikova L, Kostuchenko M, Kravchenko E, Pomerantseva E, Boikova T, Chetverina D, Parshikov A, Zobacheva P, et al. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer–promoter communication in Drosophila melanogaster. Mol. Cell. Biol. 2006;26:754–761. doi: 10.1128/MCB.26.3.754-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on the enhancer–promoter communication. Mol. Cell. Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuhn EJ, Viering MM, Rhodes KM, Geyer PK. A test of insulator interactions in Drosophila. EMBO J. 2003;22:2463–2471. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kravchenko E, Kravchuk O, Savitskaya E, Parshikov A, Georgiev P, Savitsky M. The pairing between gypsy insulators located in homologous chromosomes facilitates enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 2005;25:9283–9291. doi: 10.1128/MCB.25.21.9283-9291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- 47.Lin Q, Wu D, Zhou J. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development. 2003;130:519–526. doi: 10.1242/dev.00227. [DOI] [PubMed] [Google Scholar]
- 48.Lin Q, Chen Q, Lin L, Zhou J. The promoter targeting sequence mediates epigenetically heritable transcription memory. Genes Dev. 2004;18:2639–2651. doi: 10.1101/gad.1230004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Q, Lin L, Smith S, Lin Q, Zhou J. Multiple promoter targeting sequences exist in Abdominal-B to regulate long-range gene activation. Dev. Biol. 2005;286:629–636. doi: 10.1016/j.ydbio.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 50.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 51.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 52.Namciu SJ, Blochlinger KB, Fournier REK. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol. 1998;18:2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Namciu SJ, Fournier REK. Human matrix attachment regions are necessary for the establishment but not the maintenance of transgene insulation in Drosophila melanogaster. Mol. Cell. Biol. 2004;24:10236–10245. doi: 10.1128/MCB.24.23.10236-10245.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl Acad. Sci. USA. 2003;100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez J, Muller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome–chromosome interactions in Drosophila. Mol. Biol. Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]