Abstract
The bacteriophage ø29 DNA packaging motor that assembles on the precursor capsid (prohead) contains an essential 174-nt structural RNA (pRNA) that forms multimers. To determine the structural features of the CE- and D-loops believed to be involved in multimerization of pRNA, 35- and 19-nt RNA molecules containing the CE-loop or the D-loop, respectively, were produced and shown to form a heterodimer in a Mg2+-dependent manner, similar to that with full-length pRNA. It has been hypothesized that four intermolecular base pairs are formed between pRNA molecules. Our NMR study of the heterodimer, for the first time, proved directly the existence of two intermolecular Watson–Crick G–C base pairs. The two potential intermolecular A–U base pairs were not observed. In addition, flexibility of the D-loop was found to be important since a Watson–Crick base pair introduced at the base of the D-loop disrupted the formation of the intermolecular G–C hydrogen bonds, and therefore affected heterodimerization. Introduction of this mutation into the biologically active 120-nt pRNA (U80C mutant) resulted in no detectable dimerization at ambient temperature as shown by native gel and sedimentation velocity analyses. Interestingly, this pRNA bound to prohead and packaged DNA as well as the wild-type 120-nt pRNA.
INTRODUCTION
Bacteriophage ø29, a Bacillus subtilis double-stranded DNA (dsDNA) virus, packages its genomic DNA into a preformed protein shell (prohead) by the use of a complex molecular motor (1,2). An unusual component of this motor is a ring structure of 174-nt prohead RNA (pRNA) molecules (3,4) linked by intermolecular base pairing, and this interaction between pRNAs is essential for packaging motor function (5,6). The motor is situated at the unique portal vertex of the prohead, with the pRNA multimer bridging two protein components, the head–tail connector and the packaging ATPase (2). The head–tail connector, a dodecameric ring structure comprised of the viral gene product 10 (gp10), is imbedded within the portal vertex and has a central channel through which DNA passes during packaging and ejection (2,7). pRNA binds to the N-terminus of gp10 (8,9), forming a pentameric (2,10,11) or hexameric (5,6,12,13) ring that fits around the narrow end of the connector, with extensions that contact the capsid (10). The packaging ATPase, gp16, likely present in the same number of copies as pRNA, binds to the pRNA ring to complete the assembly of the motor (2,14). pRNA and gp16 are transient components of the motor (3) and are released from the DNA-filled head, probably during neck/tail assembly.
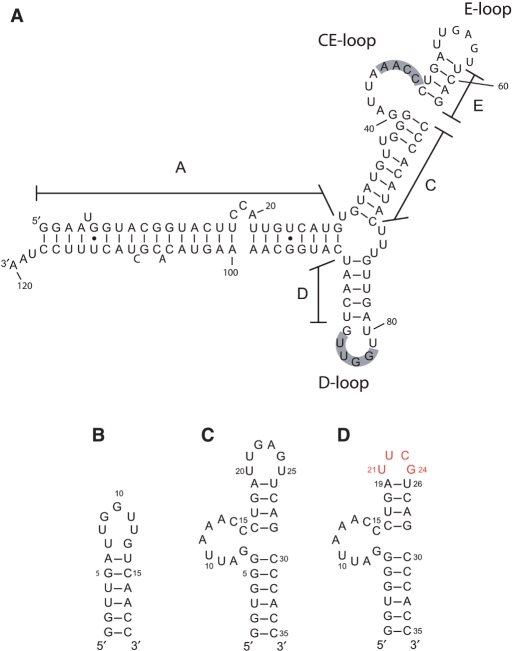
pRNA is a 174-nt transcript from the extreme left-end of the ø29 genome, and a truncated 120-nt (Figure 1A) form has full biological activity (3,4). The secondary structures of the pRNAs of ø29 and of several relatives have been determined by phylogenetic analysis and nuclease digestion studies (15). Ribonuclease footprinting and mutational analyses have identified the pRNA domain involved in prohead binding (16–18), and this domain contains the CE- and D-loops believed to be involved in pRNA oligomerization. Previous mutation studies also demonstrated that the pRNA A-helix (Figure 1A) is involved in DNA translocation rather than prohead binding (16,17,19,20), and the A-helix is the site for binding of the gp16 ATPase (14).
Figure 1.
Predicted secondary structures of ø29 pRNA and model RNAs. (A) The 120-nt form of wild-type pRNA. The shaded residues in the CE-loop and the D-loop are considered to form intermolecular base pairs. (B) The 19-mer that contains the D-loop and the D-helix. The G–U base pair at the end of the D-helix was changed to G–C, and an additional G–C base pair added in order to stabilize the helix. (C) The 35-mer that contains a part of the C-helix, the CE-loop, the E-helix and the E-loop. A G–C base pair was added at the end of the C-helix in order to stabilize the helix, and the U-bulge in the C-helix has been removed. (D) 35 -uucg in which the UUCG tetraloop (colored in red) is substituted for the E-loop (bases 20–25).
A novel and necessary aspect of pRNA structure and function is the potential intermolecular base pairing of the CE- and D-loops in neighboring pRNA molecules to form a homo-oligomeric ring (5,6). The pRNAs of ø29 relatives also have the potential for forming this intermolecular base pairing (16). Mutational analyses have suggested that residues 45AACC48 of the CE-loop base pair with residues 82 GGUU85 of the D-loop in an adjacent pRNA, and this interaction is required for DNA packaging (5,6). Only pRNAs that have the capability to form intermolecular base pairs bind efficiently to the prohead (21). Furthermore, the ATPase gp16 is pRNA dependent (22), and if the ATPase subunits prove to be gp16-pRNA heterodimers as suggested (1), the pRNA intermolecular interaction may have a role in communication between the ATPase ‘subunits’ in the ring structure of the packaging motor. Numerous biochemical studies have provided insight into the structural features of pRNA (1,23) and constraints for several models of oligomeric and dimeric pRNA that have been proposed (2,5,24,25). However, no direct observation that proves the presence and confirms the identity of the intermolecular base pairs has been reported.
In order to study the intermolecular interaction between pRNA molecules using NMR, we designed 35- and 19-nt RNA molecules that contain the CE-loop or D-loop of pRNA, respectively, the two loops hypothesized to be involved in the intermolecular interaction. In this report, we prove the existence of intermolecular base pairs between these two RNA molecules and identify residues involved in the bonds. Furthermore, we show that a new variant of pRNA that has no detectable multimerization capacity in solution at ambient temperature is fully functional in prohead binding and DNA packaging.
MATERIALS AND METHODS
RNA preparation
All RNAs were synthesized enzymatically from synthetic DNA templates using the AmpliScribe T7 transcription kit (Epicentre). The RNAs were purified by denaturing 15–20% polyacrylamide gel electrophoresis, identified by UV absorbance and excised from the gel. RNAs were eluted from the gel slice in water and desalted using SepPak columns (Waters). The RNAs were concentrated using Centricon YM-3 and exchanged into 50 mM Na-phosphate (pH6.5), 10 mM NaCl, 5 mM MgCl2. Samples for exchangeable proton detection were lyophilized, and then H2O was added to achieve a final solution of 95% H2O/5% D2O. Samples for non-exchangeable proton detection were lyophilized and then D2O was added. Before NMR experiments, the RNAs were heated to 80°C for 2min and cooled in ice water.
NMR spectroscopy
NMR data were acquired either at Varian Unity Plus Inova 600 MHz, 800 MHz equipped with a cryogenic-probe, or 900MHz equipped with a cryogenic-probe, processed with NMRPipe/NMRDraw (26) and analyzed with NMR View (27). Assignments for exchangeable protons were obtained using 15N-HSQC and ssNOESY (mixing time of 200 ms) experiments (28) in 95% H2O/5% D2O buffer at 10°C. Hydrogen bonding patterns of base pairs were determined from analysis of the NOESY spectra, and directly from J(N,N)-HNN COSY experiments (29,30) performed in 95% H2O/5% D2O buffer at 10°C and 20°C. Assignment for non-exchangeable protons were obtained from 13C-HSQC, 2D-NOESY (mixing time of 150 ms), DQF-COSY, 2D HCN-H1′ and 2D HCN-H6/H8 experiments (31) in D2O buffer at 25°C. The H2 protons of adenine bases were assigned unambiguously for the labeled RNAs by correlation between H2 and H8 resonances in 2D HCCH TOCSY experiments (32,33).
Native gel analysis
Native gel analysis of heterodimer formation of the 19-mer variants and 35-mer variants was performed as follows: two RNAs were mixed in the same buffer (50 mM Na-phosphate and 5 mM MgCl2), and the samples were heated at 80°C for 2 min and immediately cooled on ice water. Sucrose was added to the solutions to a final concentration of 8%, and the samples were run in 15% native PAGE gels in TBM-buffer (89 mM Tris, 89 mM borate and 5 mM of MgCl2) at 4°C. Annealing and electrophoresis buffers that lacked MgCl2 were used to investigate the effect of Mg2+ ion.
In vitro DNA packaging assay
DNA-gp3, proheads, and the packaging ATPase gp16 were purified as described previously (34,35). pRNA was made by in vitro transcription and purified by denaturing-urea PAGE, as described previously (18). DNA packaging assay was performed as described by Grimes and Anderson (35). Briefly, purified proheads (1 × 1011) reconstituted with either wild-type pRNA or mutant pRNA (in the ratio of 1:10) were mixed with purified DNA-gp3 (5 × 1010) and gp16 (2 × 1012) in 0.5 × TMS buffer (25 mM Tris–HCl, pH 7.8, 5 mM MgCl2, 50 mM NaCl) containing 0.5 mM ATP and incubated at room temperature for 15 min. Unpackaged DNA was digested with 1 μg/ml DNase I at room temperature for 10 min. The reaction mixture was treated with 25 mM EDTA and 250 μg/ml proteinase K for 30 min at 65°C to inactivate the DNase I and release the packaged DNA from particles. The packaged DNA was observed by agarose gel electrophoresis.
Sedimentation Velocity Analysis
The pRNA solutions, including F6, F7, U80C and the F6/F7 mixture, were used in a sedimentation velocity run, using an 8-hole rotor at 20°C and 44 000 r.p.m. The rotor and cells were pre-equilibrated at 20°C, and the samples were kept on ice during the cell loading procedure. The buffer supplied for the sample was 10 mM sodium phosphate, 50 mM NaCl and 5 mM Mg2+, at pH 6.5. Standard double sector cells were loaded with 430 μl of buffer and 420 μl of the appropriate sample solution. The rotor and cells were then equilibrated under vacuum at 20°C, and after a period of ∼1 hr at 20°C the rotor was accelerated to 44 000 r.p.m. Absorbance scans at a wavelength of 260 nm were acquired at ∼4.5 min intervals for 4 h. The buffer density and viscosity were calculated to be 1.00207 g/ml and 0.01014 poise at 20°C, respectively, using Sednterp (36). A value of 0.56 ml/g was used for the partial specific volume for the RNAs.
The data for each sample was first analyzed using the program DcDt+ (version 2.0.7). Furthermore, the g(s*) plots were fitted to a theoretical model with the DcDt+ program (37,38).
Native gel analysis of U80C and other 120-nt pRNA forms
pRNA (5 × 1013) in 45 µl water was heated to 80°C for 3 min. After the sample was removed from heat, 5 µl of 10 × TM buffer (250 mM Tris–HCl, pH 7.6, 50 mM MgCl2) was added and the sample was cooled to room temperature. A portion of the pRNA sample (7.5 × 1012) was mixed with 40% (w/v) sucrose and run on a 6% polyacrylamide gel buffered with 100 mM Tris–Hepes, pH 7.8, 10 mM MgCl2 and 0.1 mM EDTA at 2.5W at 4°C or ambient temperature. For the 4°C experiment, the gel and buffer were pre-cooled.
Prohead binding assay
RNA-free proheads were prepared as described previously (4) and purified by sucrose density centrifugation. pRNA was heated and mixed with TM buffer as described above. RNA-free proheads (1.8 × 1012) were incubated with an excess of pRNA (2.2 × 1013) in 25 µl in 0.5 × TMS buffer at ambient temperature for 10 min. Samples were diluted to 100 µl with 0.5 × TMS and layered on top of 5 ml of 5% (w/v) sucrose. The prohead–pRNA complexes were isolated by pelleting through the sucrose cushion at 35 000 r.p.m. in an SW55 rotor at 4°C for 2.5 h. The pellet containing the prohead–pRNA complexes was resuspended in 25 µl 0.5 × TMS overnight at 4°C. The sample was analyzed for RNA content on a 15% denaturing urea–PAGE and protein content by SDS–PAGE.
RESULTS
Design of RNA fragments in order to study the intermolecular interaction of pRNA
In order to analyze the intermolecular interaction, we designed two RNA molecules, a 19-mer and a 35-mer, which contain the D-loop or the CE-loop, respectively (Figure 1). Designing the loops in separate molecules prevented the formation of heterogeneous multimers and intramolecular loop–loop interactions that may occur in the full-length pRNA. Furthermore, this heterodimer system has an advantage in NMR analysis compared with a homodimer system, because we can selectively label one molecule using stable isotopes and can thus easily distinguish NMR signals from intra- and intermolecular interactions. In addition, to simplify NMR spectra, a bulge U in the C-helix of pRNA was deleted in the 35-mer (Figure 1C), a change which does not affect the biological activity of the pRNA (16,39).
In other attempts to facilitate the NMR analysis by reducing the number of signals, we designed a smaller 29-mer CE-loop, in which three base pairs at the end of the C-helix were removed from the 35-mer. However, the NMR spectra of the 29-mer showed that this RNA did not fold into a stable secondary structure (data not shown). In addition, we produced another RNA (35 -uucg) in which the UUCG tetraloop was substituted for the E-loop (Figure 1D). The UUCG tetraloop is a well-studied structural component of RNA (40) that is often used to stabilize the secondary structure of RNAs for NMR analysis (40–42). Furthermore, because the UUCG residues have unique NMR signals that are easily assigned, the substitution of the UUCG tetraloop facilitates analysis of NMR spectra of the 35-mer.
Native gel analysis of heterodimer formation
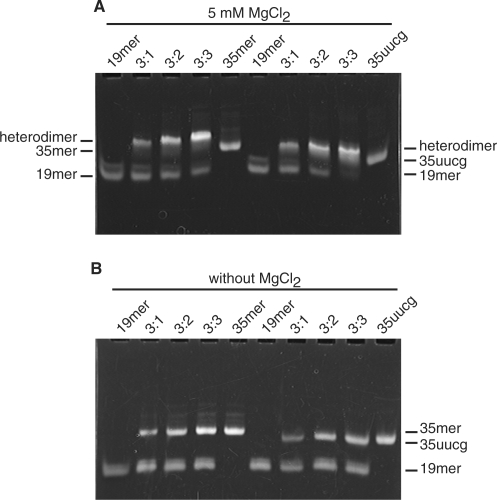
Prior to NMR structural analysis, heterodimer formation in a mixture of the 19-mer and 35-mer RNAs was confirmed using native polyacrylamide gel electrophoresis. In order to examine the Mg2+ dependence of the heterodimer formation, the RNAs were analyzed using gels and buffer either with or without Mg2+. Both the 35-mer and 35 -uucg form a 1:1 heterodimer with 19-mer only in the presence of Mg2+ (Figure 2), similar to the Mg-dependent oligomerization of 120-nt pRNA (5,6,21).
Figure 2.
Non-denaturing PAGE analysis of RNAs (A) with 5 mM MgCl2 and (B) without MgCl2. The 19-mer was mixed with varying ratios of 35-mer or 35 -uucg and heterodimer formation was assessed by native gel electrophoresis. The lanes in each gel contain: (1) 0.45 nmol 19-mer; (2) 0.45 nmol 19-mer and 0.15 nmol 35-mer; (3) 0.45 nmol 19-mer and 0.30 nmol 35-mer; (4) 0.45 nmol 19-mer and 0.45 nmol 35-mer and (5) 0.45 nmol 35-mer. In lanes (6–10), 35 -uucg was used instead of 35-mer.
NMR secondary structure analysis of the 19-mer, 35-mer and 35 -uucg monomers
Prior to structural analysis of heterodimers, we analyzed the secondary structures of the 19-mer, 35-mer and 35 -uucg monomers. NOEs from imino proton resonances to H2 resonances in adenosines or to amino proton resonances in cytosines indicated the formation of Watson–Crick base pairs. These NOEs were observed within all the base pairs predicted in the 19-mer, 35-mer and 35 -uucg. Imino proton and H2 resonances and their NOE signals of 35-mer indicated that the same Watson–Crick base pairs were formed as in 35 -uucg (data not shown), indicating that the 35-mer and 35 -uucg have the same secondary structure.
Sequential NOEs between H8/H6 and H1′ protons are observed from G2 to U7 and from U14 to C19 in the 19-mer as well as from G1 to G7, C15 to U21, C27 to G29 and C30 to C34 in 35 -uucg in 1H-1H 2D-NOESY spectra (data not shown). These NOEs imply formation of helices in these regions. The observation of sequential NOEs and the NOEs characteristic to Watson–Crick base pairs confirmed the formation of the D-helix in the 19-mer and the C- and E-helices in 35 -uucg as predicted (Figures 1B and D). In addition, NMR signals that are typical of the UUCG tetraloop were observed in the spectra of 35 -uucg, which includes the upfield shifted G24 imino proton resonance, 2′-OH resonance of U21 and a strong NOE signal between H8 and H1′ of G24 that suggests a syn conformation of this residue (40). These NMR signals indicated the proper formation of the UUCG tetraloop in 35 -uucg.
NMR secondary structure analysis of the 19-mer/35 -uucg and the 19-mer/35-mer heterodimers
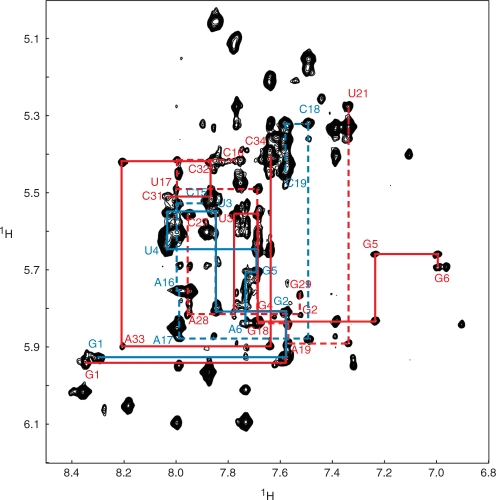
After identifying the secondary structures of the 19-mer and 35 -uucg monomers, we investigated the structure of the heterodimers. The NOE signals indicating Watson–Crick base pairs within the 19-mer and 35 -uucg monomers were preserved in the 2D 1H-1H NOESY spectra of the 19-mer/35 -uucg heterodimer (Supplementary Figure S1). The sequential NOEs between H8/H6 and H1′ in the monomers in 35 -uucg were also preserved in the 19-mer/35 -uucg heterodimer except for C15–C16, likely due to participation of C15 in the intermolecular interaction (Figure 3). Taken together, these data showed that the secondary structures of the 19-mer and 35 -uucg monomers were maintained in the heterodimer.
Figure 3.
Sequential assignment of the 19-mer/35 -uucg heterodimer. H6/H8-H1′ region of the 2D-NOESY spectrum of the 19-mer/35 -uucg heterodimer is shown. The red solid lines indicate the sequential NOEs from G1 to G6 and from C31 to C34 in 35 -uucg. The red dashed lines indicate the sequential NOEs from C16 to U21 and from C27 to A29 in 35 -uucg. The blue solid lines indicate the NOEs from G1 to A6 in the 19-mer. The blue dashed lines indicate the NOEs from C15 to C19 in the 19-mer.
15N-HSQC spectra of the labeled 19-mer/non-labeled 35-mer heterodimer and that of the labeled 19-mer/non-labeled 35 -uucg heterodimer showed the imino proton resonances at the identical chemical shifts (Supplementary Figure S2). This indicated that the 19-mer in the two heterodimers had the same structure. Furthermore, sequential NOE signals between H1′ and aromatic protons, as well as NOE signals from imino protons, indicated that the secondary structure of the 19-mer/35-mer heterodimer is the same as that of the 19-mer/35 -uucg heterodimer. In addition, the NMR signals derived from loop regions are also preserved in the 19-mer/35-mer heterodimer. This, taken together with our native gel analysis of heterodimer formation, indicated that the overall structure of the 19-mer/35-mer heterodimer and the 19-mer/35 -uucg heterodimer were the same. Therefore, we began the NMR analysis of pRNA intermolecular base pairs using the 19-mer/35 -uucg heterodimer that showed less-complex NMR spectra.
Intermolecular base pairs between the D-loop and the CE-loop
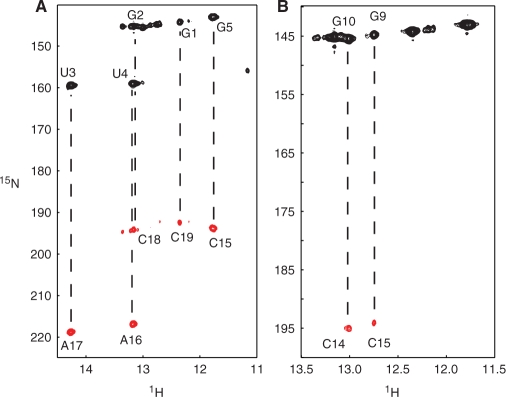
The intramolecular hydrogen bonds in the 19-mer were identified using an HNN-COSY spectrum of the 19-mer/35 -uucg heterodimer consisting of labeled 19-mer and non-labeled 35 -uucg (Figure 4A). We confirmed five hydrogen bonds in Watson–Crick base pairs in the 19-mer, from G1-C19 to G5-C15, whereas the N1–N3 correlation through the hydrogen bond of A6-U14 was not observed in the HNN-COSY spectrum due to relatively weak signal intensity of the U14 imino proton. Although it was not observed in the HNN-COSY experiment, the formation of Watson–Crick base pairs between A6 and U14 was indicated by the NOE signal between the imino proton of U14 and the H2 proton of A6 NOE.
Figure 4.
Identification of intermolecular base pairs. (A) HNN-COSY spectrum of the 15N-labeled 19-mer/non-labeled 35 -uucg heterodimer, in which only intramolecular hydrogen bonds in the 19-mer are observed. Lines indicate the intramolecular correlation between imino groups and acceptor nitrogens. (B) HNN-COSY spectrum of the G-labeled 19-mer/C-labeled 35 -uucg heterodimer, in which hydrogen bonds within the intermolecular G–C base pairs are observed.
In order to observe the intermolecular hydrogen bonds between the 19-mer and 35 -uucg, we prepared a G-labeled 19-mer/C-labeled 35 -uucg heterodimer that would show correlations only within intermolecular G–C base pairs. In addition, we prepared a U-labeled 19-mer/A-labeled 35 -uucg heterodimer in order to observe intermolecular A–U base pair correlations. Two hydrogen bonds between H1 of guanines and N3 of cytosines in the G-labeled 19-mer/C-labeled 35 -uucg heterodimer were detected in an HNN-COSY experiment (Figure 4B). However, the HNN-COSY spectrum of the U-labeled 19-mer/A-labeled 35 -uucg heterodimer showed no signal derived from intermolecular A–U base pairs (data not shown).
Imino protons that are rapidly exchanging with water would not be observed in HNN-COSY experiments (29). Therefore, in order to verify the absence of correlation signal derived from intermolecular A–U base pairs, we conducted a modified HNN-COSY experiment in which we tested hydrogen-bond correlations using the non-exchangeable H2 proton of adenosine instead of the imino proton of uridine (30). The result of this modified HNN-COSY experiment was also negative as we did not observe any signal indicating intermolecular A–U base pairing using the U-labeled 19-mer/A-labeled 35 -uucg heterodimer (data not shown). These results indicated that only two G–C base pairs are stably formed between the D-loop and the CE-loop, in contrast to the hypothesis that four base pairs are formed between these loops (5,6,43,44).
We assigned all cytidine residues in the helices of 35 -uucg using conventional NMR spectra, and the two cytidine residues involved in the intermolecular hydrogen bonds were identified as C14 and C15. In order to unambiguously assign the two guanosines involved in the intermolecular hydrogen bonds, we produced a series of 19-mer variants.
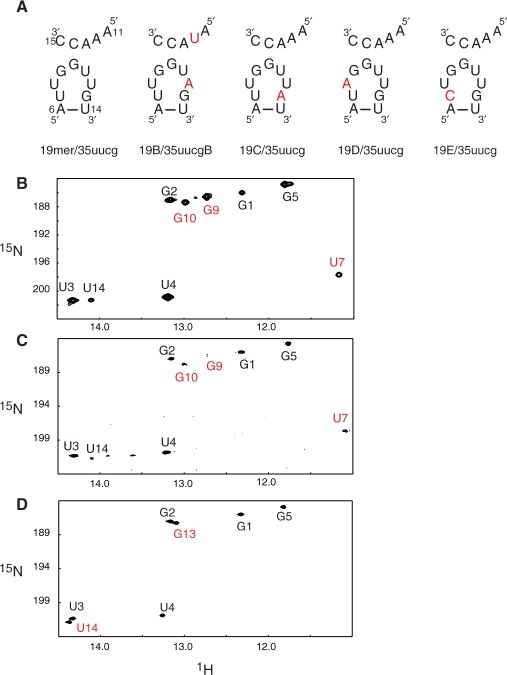
Assignment of the D-loop and the CE-loop
We made four variants of the 19-mer to complete assignment of imino proton signals derived from the D-loop (Figure 5A). The uridine imino proton signal at 11.2 p.p.m. disappeared in the 1H-15N HSQC spectrum of the labeled 19E/non-labeled 35 -uucg heterodimer (Figure 5D), while a new guanosine imino proton signal appeared at 13.1 p.p.m. We confirmed that this new guanosine imino proton forms an intramolecular G–C base pair using an HNN-COSY experiment (data not shown). Thus, this G–C base pair was assigned as C7-G13 in the 19E RNA. The spectra of the heterodimers of 19B/35 -uucg and 19D/35 -uucg (data not shown), in which uridines in the D-loop are exchanged with adenosines, did not show any disappearance of uridine imino proton signals. Therefore, the uridine imino proton resonance at 11.2 p.p.m., which disappeared in the 19E/35 -uucg spectrum, was assigned to U7 of the 19-mer. These results also demonstrated that the imino protons of U8, U11 and U12 of the 19-mer were not observed in the NMR spectra. The absence of these signals can be attributed to the rapid exchange of these protons with water, suggesting that these residues are not in a base paired conformation.
Figure 5.
19-mer variants. (A) Sequence of the RNA variants of the D-loop of the 19-mer and the CE-loop of 35 -uucg. Substituted residues are shown in red. (B-D) Imino region of 1H-15N HSQC spectra of (B) the labeled 19-mer/non-labeled 35 -uucg heterodimer, of (C) the labeled 19C/non-labeled 35 -uucg mixture and of (D) the labeled 19E/non-labeled 35 -uucg mixture.
The guanosine imino proton signals observed at 13.0 p.p.m. and 12.7 p.p.m. were shown to be involved in the intermolecular base pairing (Figure 4B). These signals remained in the 1H-15N HSQC spectrum of the labeled 19C/non-labeled 35 -uucg heterodimer (Figure 5C), in which an adenosine was substituted for the G13 of the 19-mer (Figure 5A 19C/35 -uucg). This result showed that these two guanosine imino proton signals were not derived from G13 and therefore were derived from G9 and G10.
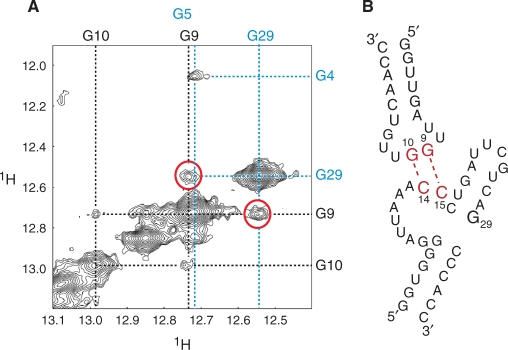
A 2D-NOESY spectrum of the 19-mer/35 -uucg heterodimer shows a NOE cross peak between the imino proton of G29 of 35 -uucg and the guanosine imino proton at 12.7 p.p.m. (Figure 6A) that was one of the two guanosine imino protons involved in the intermolecular hydrogen bond. Because these imino protons have unique chemical shifts, this NOE cross peak was unambiguously assigned as the intermolecular cross peak between G29 of 35 -uucg and G9 of the 19-mer. Thus, the other imino proton signal at 13.0 p.p.m. was assigned to G10. Taken together, our study unambiguously identified the intermolecular base pairs between G9 (D-loop) and C15 (CE-loop), and between G10 (D-loop) and C14 (CE-loop), respectively (Figure 6B).
Figure 6.
Intermolecular NOE between G29 and G9. (A) Imino–imino region of 2D-NOESY of the 19-mer/35-uucg. Dashed black lines indicate imino proton resonances derived from the 19-mer. Dashed blue lines indicate resonances derived from 35 -uucg. NOEs between G29 of 35 -uucg and G9 in the 19-mer are indicated by red circles. (B) Schematic representation of the intermolecular base pairs between the 19-mer and 35 -uucg.
pRNA dimerization and DNA packaging function
The 15N-HSQC spectrum of the labeled 19E/non-labeled 35 -uucg mixture, in which a cytidine was substituted for U7 of the 19-mer (Figure 5A 19E/35 -uucg), showed the disappearance of the G9 and G10 imino proton signals of 19E (Figure 5D), which implied the disappearance of intermolecular G–C base pairs. Subsequent native gel analysis showed that 19E and 35 -uucg did not form a heterodimer (Figure 7). To assess the effect of this alteration in D-loop structure in the context of the 120-nt pRNA, a U80C mutant was generated and tested for oligomerization and DNA packaging activity.
Figure 7.
Dimerization analysis of the E variant 19-mer. The 19-mer or the E variant (19E) was mixed with varying ratios of 35 -uucg and heterodimer formation assessed by native gel electrophoresis. The lanes contain: (1) 0.45 nmol 19-mer; (2) 0.45 nmol 19-mer and 0.15 nmol 35 -uucg; (3) 0.45 nmol 19-mer and 0.30 nmol 35 -uucg; (4) 0.45 nmol 19-mer and 0.45 nmol 35 -uucg. In lanes (5–8), 19E was used instead of 19-mer. (9) 0.45 nmol 35 -uucg.
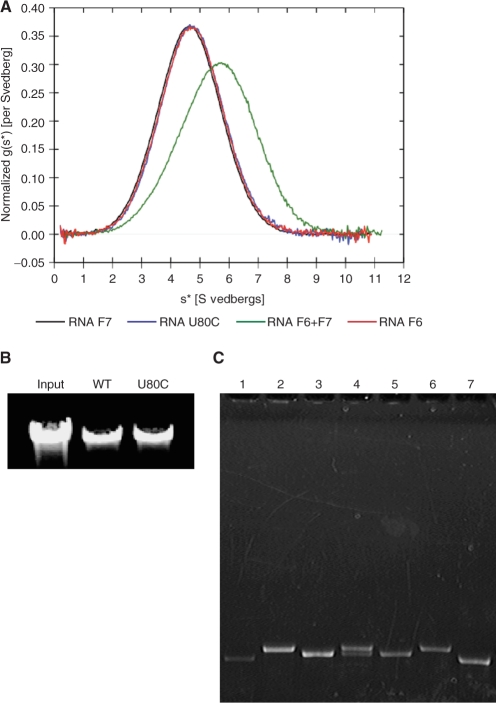
Native gel analysis of the 120-nt U80C was done at 4°C and at ambient temperature (Supplementary Figure S3). At 4°C, U80C migrated a little slower than the monomeric F6 RNA, in which the CE-loop was mutated to eliminate intermolecular base pair formation (Supplementary Figure S5A) (5), but faster than the dimeric form of wild-type 120 nt pRNA. Native gels run at ambient temperature showed U80C migrating at the same position as the F6 monomer. Sedimentation velocity analysis at 20°C showed that the U80C 120-nt pRNA was monomeric, as its sedimentation profile was virtually identical to the monomeric F6 or F7 pRNA mutants in which the CE- and D-loop, respectively, were mutated to prevent intermolecular base pair formation (Supplementary Figure 5A and B) (5). When the individual data sets were fitted to the model of a single ideal species, the values for the molecular weights were consistent with each of these RNAs being monomeric (Table 1). The sedimentation profile of a mixture of F6 and F7, which form heterodimers by their complementary mutant loops that restore their intermolecular base pairing, displayed a mixture of 52% dimer and 48% monomer (Figure 8A and Supplementary Figure S4). Taken together, we conclude that U80C has dramatically reduced capability to form stable oligomers in solution. Interestingly, however, proheads reconstituted with U80C 120-nt pRNA packaged DNA as well as the wild-type pRNA (Figure 8B).
Table 1.
Sedimentation analysis of the U80C variant
| RNA | Sedimentation coefficient (Svedbergs) | Molecular weight (kDa) | RMS error (OD260nm) |
|---|---|---|---|
| F6 | 4.75 | 42.9 ± 0.4 | 0.0094 |
| F7 | 4.69 | 42.4 ± 0.4 | 0.0081 |
| U80C | 4.77 | 41.2 ± 0.4 | 0.0110 |
The confidence intervals shown for the values of the molecular weights are the 95% confidence limits.
Figure 8.
Analysis of the U80C mutant. (A) Ultracentrifugation sedimentation velocity analysis. Overlay of the normalized g(s*) plots from DcDt+ analysis (37,38). F6 and F7 are monomeric 120-nt pRNA variants that form a dimer only when they are mixed (5). The F6 + F7 sample appears to be a mixture of 48% monomer and 52% dimer (Supplementary Figure S4). (B) DNA packaging assay. DNA packaged by proheads with pRNA or proheads reconstituted with U80C was extracted and analyzed by agarose gel electrophoresis. The negative control (no ATP) showed no packaging (data not shown). (C) 120-nt U80C pRNA binding to RNA-free proheads. pRNAs were mixed with RNA-free proheads, the prohead–pRNA complexes isolated and the RNA content analyzed by denaturing urea–PAGE. The lanes contain: (1) the monomeric mutant pRNA F6 bound to proheads; (2) U80C mutant pRNA bound to proheads; (3) ΔCCA pRNA bound to proheads; (4) U80C and ΔCCA pRNA mixture bound to proheads; (5) ΔCCA pRNA control; (6) U80C pRNA control and (7) F6 pRNA control. Lanes (1–4) contain equivalent amounts of proheads, thus revealing the relative binding efficiency of the RNAs.
Since pRNA dimerization has been associated with efficient prohead binding (21), the 120-nt U80C mutant was tested for its ability to bind proheads (Figure 8C). RNA-free proheads were mixed with an excess of pRNA and the resulting prohead–pRNA complexes isolated by layering the sample and pelleting through 5% sucrose; free pRNA stays near the top of the tube. Mutant 120-nt pRNA F6, a monomeric form of pRNA due to mutation in the CE-loop that prevents multimerization, bound to the prohead with about 25% the efficiency of U80C (Figure 8C, lane 1), consistent with it's low competitor activity (16) and it's inability to form a dimer (5,21). In contrast, the 120-nt U80C mutant pRNA bound to proheads in an amount similar to the 117-nt ΔCCA variant, which has wild-type binding activity (Figure 8C, lanes 2 and 3; Supplementary Figure S5C) (17). [Deletion of the three bases (18CCA20) yields a 117-nt RNA that allows it to be discriminated from 120-nt pRNAs in mixtures on denaturing gels.] When 120-nt U80C was mixed with the 117-nt ΔCCA immediately prior to addition to the proheads (Figure 8C, lane 4), both RNAs were found on these proheads, with a combined total RNA the same as U80C or ΔCCA alone. This indicates that U80C was capable of competing with ΔCCA pRNA, which has wild-type binding affinity.
DISCUSSION
Contribution of the pRNA E-loop to the intermolecular interaction
Our 35 -uucg construct contains the CE-loop and has a UUCG tetraloop substitution to replace the pRNA E-loop (bases 20–25, Figure 1C). Native gel analyses showed that 19-mer forms a heterodimer with 35 -uucg with a similar efficiency to the 35-mer containing the wild-type E-loop (Figure 2). In addition, the 2D 1H-1H NOESY spectra of the 19-mer/35 -uucg heterodimer are the same as those of the 19-mer/35-mer heterodimer, except for the signals derived from residues within the UUCG tetraloop (data not shown). Whereas previous mutagenesis studies have shown that the E-loop is important for the DNA packaging activity (17,43) and chemical modification show that it becomes protected during oligomerization (45), our current studies suggest that the E-loop is not directly involved in the intermolecular interaction of pRNA in solution.
Intermolecular base pair formation between the D-loop and the CE-loop
Previous genetic and biochemical experiments indicated the existence of intermolecular hydrogen bonding of pRNA (5,6,43,44). Our NMR analysis clearly demonstrated that two G–C base pairs are formed between the D-loop and the CE-loop in ø29 pRNA. Although four bases can potentially base pair in heterodimer formation, we did not observe intermolecular A–U base pairs in the original HNN-COSY experiment (29), nor in a modified HNN-COSY experiment (30) that would detect A–U base pairs even under the situation where imino protons are rapidly exchanging with water. Therefore, we conclude that there is no stable intermolecular A–U base pair within the 19-mer/35 -uucg heterodimer, and the intermolecular G-C base pairs are responsible for its formation. This result is consistent with previous biochemical studies that demonstrated that only two G-C base pairs between the D-loop and the CE-loop were required for the DNA packaging function (5,6,43,44). Thus, the two G–C base pairs demonstrated here are necessary and sufficient in pRNA multimerization and DNA packaging.
pRNA dimerization and DNA packaging function
Our NMR analyses of the 19-mer and 19-mer/35 -uucg heterodimer clearly demonstrated that the 19-mer U7 imino proton was protected from exchanging with water and was therefore in some stable conformation. However, the analysis of 19-mer/35 -uucg indicated that U7 and G13 did not form a Watson–Crick type base pair, suggesting that U7 was involved in some other type of intramolecular interaction within the D-loop. The HNN-COSY spectrum of a 19E/35 -uucg mixture, in which a cytidine was substituted for U7 of the 19-mer showed the formation of a new intramolecular C7-G13 Watson–Crick base pair. The 15N-HSQC spectrum of a labeled 19E/non-labeled 35 -uucg mixture showed the disappearance of the G9 and G10 imino proton signals (Figure 5D) demonstrating the absence of intermolecular G–C base pairs. Native gel analysis showed no dimerization of this pair (Figure 7). Furthermore, in the 15N-HSQC spectrum of the sample containing labeled 19C and non-labeled 35 -uucg, in which an adenine was substituted for G13 of the 19-mer (Figure 5A), intensities of the imino proton signals from G9 and G10 were reduced but not eliminated (Figure 5C). The 15N-HSQC and HNN-COSY spectra of this pair indicated the presence of two conformations of 19C; one in which a U7-A13 base pair was formed and one in which it was not. Based on these observations, the reduction or elimination of the G9 and G10 signal intensities in the 19C/35 -uucg or the 19E/35 -uucg mixtures, respectively, was attributed to the formation of a base pair that makes the D-loop smaller. Therefore, formation of a base pair at this position in the D-loop prevented the intermolecular interaction between the D-loop and the CE-loop in these RNA constructs.
In the context of the functional 120 -nt pRNA, U80C represents a new class of mutants. Extensive mutational analysis to change bases involved in the intermolecular interaction has been conducted to discern the requirements for oligomerization (5,6,21,44). This mode of disruption of pRNA oligomerization greatly impacts assembly and function of the DNA packaging motor. In contrast, oligomerization of U80C was dramatically affected by constriction of the D-loop, although the bases involved in the intermolecular interaction were unaltered. Oligomerization of U80C pRNA in solution was not detected by native gel analysis at ambient temperature (Supplementary Figure S3) or by ultracentrifugation at 20°C (Figure 8A), yet the RNA was fully active in DNA packaging (Figure 8B). This was unexpected as the intermolecular interaction of free pRNA in solution has been shown to be needed for efficient prohead binding (21). The prohead binding results showed that U80C pRNA can bind with wild-type efficiency to the prohead and compete for binding with the ΔCCA variant (Figure 8C). In contrast, prohead binding by mutant pRNA F6, which has low competitor activity (16), was only ∼25% that of U80C. This difference suggests that, although U80C may interact with the prohead initially as a monomer, it has the capacity to form the intermolecular interaction that locks the full complement of pRNA to the prohead. In contrast, the monomeric F6 mutant that lacks the complementary CE- and D-loops cannot form the intermolecular interaction on the prohead and is inefficiently retained. The intermolecular interaction may also be needed for post-binding functions of pRNA in the DNA packaging motor, such as mediating communication of the pRNA-dependent ATPase subunits in the ring. Although the U80C mutant appears to bind as a monomer and then link with neighboring pRNAs on the prohead, wild-type pRNA, which has the potential to form multimers in solution, may not follow this pathway. Complete structural solution of pRNA by NMR spectroscopy and other techniques is underway and will most certainly reveal additional details as to the assembly and function of this packaging motor component.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank Dr Wei Zhao and Nick Berge for technical assistance and Dr Jeffrey Lary for performing ultracentrifugation of RNAs and data analysis. Ultracentrifugation was performed at the University of Connecticut Analytical Ultracentrifugation Facility in Storrs, CT (James L. Cole, Director). Funding support has been given by National Institutes of Health (GM-059604 to S.G.; P41RR02301, P41GM66326 RR02781 and RR08438 to the National Magnetic Resonance Facility at Madison); Minnesota Medical Foundation (3484-9221-05 to H.M.); National Science Foundation (BIR-961477 to the NMR facility at University of Minnesota, DMB-8415048, OIA-9977486 and BIR-9214394 to the National Magnetic Resonance Facility at Madison). Funding to pay the Open Access publication charges for this article was provided by NIH grant number GM-059604.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grimes S, Jardine PJ, Anderson D. Bacteriophage phi 29 DNA packaging. Adv. Virus Res. 2002;58:255–294. doi: 10.1016/s0065-3527(02)58007-6. [DOI] [PubMed] [Google Scholar]
- 2.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo PX, Erickson S, Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987;236:690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- 4.Wichitwechkarn J, Bailey S, Bodley JW, Anderson D. Prohead RNA of bacteriophage phi 29: size, stoichiometry and biological activity. Nucleic Acids Res. 1989;17:3459–3468. doi: 10.1093/nar/17.9.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Lemieux S, Wu X, St-Arnaud D, McMurray CT, Major F, Anderson D. Function of hexameric RNA in packaging of bacteriophage phi 29 DNA in vitro. Mol. Cell. 1998;2:141–147. doi: 10.1016/s1097-2765(00)80123-9. [DOI] [PubMed] [Google Scholar]
- 6.Guo P, Zhang C, Chen C, Garver K, Trottier M. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 7.Guasch A, Pous J, Ibarra B, Gomis-Ruth FX, Valpuesta JM, Sousa N, Carrascosa JL, Coll M. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage phi29 connector particle. J. Mol. Biol. 2002;315:663–676. doi: 10.1006/jmbi.2001.5278. [DOI] [PubMed] [Google Scholar]
- 8.Xiao F, Moll WD, Guo S, Guo P. Binding of pRNA to the N-terminal 14 amino acids of connector protein of bacteriophage phi29. Nucleic Acids Res. 2005;33:2640–2649. doi: 10.1093/nar/gki554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atz R, Ma S, Gao J, Anderson DL, Grimes S. Alanine scanning and Fe-BABE probing of the bacteriophage o29 prohead RNA-connector interaction. J. Mol. Biol. 2007;369:239–248. doi: 10.1016/j.jmb.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morais MC, Choi KH, Koti JS, Chipman PR, Anderson DL, Rossmann MG. Conservation of the capsid structure in tailed dsDNA bacteriophages: the pseudoatomic structure of phi29. Mol. Cell. 2005;18:149–159. doi: 10.1016/j.molcel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Morais MC, Tao Y, Olson NH, Grimes S, Jardine PJ, Anderson DL, Baker TS, Rossmann MG. Cryoelectron-microscopy image reconstruction of symmetry mismatches in bacteriophage phi29. J. Struct. Biol. 2001;135:38–46. doi: 10.1006/jsbi.2001.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibarra B, Caston JR, Llorca O, Valle M, Valpuesta JM, Carrascosa JL. Topology of the components of the DNA packaging machinery in the phage phi29 prohead. J. Mol. Biol. 2000;298:807–815. doi: 10.1006/jmbi.2000.3712. [DOI] [PubMed] [Google Scholar]
- 13.Shu D, Zhang H, Jin J, Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee TJ, Guo P. Interaction of gp16 with pRNA and DNA for genome packaging by the motor of bacterial virus phi29. J. Mol. Biol. 2006;356:589–599. doi: 10.1016/j.jmb.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 15.Bailey S, Wichitwechkarn J, Johnson D, Reilly BE, Anderson DL, Bodley JW. Phylogenetic analysis and secondary structure of the Bacillus subtilis bacteriophage RNA required for DNA packaging. J. Biol. Chem. 1990;265:22365–22370. [PubMed] [Google Scholar]
- 16.Reid RJ, Zhang F, Benson S, Anderson D. Probing the structure of bacteriophage phi 29 prohead RNA with specific mutations. J. Biol. Chem. 1994;269:18656–18661. [PubMed] [Google Scholar]
- 17.Reid RJ, Bodley JW, Anderson D. Identification of bacteriophage phi 29 prohead RNA domains necessary for in vitro DNA-gp3 packaging. J. Biol. Chem. 1994;269:9084–9089. [PubMed] [Google Scholar]
- 18.Reid RJ, Bodley JW, Anderson D. Characterization of the prohead-pRNA interaction of bacteriophage phi 29. J. Biol. Chem. 1994;269:5157–5162. [PubMed] [Google Scholar]
- 19.Zhang C, Tellinghuisen T, Guo P. Confirmation of the helical structure of the 5′/3′ termini of the essential DNA packaging pRNA of phage phi 29. RNA. 1995;1:1041–1050. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C, Lee CS, Guo P. The proximate 5′ and 3′ ends of the 120-base viral RNA (pRNA) are crucial for the packaging of bacteriophage phi 29 DNA. Virology. 1994;201:77–85. doi: 10.1006/viro.1994.1267. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Sheng S, Shao Z, Guo P. A dimer as a building block in assembling RNA. A hexamer that gears bacterial virus phi29 DNA-translocating machinery. J. Biol. Chem. 2000;275:17510–17516. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 22.Grimes S, Anderson D. RNA dependence of the bacteriophage phi 29 DNA packaging ATPase. J. Mol. Biol. 1990;215:559–566. doi: 10.1016/s0022-2836(05)80168-8. [DOI] [PubMed] [Google Scholar]
- 23.Guo P. Structure and function of phi29 hexameric RNA that drives the viral DNA packaging motor: review. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:415–472. doi: 10.1016/s0079-6603(02)72076-x. [DOI] [PubMed] [Google Scholar]
- 24.Bourassa N, Major F. Implication of the prohead RNA in phage phi29 DNA packaging. Biochimie. 2002;84:945–951. doi: 10.1016/s0300-9084(02)01444-x. [DOI] [PubMed] [Google Scholar]
- 25.Hoeprich S, Guo P. Computer modeling of three-dimensional structure of DNA-packaging RNA (pRNA) monomer, dimer, and hexamer of Phi29 DNA packaging motor. J. Biol. Chem. 2002;277:20794–20803. doi: 10.1074/jbc.M112061200. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson BA, Blevins RA. NMRview: a computer program for the visualization and analysis for NMR data. J. Biomol. NMR. 1994;4:603. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Smallcombe SH. Solvent suppression with symmetrically-shifted pulses. J. Am. Chem. Soc. 1993;115:4776–4785. [Google Scholar]
- 29.Dingley AJ, Grzesiek S. Direct observartion internucleotide 2JNN couplings. J. Am. Chem. Soc. 1998;120:8293–8297. [Google Scholar]
- 30.Hennig M, Williamson JR. Detection of N-H … N hydrogen bonding in RNA via scalar couplings in the absence of observable imino proton resonances. Nucleic Acids Res. 2000;28:1585–1593. doi: 10.1093/nar/28.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marino JP, Diener JL, Moore PB, Griesinger C. Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly 13C-labeled RNA. J. Am. Chem. Soc. 1997;119:7361–7366. [Google Scholar]
- 32.Legault P, Farmer IBT, Mueller L, Pardi A. Through-bond correlation of adenine protons in a 13C-labeled ribozyme. J. Am. Chem. Soc. 1994;116:2203–2204. [Google Scholar]
- 33.Marino JP, Prestegard JH, Crothers DM. Correlation of adenine H2/H8 resonances in uniformly 13C labeled RNAs by 2D HCCH-TOCSY: a new tool for 1H assignment. J. Am. Chem. Soc. 1994;116:2205–2206. [Google Scholar]
- 34.Grimes S, Anderson D. In vitro packaging of bacteriophage phi 29 DNA restriction fragments and the role of the terminal protein gp3. J. Mol. Biol. 1989;209:91–100. doi: 10.1016/0022-2836(89)90172-1. [DOI] [PubMed] [Google Scholar]
- 35.Grimes S, Anderson D. The bacteriophage phi29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 1997;266:901–914. doi: 10.1006/jmbi.1996.0843. [DOI] [PubMed] [Google Scholar]
- 36.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. In: Harding SE, Rowe AJ, Horton JC, editors. Cambridge, England: Royal Society of Chemistry; 1992. ISBN 0851863450. [Google Scholar]
- 37.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 38.Philo JS. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- 39.Zhang C, Tellinghuisen T, Guo P. Use of circular permutation to assess six bulges and four loops of DNA-packaging pRNA of bacteriophage phi29. RNA. 1997;3:315–323. [PMC free article] [PubMed] [Google Scholar]
- 40.Allain FH, Varani G. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 41.Kitamura A, Muto Y, Watanabe S, Kim I, Ito T, Nishiya Y, Sakamoto K, Ohtsuki T, Kawai G, et al. Solution structure of an RNA fragment with the P7/P9.0 region and the 3′-terminal guanosine of the tetrahymena group I intron. RNA. 2002;8:440–451. doi: 10.1017/s1355838202026043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molinaro M, Tinoco I., Jr Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F, Anderson D. In vitro selection of bacteriophage phi29 prohead RNA aptamers for prohead binding. J. Biol. Chem. 1998;273:2947–2953. doi: 10.1074/jbc.273.5.2947. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Zhang C, Guo P. Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA. 1999;5:805–818. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trottier M, Mat-Arip Y, Zhang C, Chen C, Sheng S, Shao Z, Guo P. Probing the structure of monomers and dimers of the bacterial virus phi29 hexamer RNA complex by chemical modification. RNA. 2000;6:1257–1266. doi: 10.1017/s1355838200992501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.