Abstract
Loss of function of the RNA helicase maleless (MLE) in Drosophila melanogaster leads to male-specific lethality due to a failure of X chromosome dosage compensation. MLE is presumably involved in incorporating the non-coding roX RNA into the dosage compensation complex (DCC), which is an essential but poorly understood requirement for faithful targeting of the complex to the X chromosome. Sequence comparison predicts several RNA-binding domains in MLE but their properties have not been experimentally verified. We evaluated the RNA-binding characteristics of these conserved motifs and their contributions to RNA-stimulated ATPase activity, to helicase activity, as well as to the targeting of MLE to the nucleus and to the X chromosome territory. We find that RB2 is the dominant, conditional RNA-binding module, which is indispensable for ATPase and helicase activity whereas the N-terminal RB1 motif does not bind RNA, but is involved in targeting MLE to the X chromosome. The C-terminal domain containing a glycine-rich heptad repeat adds potential dimerization and RNA-binding surfaces which are not required for helicase activity.
INTRODUCTION
The gene that encodes the RNA helicase maleless (MLE) was originally discovered in a screen for male-specific lethal mutations that revealed genes crucial for dosage compensation in male Drosophila melanogaster (1). This system serves to increase the transcription from the single X chromosome in male fruit flies to match the cumulative expression from the two female X chromosomes (2–4). Failure of this activation of transcription in the 2-fold range is lethal for male flies. The function of MLE in dosage compensation is not known but it is presumably involved in mediating the effects of two non-coding roX (RNA-on-the-X) RNAs, roX1 and roX2, which are obligatory for dosage compensation. These two RNAs reside in a regulatory ribonucleoprotein complex, the dosage compensation complex (DCC; also known as Male-specific lethal or MSL complex). The DCC presumably first assembles at the site of roX transcription, from which it distributes to associate with many sites on the X chromosome, most prominently the coding regions of target genes (5,6). In the absence of roX the MSL proteins will only bind to a reduced number of sites on the X chromosome (7).
So far, three proteins of the DCC are known to interact with RNA: the histone acetyltransferase MOF (8), male-specific lethal-3 (MSL3) (9,10) and MLE (1,11,12). Since MLE is maternally provided to the Drosophila egg, it is the first protein to interact with and to stabilize the roX1 RNA, which is transcribed 2 h after egg laying (13). In the absence of MLE, roX RNA is not incorporated into the DCC and can only be seen at the site of transcription in polytene chromosomes (14). The ATPase/helicase activity of MLE is required for its function in dosage compensation (11,15). Recently, Lucchesi and colleagues generated mutations in MLE that separate ATPase and helicase activities and found that the ATPase activity was sufficient for MLE's role in transcriptional activation, whereas the helicase activity is necessary for the spreading of the complex along the X chromosome (16). roX RNA may play a transient role in targeting the DCC to the X chromosome (17), which suggests that its interaction with the complex is dynamic. Accordingly, MLE is not an integral member of the DCC, but peripherally associated, which leads to its loss during purification of the complex (18,19).
Although it is usually assumed that roX RNAs are the crucial targets of MLE, this has not been confirmed. In fact, MLE has roles outside dosage compensation that are not reflected by the male-specific lethal phenotype of its loss-of-function mutant. One particular temperature sensitive (ts) MLE allele, MLEnapts (nap stands for ‘no action potential’), is characterized by a reduced expression of the para gene, which encodes a Na+ channel of the nervous system (20). The data are consistent with the idea that the MLE helicase activity is required to unwind a secondary structure of the para primary transcript to permit faithful splicing. Other possibilities should not be excluded since RNA helicase A (RHA) (12,21), the MLE ortholog in vertebrates, has been implicated in various aspects of RNA metabolism, including transcription, processing and translation (22). Most recently, RHA was shown to be involved in the loading of small interfering RNAs (siRNA) into RISC (RNA-induced silencing complex) (23). Following the idea that dosage compensation mechanisms adapt components of other nuclear processes to fine-tuning chromatin structure (3) leads to speculations that MLE activity may affect the secondary structure of roX RNAs to facilitate productive interactions with the MSL proteins.
Currently, all our knowledge about MLE as an enzyme stems from the pioneering study of Lee et al. (11) who documented that recombinant MLE has ATPase and helicase activities and binds single-stranded nucleic acids. In the current work we extended the biochemical analysis significantly, by subjecting MLE to a thorough structure-function analysis, which clarified the domain requirements for RNA interaction, helicase activity and localization to the X chromosomal territory. In addition we analyzed the effects of nucleotides on RNA-binding as a first step towards a mechanistic understanding of the nucleotide cycle of MLE's helicase activity.
MATERIALS AND METHODS
Monoclonal anti-MLE antibodies
The NcoI/HindIII DNA fragment, corresponding to the first 265 amino acids (aa) of MLE was cloned from the pBB-6X-HisMLE (a gift from M. Kuroda) into the pGEX-2KG expression vector (Amersham). This construct was used to express and purify GST-MLE1–265 from Escherichia coli BL21 using standard conditions. Monoclonal antibodies were raised and MLE1–265 specific antibodies were screened by ELISA. Hybridoma 6E11 was subcloned to obtain monoclonal antibodies.
Expression and purification of proteins from Sf9 cells
Sf9 cells were kept at 26°C in Sf-900 II medium (Invitrogen) supplemented with penicillin and streptomycin. Recombinant baculoviruses expressing MLE derivatives were produced using the Bac-to-Bac expression system (Invitrogen). MLE full length was expressed with a C-terminal flag-tag or with an N-terminal His6-tag. MLE deletion mutants were all C-terminally flag-tagged. The RB1, RB2 and RB1-2 domains were expressed with an N-terminal His6-tag, whereas the G-domain was flag-tagged at the C-terminus.
Sf9 cells were infected under standard conditions and the optimal amount of each virus was determined empirically. The cells (∼3 × 107) were collected 2 days post infection and lysed in 1 ml lysis buffer [50 mM Hepes pH 7.6, 0.3 M KCl, 0.5 mM EDTA, 0.1% NP40, 1 mM DTT, protease inhibitors (PMSF, leupeptine, aprotinin, pepstatin) and incubated for 15 min on ice. The suspension was sonicated by three pulses of 15 sec at 15% amplitude (Branson digital sonifier model 250-D), and centrifuged for 30 min at 14 k.r.p.m. at 4°C. Supernatants were incubated for 2 h on a rotating wheel with anti-flag M2 (Sigma) affinity beads (50 µl of a 1:1 slurry in lysis buffer). The beads were washed five times with 1 ml of each of the three following buffers: Buffer I: 0.3 M KCl, 50 mM Hepes pH 7.6, 0.5 mM EDTA, 1% NP40, 0.5 mM DTT, protease inhibitors; Buffer II: Buffer I with 1 M KCl; Buffer III: 0.15 M KCl, 10 mM Hepes pH 7.6, 0.5 mM EDTA, 0.1% NP40, 10% glycerol, 0.5 mM DTT, protease inhibitors. Proteins were eluted (two times for 2 h on a rotating wheel at 4°C) in 100 µl of Buffer III supplemented with 5–10 µg/ml flag peptide. Aliquots of the eluted proteins were analyzed by SDS-PAGE and Coomassie staining and the remainder was stored at −80°C.
Pull down experiments
To study MLE dimerization in vitro, Sf9 cells were coinfected with a virus expressing His6-MLE in combination with flag-tagged MLE derivatives. Total extracts were prepared and diluted to 0.15 M KCl before incubation with anti-flag M2 affinity beads for 2 h at 4°C. Beads were then washed five times with 1 ml of Lysis Buffer (0.15 M KCl) and bound proteins were analyzed by SDS-PAGE and western-blot.
Glycerol gradient
A 5–16% (w/v) glycerol gradient was prepared in a buffer containing 50 mM Hepes pH 7.6, 0.3 M KCl, 0.5 mM EDTA, 0.1% NP40, 1 mM DTT and protease inhibitors, using a Gradient Master 105/106 (BioComp) set at 2.40 min/81.5 deg/15 r.p.m. A 500 µl sample containing 15 µg of MLE-flag purified from Sf9 cells and 10 µg of aldolase and catalase standard markers (Amersham) was loaded on top of the gradient. Centrifugation was performed using a SW41 rotor (Beckman) at 35 k.r.p.m. for 20 h at 4°C. Fractions of 0.5 ml were collected and analyzed by 8% SDS-PAGE and western blot.
Preparation of RNA substrates
The SP6.roX2 reverse primer (5′CGATTTAGGTGACACTATAGAAATA-TTTG-CTTAATTTGC3′), containing a SP6 promoter and the T7.roX2 forward primer (CGTTAATACGACTCACTATAGGGAGA-CGTG-TAAAA-TGTT-3′), containing a T7 promoter were used to amplify a 75 bp roX2 DNA sequence, predicted not to form double strand RNA structures (RNA fold program; http://rna.tbiunivie.ac.at/cgi-bin/RNAfold.cgi) using a roX2.78.2.2 cDNA) template (a gift from Kuroda M.). The PCR product was purified on MinElute Spin Columns (Qiagen). 1 µg of the PCR product was transcribed in vitro in the presence of ATP, GTP, UTP (2.5 mM each), CTP (100 µM) supplemented with α32P-CTP, 1 mM DTT, 1 µl of T7 RNA Polymerase (Promega), 1 U/ml RNAsin (Promega) and 1 × transcription buffer (Promega) in 25 µl of final volume. The reaction mixture was incubated at 37°C for 2 h and the radiolabeled ssRNA was purified on miniQuickSpin RNA columns (Roche) and obtained in 100 µl of DEPC-treated water. The RNA concentration was measured spectrophotometricaly. In most cases, the specific activity of the labeled ssRNA substrate was 102 cpm/fmole. The dsRNA substrate was prepared by annealing the radiolabeled ssRNA with the corresponding cold complementary ssRNA transcribed from the same PCR product with SP6 RNA Polymerase. The resulting partial RNA duplex molecule consisted of 40 bp dsRNA flanked by 18 bp and 19 bp ssRNA 3′overhanging sequences in the top and the bottom strand, respectively (Figure 1B). The annealing reaction (200 µl) containing equimolar amounts of the radioactive ssRNA and the cold complementary RNA in hybridization buffer (20 mM HEPES-KOH, pH 7.6, 0.5 M NaCl, 1 mM EDTA, 0.1% SDS), was incubated 5 min at 100°C, 30 min at 65°C and 4 h at room temperature. The dsRNA was then purified by electrophoresis on a native 10% polyacrylamide (30:1) gel in TBE. The gel slice, containing the dsRNA transcript (located by autoradiography) was ground with a micropestle and eluted twice in 0.4 ml of elution buffer (0.5 M ammonium acetate, pH 7, 0.1% SDS, 10 mM EDTA) for 2 h at room temperature in a shaking incubator. After a brief centrifugation the supernatant was extracted with phenol/chloroform, ethanol precipitated and resuspended in the appropriate volume of DEPC-treated water.
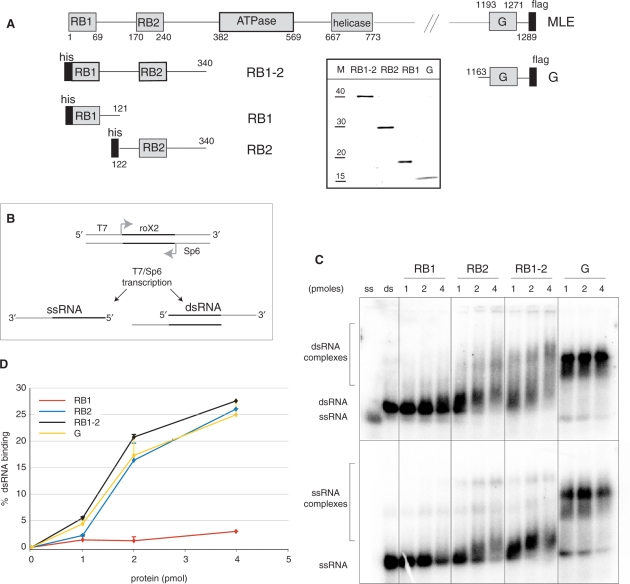
Figure 1.
Predicted RNA-binding domains in MLE. (A) Schematic representation of the predicted features of MLE and of the expressed fragments containing individual domains that were analyzed for RNA-binding. All proteins were tagged, either with a C-terminal flag-tag or an N-terminal His6-tag (black bars). The numbers represent the MLE aa that delineate the domains. Insert: The purified recombinant MLE derivatives were separated by polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie Blue. Lane M displays a size marker with indicated molecular weights in kDa. (B) Scheme of the ssRNA and dsRNA substrates. The sequence of roX2 is depicted in black, Sp6 promoter and T7 promoter are shown in grey. (C) Electrophoretic mobility shift assay (EMSA). The indicated amounts of the predicted RNA-binding domains (1–4 pmoles/50–200 nM) were incubated on ice in the presence of 25 fmol (1.25 nM) radiolabeled dsRNA or ssRNA. RNA–protein complexes (bracketed regions) were resolved on a 1.8% agarose gel and visualized by PhosphoImager analysis of the dried gel. Positions of ssRNA and dsRNA are indicated. (D) Filter-binding assay. Binding of the predicted RNA-binding domains to dsRNA was analyzed by filter-binding as described in Materials and Methods section. Each sample was analyzed in duplicates and error bars reflect the experimental variation.
ATPase assay
The ATPase activity of MLE-flag (25 fmol, 1.25 nM) and the corresponding mutant proteins was measured in a 20 µl reaction containing 6.6 mM HEPES pH 7.6, 0.66 mM EDTA, 0.66 mM 2-mercaptoethanol, 0.033% NP40, 1.1 mM MgCl2, 33 µM ATP, 5 µCi [γ-32P] ATP (3000 Ci mmol−1, NEN) in the presence of the indicated amounts of cold ssRNA or dsRNA. [γ-32P] ATP was added only after 5 min incubation at 26°C and 1 µl aliquots were spotted onto PEI thin layer chromatography plates (Merck) at various time points as previously described (24). Usually the reaction was linear during the 20 min reaction.
Helicase assay
RNA helicase activity was measured in a 20 µl reaction containing 20 mM Hepes-NaOH pH 7.5, 2 mM DTT, 3 mM MgCl2, 1 mM ATP (when indicated), 0.1 mg/ml BSA (NEBiolabs) 1 U/ml RNAsin (Promega), 25 fmoles of dsRNA substrate and the indicated amounts of purified proteins for 30 min at 37°C and then stopped by the addition of 5 µl of 5 × Stop Buffer (0.1 M Tris–Cl pH 7.5, 20 mM EDTA, 0.5% SDS, 0.1% NP40, 0.1% bromophenol blue, 0.1% xylene cyanol and 50% glycerol). Aliquots (10 µl) of each reaction were loaded onto a 10% polyacrylamide (30:1) gel in TBE and electrophoresed at 20 mA for 2–3 h. RNA was visualized by autoradiography of the dried gel.
Electrophoretic mobility shift assay
RNA-binding reaction (20 µl) contained radiolabeled ssRNA or dsRNA (25 fmol/1.25 nM) in Band-shift Buffer (20 mM Hepes-KOH pH 7.6, 3 mM MgCl2, 10% glycerol, 1 mM DTT, 0.1 mg/ml BSA (NEB)). 1 mM ATP, AMP-PNP, AMP-PCP, ATPγS or ADP were added when indicated and kept on ice to avoid unwinding. After incubation for 5 min on ice, 10 µl of each reaction were electrophoresed on a 0.8% agarose gel in 0.3 X TBE at 20 mA for about 1.5 h. Gels were dried and the RNA–protein complexes visualized by PhosphoImager.
Filter-binding assay
The radiolabeled dsRNA (25 fmol) was mixed on ice with MLE and derivatives in 40 µl of Band Shift Buffer in the absence or presence of 1 mM of the indicated NTPs. After 5 min on ice 15 µl of each reaction were filtered in duplicates through a BA85 Protran nitrocellulose membrane (Schleicher & Schuell) with an underlying nylon membrane (N+ Amersham) and two sheets of 3MM Whatman paper under vacuum, using a Dot Blot apparatus (Schleicher & Schuell). Both membranes and the sheets of paper were equilibrated for 15 min in binding buffer before assembling. The trapped RNA–protein complexes were washed once with 200 µl of binding buffer. The radioactivity retained on both membranes were quantified by PhosphoImager and the percentage of dsRNA retained on the nitrocellulose membrane were calculated and plotted after subtraction of background from control samples prepared in the absence of proteins.
Because of the low retention efficiency of the nitrocellulose membrane for ssRNA–protein complexes, the filter-binding assay was performed only with dsRNA.
Immunofluorescence on SF4
Drosophila SF4 cells were obtained from D. Arndt-Jovin (MPI for Biophysical Chemistry, Gottingen, Germany). Cells were kept at 26°C in Schneider's Drosophila medium (Invitrogen), supplemented with penicillin, streptomycin, glutamine and 10% fetal calf serum (FCS). The cell density was maintained below 4 × 106 cell/ml.
Transient transfections were performed using the Effectene Reagent Kit (Qiagen), following the manufacturer's instructions. The cells were processed for immunofluorescence as described (25) after 48 h of transfection. Anti-MSL1 antibodies (a gift of M. Kuroda) were used to mark the X chromosome territory and mouse anti-GFP antibodies (Molecular Probes) were used to visualize the fusion proteins. Pictures were taken at 1200 × magnification using a Zeiss AxiopHot microscope coupled to a Retiga Exi CCD Camera (Qimaging, Burnaby, Canada). Images were processed using Adobe Photoshop CS.
RESULTS
RNA-binding domains of MLE
MLE and its ortholog RHA belong to a group of DExH-type helicases that are characterized by two doubled-stranded (ds) RNA-binding (RB) motifs at their N-termini [Figure 1A, Supplementary Figure 1; (1,21,26)]. In keeping with their proposed RNA-binding function they are positively charged at neutral pH, with pIs of 9.6 and 10.0 for RB1 and RB2, respectively. A further feature is a 9-fold imperfect repetition of the sequence GGGYGNN in the C-terminus of MLE (‘G’ in Figure 1A) (1,12), where a tyrosine occupies every seventh position within the repeat (Supplementary Figure 2). Such a pseudo-heptad arrangement could indicate an amphipatic helix, with a hydrophobic surface that may be employed in interactions. The C-terminus of MLE (from aa 1163 onwards) has an overall pI of 11.0 and hence is a strong candidate for an RNA-binding domain.
In order to experimentally evaluate the RNA-binding potential of these domains we expressed tagged versions of RB1, RB2, a fragment consisting of RB1 and RB2, as well as the glycine-rich C-terminus in insect cells (Figure 1A) and purified them to apparent homogeneity using tag affinity chromatography (Figure 1A, insert). Since any complex RNA consists of single- (ss) and double-stranded (ds) RNA segments we characterized the interaction of the isolated MLE domains with both types of substrates. Although the RNA substrates we tested were derived from roX2 RNA the interaction with these substrates under our conditions was non-specific [electrophoretic mobility shift assay (EMSA) data not shown]. In order to produce a truly ssRNA substrate we amplified a 75 bp piece of roX2 cDNA which was predicted not to form any secondary structure when transcribed into RNA (see Methods section). The PCR product was transcribed with radiolabeled nucleotide precursors to produce ssRNA. Annealing of this RNA with unlabeled complementary RNA, transcribed from the other strand of the DNA template, yielded a substrate with a dsRNA core (dsRNA, Figure 1B). The ss and ds RNAs can be separated by gel electrophoresis (Figures 1C, 2C lanes ‘ss’, ‘ds’) and were used for EMSA. We observed robust binding of RB2, yielding a continuum of complexes with ssRNA as well as dsRNA (Figure 1C). Surprisingly, RB1 bound either RNA substrate much worse than RB2. Both domains together were only slightly more efficient in the EMSA than RB2 alone, pointing to a very small contribution of RB1 to RNA interactions in this context. The G-rich C-terminus proved to be the best binder and yielded a well-defined complex with both RNA substrates.
In light of the poor RNA interaction of RB1 we were concerned that EMSA was too stringent to reveal all RNA interaction potential. We therefore employed filter-binding as a second assay. The different MLE domains were allowed to bind radiolabeled dsRNA and the reactions were then filtered through a nitrocellulose membrane with an underlying positively charged nylon membrane. In the absence of protein dsRNA runs through the nitrocellulose membrane and is retained on the nylon. Proteins, however, bind to nitrocellulose and trap associated RNA on this layer of the blot. The percentage of input dsRNA retained on the nitrocellulose membrane was calculated and plotted after subtraction of background from control samples prepared in the absence of proteins. The filter-binding assay essentially confirmed the poor RNA-binding potential of RB1 relative to RB2 and the C-terminus (Figure 1D).
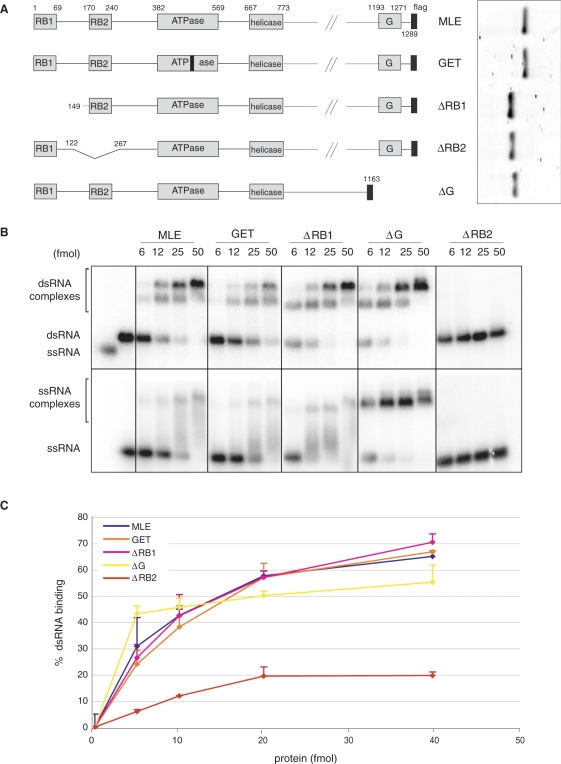
In order to assess the functions of these domains in the context of full length MLE we generated a series of MLE derivatives in which individual domains were deleted (Figure 2A). These proteins were subjected to the same RNA-binding assays and showed a much stronger RNA-binding capacity, since binding is observed at a stoichiometry of about 1:1 (as compared to 50–100 fold excess used for the isolated domains). Binding of intact MLE to dsRNA yielded two retarded bands, which we interpret as one and two molecules of MLE interacting with the RNA, respectively (Figure 2B). Interaction of MLE with ssRNA led to a clearly defined ssRNA–MLE complex as well as a smear of retarded bands, conceivably due to heterogeneity of RNA conformations. Inactivating the ATPase of MLE by converting lysine 413 to glutamic acid (K413E) (MLEGET) led to a slight reduction of dsRNA-binding, in line with previous observations (11). In agreement with the poor RNA interaction potential of RB1 (Figure 1), deletion of the N-terminal RB1 motif did not affect binding to either substrate (Figure 2B). To our surprise we found that deleting the G-rich C-terminus improved the interaction of MLE with dsRNA slightly, but led to a much better definition of the MLE–ssRNA complex. Obviously, although this fragment can interact with RNA (Figure 1), it does not provide the dominant interaction surface in the context of MLE in EMSA. Deletion of RB1 was also neutral in the context of the C-terminal truncation (Supplementary Figure 3). The dominant RNA recognition motif of the enzyme was revealed by the deletion of RB2, which essentially abolished all RNA-binding activity of MLE under those conditions.
Figure 2.
Binding of MLE and MLE derivatives to dsRNA and ssRNA. (A) Schematic representation of MLE deletion constructs analyzed in this study as in Figure 1A. The insert to the right shows the purified recombinant MLE derivatives, separated by PAGE and stained with Coomassie Blue. (B) EMSA as in Figure 1C with protein–RNA complexes (bracketed regions) resolved on a 1.8% agarose gel. Protein amounts range from 6 fmoles (0.3 nM) to 50 fmoles (2.5 nM). (C) dsRNA interactions measured by filter-binding as in Figure 1D.
When subjected to filter-binding assay increasing amounts of MLE and its derivatives led to a concentration-dependent trapping of RNA on the nitrocellulose (Figure 2C). Consistent with the EMSA, dsRNA-binding of GET, ΔRB1 and ΔG derivatives were similar to that of MLE. In this assay, an MLE derivative lacking the RB2 domain still showed significant dsRNA-binding, which may be due to the C-terminal domain and the central helicase domain itself, which is known to contact RNA in the context of other helicases (27). We conclude that EMSA mainly reveals the strong RNA interaction of RB2, but other domains, such as the helicase domain and the C-terminus, contribute to RNA interactions.
Effects of nucleotides on RNA interactions
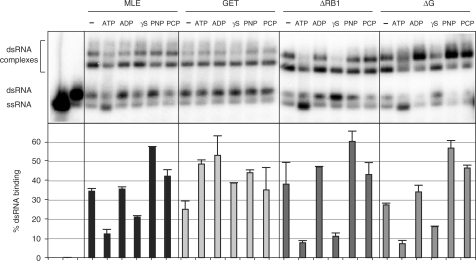
Previously, Lee and Hurwitz have analyzed the effect of ATP on the interactions of MLE with ssRNA and did not observe a major effect (11). We have now explored how the presence of ATP, ADP, AMP-PNP, AMP-PCP or the slow-hydrolyzing derivative ATPγS affect the interaction of MLE with dsRNA. The incubation of the enzyme with RNA in the presence of nucleotide was on ice in order to limit the unwinding of dsRNA in the presence of ATP. The MLE–RNA complexes were assayed by EMSA as well as filter-binding (Figure 3). Under our conditions some ATP hydrolysis and hence helicase activity still occurred (Figure 3, upper panel, ATP-lanes). The corresponding filter-binding values appear reduced since MLE binds ssRNA less stably as already shown in Figure 2.
Figure 3.
Effects of nucleotides on the RNA interactions of MLE. Binding of MLE and derivatives (12 fmoles/0.6 nM) to dsRNA (25 fmoles). Binding reactions are done in the presence of 1 mM of the indicated nucleotide and kept on ice to limit helicase activity. EMSA analysis (upper panel) and corresponding filter-binding assay (lower panel) are presented.
ATPγS also leads to reduced recovery of MLE–RNA complexes in the filter-binding reactions and furthermore in the EMSA with MLEΔRB1 and MLEΔG, however, since this effect is not seen with AMP-PNP and AMP-PCP, we cannot derive any general conclusion other than that ATPγS affects the interaction in a particular way. There is a trend for MLE derivatives to bind RNA better in the presence of non-hydrolysable ATP analogs. Conceivably, the RNA interactions are promoted by ATP-binding. However, MLE also binds RNA well in the presence of ADP. The MLEGET mutant is largely unaffected by the presence of nucleotides presumably because the mutant enzyme binds nucleotides with much reduced affinity (11).
Effect of the RNA substrate on ATPase activity
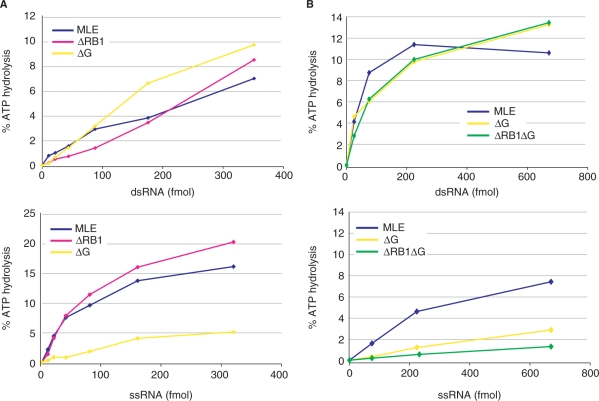
The ATPase activity of RHA and MLE is strongly stimulated by RNA, but the effect of ssRNA and dsRNA have not been compared so far. We also wished to establish whether any of the domains, besides RB2, was particularly involved in transmitting the RNA signal to the ATPase domain. We did not detect significant ATPase activity of MLEΔRB2 (data not shown), which is explained by the fact that RB2 is the major RNA interaction domain. Deleting the N-terminal RB1 (ΔRB1) or the C-terminus including the G-rich domain (ΔG) did not affect the robust, concentration-dependent stimulation of the ATPase activity by dsRNA (Figure 4A, upper panel). Interestingly, deletion of the C-terminus selectively reduced the ATPase stimulation by ssRNA (Figure 4A, B, lower panels). Combining N- and C-terminal deletions again did not affect dsRNA stimulation but selectively reduced the stimulatory effect of ssRNA (Figure 4B). Analysis of MLEΔG thus shows that RNA-binding (Figure 2) and ATPase stimulation need not necessarily correlate. A very stable interaction as observed by EMSA may indicate structural rigidity that does not support extensive ATPase cycles. Deletion of the C-terminus renders the enzyme differentially sensitive towards a potential helicase substrate (dsRNA) versus the product of a helicase reaction (ssRNA).
Figure 4.
Stimulation of MLE ATPase activity by RNA. MLE and selected derivatives were analyzed in ATPase assays. 25 fmol (1.25 nM) of the indicated protein and increasing amounts of dsRNA (upper panels) or ssRNA (lower panels) were used in the ATPase reaction as described in Materials and Methods section. In (B) different enzyme preparations from (A) were used, with slightly different specific activities.
Helicase activity of mutant enzymes
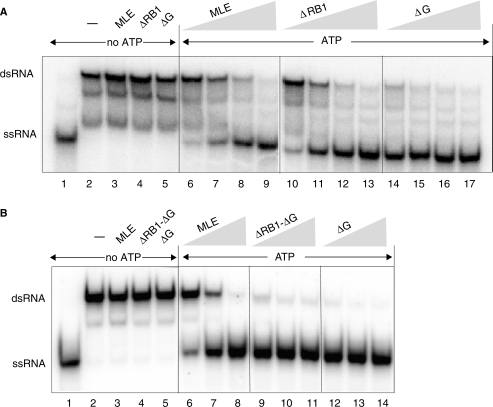
We next explored how RNA interaction and ATPase activity are translated into productive helicase activity. The dsRNA used for EMSA and ATPase stimulation contains 3′ overhangs, which renders it a substrate for the unwinding reaction of MLE (11). The radiolabeled single strand that is freed by the unwinding reaction is clearly separated from the dsRNA on a polyacrylamide gel (Figure 5). In the absence of ATP no unwinding was detected (Figure 5A, B, lanes 2–5). Catalytic amounts of full length MLE efficiently unwound the dsRNA substrate and this process was unaffected by deletion of the RB1 domain (Figure 5A, lanes 6–13). Deletion of the glycine-rich C-terminus, alone or in combination with RB1, led to a striking increase in helicase activity (Figure 5A, lanes 14–17, 5B, lanes 9–14), demonstrating that the ssRNA-binding activity of the C-terminus is not required for optimal unwinding, but conceivably for regulation of the appropriate enzyme activity in a more physiological context.
Figure 5.
Contributions of MLE domains to helicase activity. Helicase reactions were carried out with increasing amounts (1.5, 3, 6, 12 fmole/75, 150, 300, 600 pM) of MLE and derivatives and 25 fmoles (1.25 nM) of radiolabeled dsRNA as described in Materials and Methods section. Lanes 1 and 2 indicate the migration position of the ssRNA (boiled substrate) and dsRNA substrate, respectively. Lanes 3–5 correspond to unwinding reactions from which ATP was omitted. In (B) different enzyme preparations from the ones tested in (A) were used, with enzyme inputs of 6, 12, 25 fmoles.
MLE functions as a monomeric enzyme
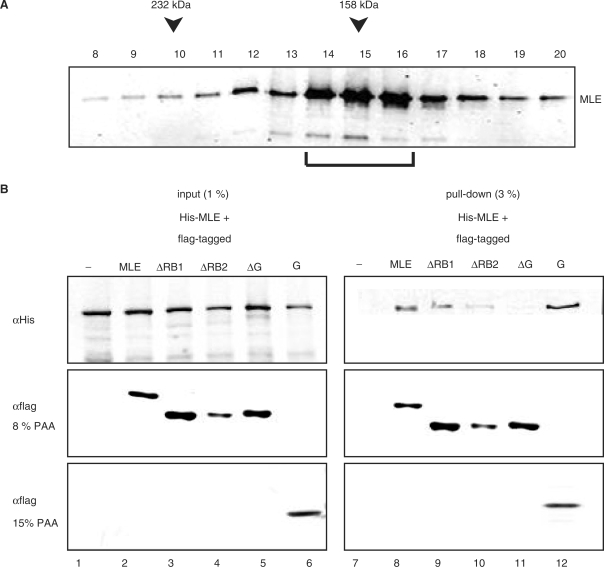
For mechanistic considerations of helicase activity it is important to establish whether MLE functions as a monomeric or as a dimeric enzyme. It has previously been suggested that monomeric helicases are characterized by multiple RNA-binding domains (26), however, our analysis shows that the predicted RB1 and the G-rich C-terminus are dispensable for helicase activity. Native and recombinant RHA migrate as monomeric enzymes on glycerol gradients (12,28). However, dimers may form transiently and conditionally. For example, the HCV helicase NS3 is a model for monomeric helicases (29,30), yet an oligomeric state was evidenced by crosslinking and has been suggested to be of functional importance (31). We subjected recombinant full length MLE to glycerol gradient sedimentation and observed a single peak of migration around 140–160 kDa (Figure 6A), which suggests that MLE is predominantly monomeric in solution and confirms earlier results (11). Addition of various RNAs (total cellular RNA, ss/ds EMSA probes) did not change this migration behavior. However, we cannot exclude that dimerization might be promoted by a very specific RNA substrate. In order to monitor more transient interactions we coinfected Sf9 cells with viruses expressing His6-MLE and various flag-tagged MLE derivatives. We monitored the expression of each derivative by western blot analysis of total extracts from infected cells (Figure 6B, ‘input’). Flag-tagged MLE derivatives were retrieved from the extracts (Figure 6B, ‘pull down’, centre and bottom panel) and probing for the his-tag revealed associated His6-MLE. This experiment documented an interaction between two differently tagged MLE proteins (Figure 6B, lane 8). Deletion of RB1 did not affect this interaction and deletion of RB2 diminished the binding to some extent (Figure 6B, lane 9). However, deleting the C-terminus (from aa 1163 onwards) abolished the interaction entirely (Figure 6B, lane 11). A C-terminal fragment containing the glycine-rich heptad repeats was able to pull down full length MLE very efficiently (Figure 6B), which revealed a potential for this peptide in MLE dimerization. However, MLEΔG is a very efficient helicase in vitro (Figure 5), thus we conclude that for mechanistic considerations MLE should be treated as a monomeric helicase. A role for the glycine-rich C-terminus in dimerization in a more physiological context cannot be excluded.
Figure 6.
Analysis of MLE dimerization. (A) Glycerol gradient sedimentation of MLE. 15 µg of MLE was centrifuged through a 5–16% glycerol gradient. 0.5 ml fractions were collected. Aliquots (10 µl) of each fraction were tested for the presence of MLE by western blotting using a specific monoclonal antibody. The sedimentation positions of the molecular weight standards (catalase, 232 kDa; aldolase, 158 kDa) are indicated. The bracket indicates MLE peak fractions. (B) MLE dimerization assessed by co-immunoprecipitation. His-tagged MLE was coexpressed in Sf9 cells with flag-tagged MLE derivatives in the combinations indicated. After flag-mediated pull down from total Sf9 extracts, bound proteins were analyzed on a 8% or 15% SDS-PAGE (for the G-box domain) and western blot using anti-His (αHis) and anti-flag (αflag) antibodies. An extract from Sf9 cells expressing only His6-MLE served as a control (lane 7).
In vivo localization
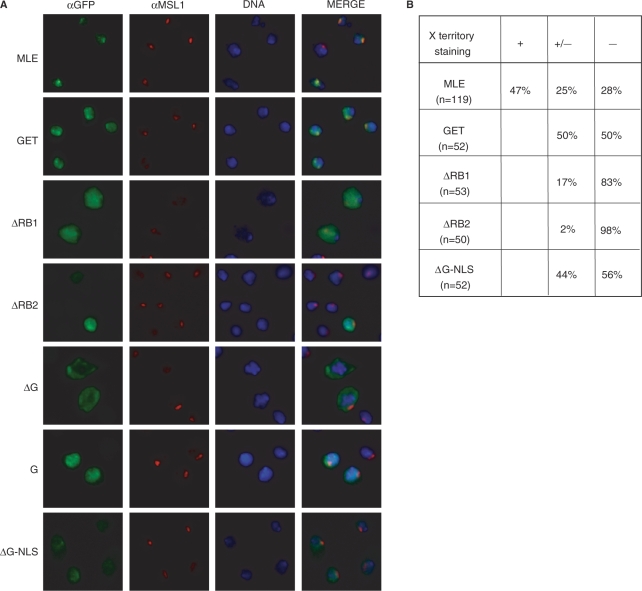
The association of MLE with the X chromosome in male Drosophila cells depends on interactions with RNA, since RNAse treatment of nuclei led to dissociation of MLE (32). In order to see which of the RNA-binding domains analyzed in this study were involved in X chromosome targeting, we transiently expressed MLE derivatives (Figure 2A) with C-terminal Green Fluorescent Protein (GFP) fusions in male SF4 cells. Successful targeting to the X chromosome was concluded from co-localization with the MSL1 marker (Figure 7). For each fusion protein at least 50 nuclei of MLE expressing cells were analyzed. Clear and selective X chromosomal staining was scored as positive (‘+’ in Figure 7B), whereas a clear enrichment of GFP at sites of MSL1-binding over a diffuse nuclear staining was scored as intermediate (‘+/−‘in Figure 7B). Examples are shown in Figure 7A. MLE-GFP fusion protein localized to the X chromosomal territory with a pattern very similar to endogenous MLE (data not shown). 73% of all nuclei showed an enrichment of the protein on the X chromosome. Inactivating the ATPase activity of MLE (MLEGET) led to a reduced X chromosomal targeting of the fusion protein. This is in agreement with the observations of Lee et al. who found that MLEGET bound fewer sites on X chromosomes and an increased number of autosomal sites in transgenic flies (11). Surprisingly, MLEΔRB1 failed to be efficiently targeted to the X chromosome. Considering that the RNA-binding and helicase activities of this protein were similar to wild type MLE, this suggests that RNA interactions are not the only targeting principles. However, RNA interactions are required for X chromosomal recognition, since deletion of RB2 abolished all recruitment activity. MLEΔG was found exclusively in the cytoplasm. This may be explained by the fact that the deletion removed a sequence preceding the G-rich heptad repeats that harbors a candidate nuclear localization sequence (NLS Supplementary Figure 2), which has been recently mapped in the context of RHA (33,34). We confirmed the presence of an NLS in this fragment of MLE by expression of the C-terminal fragment (‘G’ in Figure 1) fused to GFP in SF4 cells (‘G’ in Figure 7). As expected the fusion protein was nuclear, but did not localize to the X chromosome. Lastly, we furnished the MLEΔG with a heterologous, SV40-derived NLS. The fusion protein became nuclear by this NLS and localized to the X chromosome with an intermediate efficiency, similar to that of the MLEGET mutant. In summary, this experiment showed that the two N-terminal RB domains contribute to X chromosome targeting. Targeting via RB2 may involve RNA interactions, since RB2 provides the dominant RNA interaction surface. RB1, however, which does not bind RNA, must contribute to targeting by other types of interactions.
Figure 7.
In vivo localization of MLE and MLE derivatives. (A) Drosophila SF4 cells were transiently transfected with either MLE or the indicated MLE derivatives, all fused to GFP at the C-terminus. Localization of the fusion proteins was visualized by using anti-GFP antibodies (αGFP) and the X territory was marked with anti-MSL1 antibodies (αMSL1). The DNA was counterstained with Hoechst 33258 (DNA). (B) Quantitative analysis of the experiment. Clear and selective X chromosomal staining was scored as positive (‘+’), whereas a clear enrichment of GFP at sites of MSL1-binding over a diffuse nuclear staining was scored as intermediate (‘+/−’).
DISCUSSION
It is generally assumed that helicases make segments of ss RNA available for interactors within complex nucleoprotein assemblies. Little is known about the role of MLE in dosage compensation, although the available evidence suggests that its substrates are the long, non-coding roX RNAs, which may need to be reorganized for incorporation into productive DCCs. Whether this reorganization involves unwinding inappropriate secondary structures or destabilization of unwanted protein–RNA interactions is unknown. A prerequisite to understanding MLE's function is knowledge about the properties of its presumed RNA interaction domains, which are predicted from sequence comparison, but have not been characterized so far.
The role of the N-terminal dsRB motifs
The N-terminal RB1 and RB2 motifs of MLE are related to a family of RNA-binding domains called dsRBM (double-stranded RNA-binding motifs). Although the name suggests that they interact with dsRNA it has become clear that the term subsumes a variety of related domains with different interaction potential. This includes ssRNA-binding and even protein interactions; for some ‘orphan dsRBMs’ interactors have not been identified so far [(35) and references therein]. We now show that the most N-terminal RBM of MLE (RB1), despite of its basic nature, does not contribute to RNA-binding in vitro. At first glance this finding is at odds with the observed RNA-binding potential of the homologous domain in RHA (36). However, a more resolving analysis revealed that dsRBM1 of RHA by itself is a poor RNA binder and requires the context of a proline-rich segment between RHA aa 73–120 to bind nucleic acids, notably DNA in addition to RNA (35). This accessory sequence is not conserved in MLE, which explains the lack of RNA interaction in this context. However, it remains possible that RB1 synergizes with other structures within DCC to interact with nucleic acids. Our observation that deletion of RB1 abolished the targeting of MLE to the X chromosome territory in male cells is consistent with this hypothesis. Whether this targeting involves RNA, DNA or protein interactions remains an interesting subject for future studies.
In striking contrast to the poor RNA interacting potential of RB1, RB2 provides a dominant nucleic acid interaction surface for MLE. Deletion of RB2 abolishes those RNA interactions that resist the experimental conditions of gel electrophoresis. Notably, RB2 is required for RNA-dependent ATPase and helicase activity of MLE. RB2–RNA interactions are therefore an obligatory intermediate of MLE activity. Gibson and Thompson (26) have speculated earlier on the importance of dsRB-type accessory RNA interaction domains (i.e. those in addition to the helicase domain) for helicase activity.
The role of the G-rich C-terminus
Our studies reveal integration of various functions at the very C-terminus of MLE. This part of MLE is dominated by a 9-fold imperfect repeat of the aa heptad GGGYGNN, which is reminiscent of the wide spread RGG domains that contribute to RNA-binding in many different contexts. This repeat is preceded by a basic sequence that is conserved in vertebrate RHA, where it was shown to function as an NLS (34). Although we have not repeated the fine mapping of the NLS in the context of MLE, our data are consistent with the notion that the homologous sequence (aa 1171–1189, Supplementary Figure 2) is also an NLS. Deletion into this sequence (MLEΔG) destroys the nuclear localization of MLE and fusion of the C-terminus targets GFP to the nucleus. There are other examples where the NLS is separable from adjacent G-rich domains, such as the RGG/GAR domains (37). The basic nature of the NLS may be responsible for the observed RNA interactions in vitro. However, these interactions are not required for helicase activity; to the contrary, deletion of the entire C-terminus improves ssRNA interactions and helicase activity of MLE in vitro. The C-terminus may thus be engaged in other interactions, including transport factors, and not be available for RNA-binding. Interestingly, we observed that the C-terminus was able to mediate MLE dimerization, which is reminiscent of the glycine-rich GAR domains of hnRNPA1 that enable self-association in vitro (38). The regular heptad repeat of a tyrosyl residue may define a hydrophobic interaction surface on a presumed amphipatic helix, which may engage in corresponding interactions. These interactions may play a regulatory role including conditional RNA-binding. In the absence of these regulatory factors the C-terminus may interact with RNA and retard helicase activity, or interfere with ssRNA interactions of the helicase domain. Interestingly, the GGGYGNN heptad is not conserved even in the MLE of D.pseudoobscura and D.virilis (Supplementary Figure 2), suggesting that D.melanogaster MLE may have acquired a novel function. The G-rich domain of vertebrtate RHA is reminiscent of the well-known RGG/GAR domain found in many RNA-binding proteins (26,38,39). RGG/GAR domains can adopt a spiral structure through repeated β-turns and can bind RNA non-specifically (40).
Implications for MLE function
The above considerations rest on the assumption that MLE is a bona fide helicase whose purpose is to unwind double-stranded RNA. Such a scenario is consistent with its role in para splicing (20). Conceivably, MLE modifies the secondary structures of the complex roX RNA to facilitate the MSL protein interactions that characterize a productive DCC. Currently it is not known whether DCC corresponds to a single homogenous assembly or whether variant configurations exist. Alternate RNA structures, brought about by MLE action, may affect the arrangement and activity of the protein subunits of DCC. Interestingly, certain RNA translocases are able to displace bound protein by tracking along ssRNA, a property that is ideally suited to (re-) organize the conformations of ribonucleoprotein particles (41).
The context of DCC defines special circumstances of MLE action. First, MLE must not necessarily be a very processive enzyme in order to elicit its function. Further, its action is likely to be sequence/structure-specific. Identifying its preferred RNA substrate in the long roX RNAs poses a considerable challenge. Finally, although the association of MLE with the remaining DCC is relatively labile, its co-localization with the other MSL proteins on chromosomal target sites (1,42) suggests that it is a component of DCC. How the context of the MSL proteins affect MLE functions remains to be explored.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank F. Müller-Planitz for suggestions that improved the manuscript, C. Schwarzlose for expert tissue culture assistance, M. Kuroda for antibodies against MSL1 and the MLE cDNA. This work was supported by Deutsche Forschungsgemeinschaft through Transregio 5 and Fonds der Chemischen Industrie. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 2.Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 2006;31:526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Straub T, Becker PB. Dosage compensation: the beginning and end of generalisation. Nat. Rev. Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- 4.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 2005;39:615–51. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 5.Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 9.Morales V, Regnard C, Izzo A, Vetter I, Becker PB. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol. Cell Biol. 2005;25:5947–5954. doi: 10.1128/MCB.25.14.5947-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buscaino A, Kocher T, Kind JH, Holz H, Taipale M, Wagner K, Wilm M, Akhtar A. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol. Cell. 2003;11:1265–1277. doi: 10.1016/s1097-2765(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 11.Lee CG, Chang KA, Kuroda MI, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J. 1997;16:2671–2681. doi: 10.1093/emboj/16.10.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee CG, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J. Biol. Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 13.Meller VH. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mech. Dev. 2003;120:759–767. doi: 10.1016/s0925-4773(03)00157-6. [DOI] [PubMed] [Google Scholar]
- 14.Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI. Ordered assembly of roX RNAs into MSL complexes on the dosage-compensated X chromosome in Drosophila. Curr. Biol. 2000;10:136–143. doi: 10.1016/s0960-9822(00)00311-0. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Wei X, Pannuti A, Lucchesi JC. Targeting the chromatin remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J. 2000;19:5202–5211. doi: 10.1093/emboj/19.19.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morra R, Smith ER, Yokoyama R, Lucchesi JC. Mol. Cell Biol. 2007. The MLE subunit of the Drosophila MSL complex uses its ATPase activity for dosage compensation and its helicase for targeting. Nov 26; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X, Rattner BP, Souter S, Meller VH. The severity of roX1 mutations is predicted by MSL localization on the X chromosome. Mech. Dev. 2005;122:1094–1105. doi: 10.1016/j.mod.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reenan RA, Hanrahan CJ, Barry G. The mle(napts) RNA helicase mutation in drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 21.Sanjuan R, Marin I. Tracing the origin of the compensasome: evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol. Biol. Evol. 2001;18:330–343. doi: 10.1093/oxfordjournals.molbev.a003809. [DOI] [PubMed] [Google Scholar]
- 22.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 23.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Corona DFV, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 25.Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB. Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J. 2004;23:2258–2268. doi: 10.1038/sj.emboj.7600235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson TJ, Thompson JD. Detection of dsRNA-binding domains in RNA helicase A and Drosophila maleless: implications for monomeric RNA helicases. Nucleic Acids Res. 1994;22:2552–2556. doi: 10.1093/nar/22.13.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 28.Lee CG, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 29.Levin MK, Gurjar M, Patel SS. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 30.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin MK, Patel SS. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 32.Richter L, Bone JR, Kuroda MI. RNA-dependent association of the Drosophila maleless protein with the male X chromosome. Genes Cells. 1996;1:325–336. doi: 10.1046/j.1365-2443.1996.26027.x. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, McDonald D, Middlesworth T, Hope TJ, Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol. Cell Biol. 1999;19:3540–3550. doi: 10.1128/mcb.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aratani S, Oishi T, Fujita H, Nakazawa M, Fujii R, Imamoto N, Yoneda Y, Fukamizu A, Nakajima T. The nuclear import of RNA helicase A is mediated by importin-alpha3. Biochem. Biophys. Res. Commun. 2006;340:125–133. doi: 10.1016/j.bbrc.2005.11.161. [DOI] [PubMed] [Google Scholar]
- 35.Hung ML, Chao P, Chang KY. dsRBM1 and a proline-rich domain of RNA helicase A can form a composite binder to recognize a specific dsDNA. Nucleic Acids Res. 2003;31:5741–5753. doi: 10.1093/nar/gkg759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II (RNA helicase A) J. Biol. Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 37.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 39.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghisolfi L, Joseph G, Amalric F, Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 1992;267:2955–2959. [PubMed] [Google Scholar]
- 41.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rastelli L, Richman R, Kuroda MI. The dosage compensation regulators MLE, MSL-1 and MSL-2 are interdependent since early embryogenesis in Drosophila. Mech. Dev. 1995;53:223–233. doi: 10.1016/0925-4773(95)00438-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.