Abstract
PKR is an interferon (IFN)-induced protein kinase, which is involved in regulation of antiviral innate immunity, stress signaling, cell proliferation and programmed cell death. Although a low amount of PKR is expressed ubiquitously in all cell types in the absence of IFNs, PKR expression is induced at transcriptional level by IFN. PKR's enzymatic activity is activated by its binding to one of its activators. Double-stranded (ds) RNA, protein activator PACT and heparin are the three known activators of PKR. Activation of PKR in cells leads to a general block in protein synthesis due to phosphorylation of eIF2α on serine 51 by PKR. PKR activation is regulated very tightly in mammalian cells and a prolonged activation of PKR leads to apoptosis. Thus, positive and negative regulation of PKR activation is important for cell viability and function. The studies presented here describe human dihydrouridine synthase-2 (hDUS2) as a novel regulator of PKR. We originally identified hDUS2 as a protein interacting with PACT in a yeast two-hybrid screen. Further characterization revealed that hDUS2 also interacts with PKR through its dsRNA binding/dimerization domain and inhibits its kinase activity. Our results suggest that hDUS2 may act as a novel inhibitor of PKR in cells.
INTRODUCTION
Interferons (IFNs) are cytokines with antiviral, antiproliferative and immunomodulatory properties, which they exert by inducing synthesis of several proteins (1,2). The IFN-induced, dsRNA-activated protein kinase PKR, a serine/threonine kinase, is a major mediator of the antiproliferative and antiviral actions of IFN (3–6). Although induced at the transcriptional level by IFNs, PKR is present at a low, basal level in most cell types (7). PKR's kinase activity stays latent until it binds to one of its three known activators, double-stranded (ds) RNA, heparin and the protein activator PACT. In virally infected cells, dsRNA produced during viral replication or viral RNAs with extensive ds regions serve as PKR activators (8,9). Polyanionic agents such as heparin also activate PKR both in vitro (10) and in vivo (11). In addition, PACT is a cellular, protein activator of PKR, which heterodimerizes with PKR and activates it in the absence of dsRNA (12,13), thereby playing an important role in PKR activation in response to stress signals (14). The α-subunit of the eukaryotic protein synthesis initiation factor eIF-2 (eIF2α) is the most studied physiological substrate of PKR. Phosphorylation of eIF2α on Ser51 by PKR leads to an inhibition of protein synthesis (15,16). In addition to its central role in antiviral activity of IFNs, PKR is also involved in the regulation of apoptosis (17,18), cell proliferation (4,5), signal transduction (19–21) and differentiation (22,23).
The dsRNA-mediated activation of PKR has been characterized in detail (24–29). The dsRNA-binding domain (dsRBD) of PKR is composed of two copies of the dsRNA-binding motif (dsRBM), a sequence motif conserved in many RNA-binding proteins (30,31). Binding of dsRNA to PKR through these motifs causes a conformational change (32,33) that leads to an unmasking of the ATP-binding site in the kinase domain and results in autophosphorylation of PKR on several sites (34–36). The domains that are involved in dsRNA binding are also involved in mediating dimerization of PKR, which is essential for its kinase activity in the presence of dsRNA (37–40). Although the same domain mediates PKR's dsRNA binding and dimerization, distinct residues have been identified that contribute to one or both of these properties (40). Heparin binds to PKR at a site that is nonoverlapping with PKR's dsRBD and leads to PKR activation (41,42). Activation of PKR by heparin in vascular smooth muscle cells leads to an arrest in cell cycle progression by causing elevation in p27kip1 protein levels, inhibition of Cdk2 activity and Rb phosphorylation (43). PACT interacts with PKR by binding to its dsRBD in a dsRNA-independent manner and activates PKR in response to cellular stress (14). Stress signals lead to phosphorylation of PACT at serine 287, which causes its higher affinity association with PKR leading to PKR activation, eIF2α phosphorylation and consequent inhibition of translation (44). PACT has three copies of dsRNA binding/dimerization motifs and the two amino-terminal copies are required for high-affinity binding to PKR (45). The third, carboxy-terminal motif is required for PKR activation presumably by making a direct contact with PKR's catalytic domain (45,46).
Several cellular and viral inhibitors of PKR have been identified (47). P58IPK, the trans-activation response (TAR) RNA-binding protein (TRBP), nucleophosmin (NPM) and several virally encoded proteins inhibit PKR activity. Since PKR activation leading to eIF2α phosphorylation and inhibition of translation would be detrimental to viral replication, various viruses have developed efficient mechanisms to inhibit PKR activation (9,48,49). In this article, we report the identification of hDUS2 as a novel cellular PKR inhibitor. hDUS2 was identified as a PKR- and PACT-interacting protein using PACT as a bait protein in yeast two-hybrid screen. hDUS2 was reported recently to be the human homolog of Saccharomyces cerevisiae dihydrouridine synthase 2 (Dus2) enzyme (50). It was also shown to possess tRNA-dihydrouridine synthase (DUS) activity. hDUS2 has one copy of the conserved dsRBM and our results indicate that hDUS2 interacts with PKR and PACT via its dsRBM. Binding of hDUS2 with PKR resulted in an inhibition of PKR activity both in vitro as well as in mammalian cells. Furthermore, hDUS2 overexpression inhibited stress-induced apoptosis in HT1080 cells indicating that it acts as an important negative regulator of PKR activity in cells.
MATERIALS AND METHODS
Yeast two-hybrid screening for PACT-interacting proteins
The PACT coding region expressed as a GAL DNA-binding domain fusion protein was used as bait. A total of 3 million transformants from a human placenta matchmaker library (Clontech) were screened in the yeast strain HF7c (Clontech) and 250 colonies were recovered on triple dropout medium lacking histidine, leucine and tryptophan, out of which 22 tested positive for β-galactosidase. On further analysis, 18 of these were dependent on PACT to give a positive β-galactosidase reaction. These clones were subjected to a second screen to ensure that they gave a positive β-galactosidase reaction in a manner specific for co-expression of PACT. One of these clones (hDUS2), which did not contain a full-length cDNA insert, was analyzed further. Sequence analysis of the cDNA clones revealed that they originated from three unrelated genes. There were 14 clones isolated that coded for the PACT protein and the other three encoded a protein that has been characterized as a zinc finger protein just another zinc-finger (JAZ) that interacts with PKR.
β-Galactosidase reporter assay in yeast
HF7c yeast reporter strain containing the LacZ-Gal4-inducible gene were co-transformed with the indicated plasmids and plated on selective medium lacking tryptophan and leucine. Double transformants were streaked on selective medium, replica-lifted on nitrocellulose filters and tested for β-galactosidase activity (51).
Cloning full-length hDUS2 open reading frame
The full-length open reading frame of hDUS2 was amplified by RT–PCR using the sequence information from Genbank and the appropriate primers. The full-length open reading frame was then sub-cloned into pGBKT7 and pGADT7 (Clontech) vectors, and pCDNA3.1(–) (Invitrogen) vector. Cloning in pCDNA3.1 (–) vector was designed such that it would add a flag tag at the amino-terminus of hDUS2.
In vitro protein–protein interaction assay
In vitro translated, 35S-labeled hDUS2 protein was synthesized using the TNT T7 coupled reticulocyte system from Promega. Five microliters of the in vitro translated 35S-labeled proteins were incubated with 1 μg of pure recombinant PACT or PKR and 20 μl of Ni-agarose (Novagen) in 200 μl of binding buffer [5 mM imidazole, 20 mM Tris–HCl pH 7.9, 200 mM NaCl, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 0.5% IGEPAL (Sigma)] at 25°C for 30 min on a rotating wheel. The beads were washed four times in 500 μl of binding buffer each time and the washed beads were then boiled in 20 μl Laemmli buffer (150 mM Tris–HCl pH 6.8, 5% SDS, 5% β-mercaptoethanol, 20% glycerol) for 2 min and eluted proteins were analyzed by SDS–PAGE on a 12% gel. Fluorography was performed at −80°C with intensifying screens.
dsRNA binding assay
The in vitro translated, 35S-labeled PKR, PACT and hDUS2 proteins were synthesized using the TNT T7 coupled reticulocyte lysate system from Promega. The dsRNA-binding activity was measured by poly(I)–poly(C)–agarose binding assay. The translation products (4 μl) diluted with 25 μl of binding buffer [20 mM Tris–HCl, pH 7.5, 0.3 M NaCl (or 50 mM NaCl), 5 mM MgCl2, 1 mM DTT, 0.1 mM PMSF, 0.5% NP-40, 10% glycerol) were mixed with 25 μl of poly(I)–poly(C)–agarose (Pharmacia) beads and incubated at 30°C for 30 min with intermittent shaking. The beads were then washed four times with 500 μl of binding buffer each time. The proteins bound to beads after washing were analyzed by SDS–PAGE followed by fluorography.
Expression in mammalian cells and co-immunoprecipitation assay
HeLa cells were co-transfected in 100 mm culture dishes with 5 μg each of (i) negative control: HA-B56α/pCDNA3.1(–) and flag-hDUS2/pCDNA3.1(–), (ii) HA-PKR/pCDNA3.1(–) and flag-hDUS2/pCDNA3.1(–) or (iii) HA-PACT/pCDNA3.1(–) and flag-hDUS2/pCDNA3.1(–) using the Effectene (Qiagen) reagent. At 24 h post-transfection, cell extracts were prepared. Cells were washed in ice-cold PBS and collected by centrifugation at 600 g for 5 min. They were lyzed by addition of an equal volume of lysis buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 100 U/ml aprotinin, 0.2 mM phenylmethanesulfonyl fluoride, 20% glycerol, 1% Triton X-100). The lysates were centrifuged at 10 000 g for 5 min and the supernatants were collected as total cell extract. One hundred micrograms of total protein was used to immunoprecipitate flag-hDUS2 with anti-flag mAb–agarose in immunoprecipitation buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 100 U/ml aprotinin, 0.2 mM phenylmethanesulfonyl fluoride, 20% glycerol, 1% Triton X-100). The agarose beads were washed four times with 500 μl of IP buffer each time. The proteins bound to the beads were then analyzed by a western blot analysis with the anti-HA tag and anti-flag tag monoclonal antibodies (Sigma).
PKR kinase activity assay
PKR activity assays were performed using an anti-PKR monoclonal antibody (R & D systems; 71/10). HeLa M cells were maintained in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum. The cells were harvested when they were at 70% confluency. Cells were washed in ice-cold PBS and collected by centrifugation at 600 g for 5 min. They were lyzed by addition of an equal volume of lysis buffer (20 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 50 mM KCl, 400 mM NaCl, 2 mM DTT, 1% Triton X-100, 100 U/ml aprotinin, 0.2 mM PMSF, 20% glycerol). The lysates were centrifuged at 10 000 g for 5 min and the supernatants were assayed for PKR activity. A 100 μg aliquot of total protein was immunoprecipitated using anti-PKR monoclonal antibody (71/10) in high-salt buffer (20 mMTris–HCl, pH 7.5, 50 mM KCl, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, 100 U/ml aprotinin, 0.2 mM PMSF, 20% glycerol, 1% Triton X-100) at 4°C for 30 min on a rotating wheel. Then 10 μl of Protein A-Sepharose slurry was added and incubation was carried out for a further 1 h. The Protein A-Sepharose beads were washed four times in 500 μl of high-salt buffer and twice in activity buffer (20 mM Tris–HCl, pH 7.5, 50 mM KCl, 2 mM MgCl2, 2 mM MnCl2, 100 U/ml aprotinin, 0.1 mM PMSF, 5% glycerol). The PKR assay was performed with PKR still attached to the beads in activity buffer containing 0.1 mM ATP and 1 μCi of [γ 32P] ATP at 30°C for 10 min. The standard activator of the enzyme was 0.1 μg/ml poly(I)-poly(C) or 0.116 pmol of pure PACT protein. Purified hDUS2 protein in varying amounts as indicated was added to test its effect on PKR activity. Labeled proteins were analyzed by SDS–PAGE on a 12% gel followed by autoradiography.
Translation inhibition assay
The effect of co-transfection of pCDNA3.1(–) empty vector, TRBP/pCDNA3.1(–) and hDUS2/pCDNA3.1(–) on the reporter pGL2-Control (Promega) in HeLa cells was measured as described before (52). HeLa cells were transfected in six-well plates in triplicate with the indicated plasmids using Effectene transfection reagent. At 24 h after transfection, the cells were treated with 100 U/ml of IFN-β. Cells were harvested 48 h after transfection and assayed for luciferase activity after normalizing for the transfection efficiency by measuring the total protein.
Apoptosis assay
HT1080 cells were grown to 50% confluency on coverslips in six-well plates and co-transfected with 500 ng of the indicated effector and 100 ng of pEGFPC1 (Clontech) plasmid using the Effectene reagent. The cells were observed for green fluorescent protein (GFP) fluorescence 24 h after transfection using an inverted fluorescence microscope, and were then treated with 0.1 μg/ml tunicamycin to induce ER stress and apoptosis. At 24 and 48 h after this treatment, the cells (coverslips) were washed twice with PBS and fixed in acetone:methanol (1:1, v/v) for 1 min, and were mounted in Vectashield (Vector Laboratories) mounting medium containing DAPI (4,6-diamidino-2-phenylindole). At least 300 fluorescent (GFP-positive) cells were counted as alive or dead, based on their morphology and nuclear DAPI staining. Intense DAPI staining indicates nuclear condensation, which is a hallmark of apoptosis. The experiment was repeated three times and the counting was done in a blinded manner to ascertain the validity of obtained numbers. The cells showing a normal flat morphology were scored as alive, and cells showing cell shrinkage, membrane blebbing, rounded morphology, partial detachment from the plate and chromatin condensation (intense DAPI stain) were counted as dead. Apoptosis (%) = (fluorescent cells with intense DAPI staining in the nucleus/total fluorescent cells) × 100.
RESULTS
hDus2 interacts with both PACT and PKR
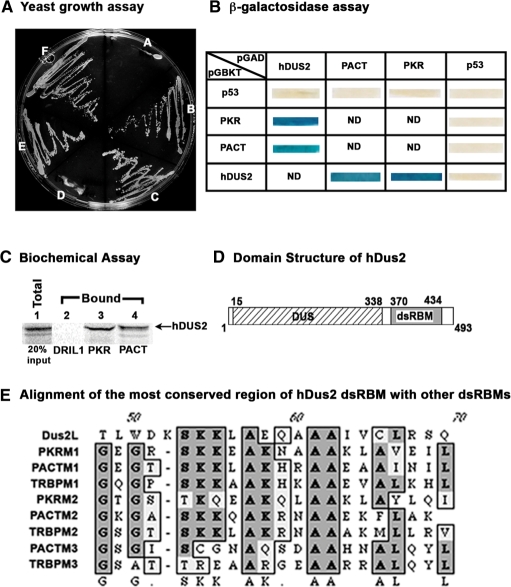
Various stress signals induce cellular apoptosis via activation of PKR (18). PACT plays a central role in this pathway by functioning as an activator of PKR (13,14). In order to identify proteins that may regulate this stress-activated apoptotic pathway, we performed a yeast two-hybrid screen using PACT as a bait. Among the cDNA clones identified as the ones that encoded PACT-interacting proteins, there were 14 isolates of PACT, 3 isolates of JAZ and 1 isolate of hDUS2 (human dihydrouridine synthase-2). PACT is expected to interact with itself to form dimers/multimers since it has three copies of dsRBM, a conserved protein motif involved in dsRNA binding as well as dimerization (45,46). JAZ has been previously shown to interact with dsRNA (53). hDUS2 encodes for a human tRNA modification enzyme, dihydrouridine synthase (50). Four dihydrouridine synthases have been identified in human cells and these show homology with the yeast enzymes that have been characterized before. hDUS2 has never before been described to interact with PKR or PACT and thus we characterized this interaction further. The original hDUS2 cDNA insert isolated from the yeast two-hybrid library was a partial cDNA clone and lacked the amino-terminal sequences that encoded the amino-terminal 320 amino acids. We obtained the full-length cDNA clone for hDUS2 based on the Genbank sequence information (FLJ20399, accession no. NM_017803) by performing RT–PCR on total RNA isolated from HeLa cells. The full-length open reading frame for hDUS2 was sub-cloned into the yeast two hybrid vectors pGBKT7 and pGADT7 to obtain GAL4 DNA-binding domain fusion and GAL4 activation domain fusion, respectively. In order to confirm that the full-length hDUS2 protein can interact with PACT and to test if hDUS2 interacts with PKR we performed the yeast growth assay on triple drop-out medium lacking three amino acids: histidine, leucine and tryptophan. Growth on the triple drop-out medium indicates positive interaction between the two proteins that are being tested. As seen in Figure 1A, hDUS2 showed positive interaction in both combinations with PKR (sectors B and E), as well as PACT (sectors C and F). The negative controls with empty vectors did not show any growth on the plate (sectors A and D). These results confirmed that full-length hDUS2 interacts with both PACT and PKR. In order to further confirm the positive interaction between hDUS2, PKR and PACT we also tested activation of another reporter, β-galactosidase. Expression of β-galactosidase in yeast cells is indicated by blue color and occurs only if the proteins encoded by pGADT7 and pGBKT7 constructs interact. As seen in Figure 1B, PKR/pGBKT7 and PACT/pGBKT7 both showed a positive interaction with hDUS2/pGADT7. The opposite combinations also showed blue color and thus confirmed the interactions. The negative controls with p53/pGBKT7 and p53/pGADT7 all showed white color indicating no interaction with hDUS2, PKR and PACT. These results confirm the specificity of the interaction between hDUS2 and PKR or PACT. The interaction of hDUS2 with PACT and PKR was further confirmed by performing a biochemical interaction assay. In vitro translated, 35S-methionine labeled hDUS2 was allowed to interact with pure, recombinant hexahistidine tagged PKR or PACT proteins and was then pulled-down using Ni-agarose beads. As seen in Figure 1C, hDUS2 showed no interaction with DRIL1 protein bound to Ni-agarose beads (lane 2). However, it showed a strong interaction with both PKR (lane 3) and PACT (lane 4) bound to Ni-agarose beads. These results further confirm the in vivo interaction detected between these proteins in yeast two-hybrid assay.
Figure 1.
Identification of hDUS2 as PKR and PACT interacting protein. (A) Yeast two-hybrid interaction assay. The indicated plasmids were transformed into yeast strain Hf7C and the transformants were streaked on triple dropout medium lacking leucine, tryptophan and histidine. A: hDUS2/pGBKT7 and empty vector pGADT7, B: hDUS2/pGBKT7 and PKR/pGADT7, C: hDUS2/pGBKT7 and PACT/pGADT7, D: empty vector pGBKT7 and hDUS2/pGADT7, E: PKR/pGBKT7 and hDUS2/pGADT7, F: PACT/pGBKT7 and hDUS2/pGADT7. (B) β-Galactosidase assay for the interactions in yeast. The indicated plasmids were transformed into yeast strain Hf7C and the transformants were streaked on double dropout medium lacking leucine, and tryptophan. After 4 days the growth was lifted on nitrocellulose membrane and β-galactosidase activity assay was performed after lysis of yeast cells on the membrane. Blue color indicates a positive interaction and white color indicates no interaction. (C) Biochemical interaction assay. In vitro-translated hDUS2 protein was allowed to interact with hexahistidine-tagged pure recombinant PKR or PACT proteins that were bound to Ni-agarose affinity beads. The bound proteins remaining on the beads were analyzed by SDS–PAGE followed by phosphorimager analysis. Lane 1: total protein from the translation mix (20% of input in lanes 2–4), lanes 2–4: hDUS2 protein bound to beads. Lane 2: protein bound to his-DRIL1 beads: negative control, lane 3: protein bound to his-PKR beads, lane 4: protein bound to his-PACT beads. (D) Domain structure of hDUS2. A schematic representation of the DUS and dsRBM domains in hDUS2 protein. The DUS domain is shown as a hatched box and the dsRBM is shown as a gray box. The amino acid numbers are indicated on the top and bottom of the boxes. (E) Alignment of hDUS2 dsRBM with dsRBMs from three other human dsRNA-binding proteins PKR, PACT and TRBP. ClustalW alignment of the dsRBMs present in PKR, PACT and TRBP is shown. The conserved residues are shown in dark gray and similarities are shown in light gray. The consensus is shown at the bottom.
hDus2 contains one copy of evolutionarily conserved dsRBM and interacts with dsRNA
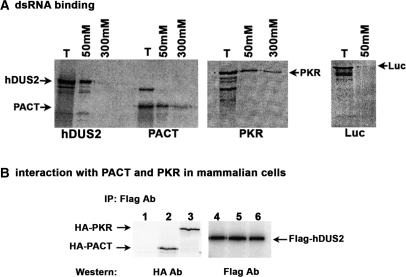
Homology searches with Genbank sequences revealed that hDUS2 contains two domains, the DUS domain that functions in the enzymatic activity in modifying the uridines in tRNAs post-transcriptionally (50). The DUS domain in hDUS2 is located between residues 15 and 338 (Figure 1D). In addition to this, one copy of the previously characterized evolutionarily conserved dsRBM (30) is present in hDUS2 between residues 370 and 434. An alignment of the dsRBM of hDUS2 with the dsRBMs present in PKR, PACT and TRBP is shown in Figure 1E. The consensus residue lysine at position 58 of the dsRBM is absent in hDUS2 motif. This lysine has previously been shown to be important for conferring a high-affinity binding to dsRNA (52,54). It is noteworthy that hDUS2 has a glutamic acid at this position, thus raising a possibility that the dsRBM present in hDUS2 may not mediate high-affinity binding to dsRNA. We examined this by comparing the binding of hDUS2, PACT and PKR to dsRNA covalently linked to agarose beads. As seen in Figure 2A, hDUS2 showed binding to dsRNA at 50 mM salt concentration but not at 300 mM salt. PACT and PKR both showed very strong binding to dsRNA at 50 and 300 mM salt concentrations. The binding of hDUS2 to dsRNA at 50 mM salt concentration is quite significant since firefly luciferase, a protein that is known not to interact with dsRNA does not show binding to dsRNA at this salt concentration. These results indicate that although hDUS2 interacts with dsRNA, its affinity for dsRNA is much lower than PACT and PKR. This could be due to the lack of the conserved lysine at position 58 of hDUS2 dsRBM, since this lysine is required for the high-affinity interaction with dsRNA (52,54).
Figure 2.
hDUS2 interaction with dsRNA, PACT, and PKR. (A) dsRNA-binding characteristics of hDUS2. Binding of hDUS2 to dsRNA was analyzed by polyI:polyC-agarose-binding assay. The binding was assayed at 50 and 300-mM salt concentration as indicated. The T lanes represent total proteins from reticulocyte lysates. The positions of hDUS2, PACT, PKR and Luciferase (Luc) are indicated by arrows. (B) hDUS2 interaction with PACT and PKR in mammalian cells. HeLa cells were transfected with (i) HA-B56α/pCDNA3.1(–) + Flag-hDUS2/pCDNA3. (-1), (ii) HA-PACT/pCDNA3.1(–) + Flag-hDUS2/pCDNA3.1(–) and (iii) HA-PKR/pCDNA3.1(–) + Flag-hDUS2/pCDNA3.1(–). Twenty-four hours after transfection the Flag-tagged hDUS2 protein was immunoprecipitated using anti-Flag monoclonal antibody conjugated to agarose. The immunoprecipitates were analyzed by western blot analysis with anti-HA monoclonal antibody. The positions of HA-PKR, HA-PACT and Flag-hDUS2 are as indicated by arrows.
hDus2 interacts with PACT and PKR in mammalian cells
In order to test if hDUS2 interacts with PACT and PKR in mammalian cells, we tested the ability of flag-tagged hDUS2 to interact with HA-tagged PKR and PACT in HeLa cells. As shown in Figure 2B, when flag-hDUS2 was immunoprecipitated with anti-flag antibody, HA-tagged PKR and PACT both co-immunoprecipitated with it indicating that hDUS2 interacts with both PKR and PACT. The negative control HA-B56α, which is an unrelated protein, did not co-immunoprecipitate with flag-hDUS2, thus demonstrating the specificity of interaction. These results demonstrate that hDUS2 forms a complex with PKR and PACT in mammalian cells.
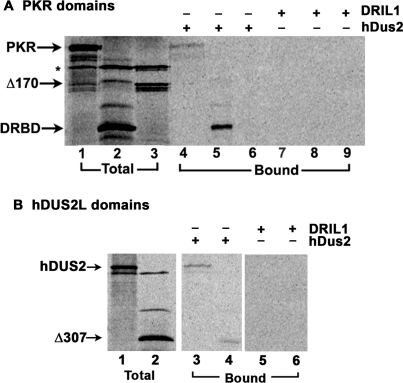
Mapping the interaction domains in PKR and hDus2
To map the domains within PKR that mediate its interaction with hDUS2, we tested the interaction of hDUS2 with two different deletion mutants of PKR using the in vitro interaction assay. In vitro translated 35S-methionine labeled PKR proteins were mixed with pure, hexahistidine-tagged hDUS2 recombinant protein and Ni-agarose beads and the proteins that bound to hDUS2 were analyzed by SDS–PAGE. The DRBD mutant lacks the carboxy-terminal residues 171–551 and the Δ170 mutant lacks the amino-terminal residues 1–170 (12). As seen in Figure 3A, wild-type PKR showed interaction with hDUS2 (lane 4) and DRBD showed a very strong interaction with hDUS2 (lane 5). However, Δ170 did not show any interaction with hDUS2 (lane 6). These results establish that PKR interacts with hDUS2 via its DRBD. This region contains both the dsRBMs of PKR, which are known to mediate protein–protein interactions. When an unrelated protein DRIL1 was immobilized on the Ni-agarose beads, none of the in vitro translated protein showed any interaction with it, thereby demonstrating specificity of the interactions between hDUS2 and PKR. In order to map the domain within hDUS2 that mediates the interaction with PKR, similar experiments were done with a deletion mutant of hDUS2. Δ307 lacks the amino-terminal residues 1–307. In vitro-translated 35S-methionine-labeled hDUS2 proteins were mixed with pure, hexahistidine-tagged PKR recombinant protein and Ni-agarose beads and the proteins that bound to PKR were analyzed by SDS–PAGE. As seen in Figure 3B, full-length hDUS2 and Δ307 both showed interaction with PKR (lanes 3 and 4). The Δ307 mutant of hDUS2 lacks the amino-terminal DUS domain but retains the single dsRBM present in hDUS2, thus indicating that its interaction with PACT and PKR is mediated via its dsRBM. The negative control protein DRIL1 did not show any binding to hDUS2, thus demonstrating that the interaction between PKR and hDUS2 is specific.
Figure 3.
Mapping the hDUS2 interaction domain within PKR. (A) PKR and its deletion mutants DRBD and Δ170 were in vitro translated. Hexahistidine-tagged, pure recombinant hDUS2 protein was incubated with 5 μl of the reticulocyte lysates before pull-down with Ni-charged affinity resin. As a negative control, his-tagged DRIL1 protein that does not interact with PKR was immobilized on Ni-agarose beads in lanes 7–9. The proteins that associate with recombinant hDUS2 are also pulled down with the beads and were analyzed by SDS–PAGE followed by phosphorimager analysis. Lanes 1–3 represent total proteins from the translation mix (20% of input in lanes 4–9), lanes 4–6 represent proteins pulled down with hDUS2 bound to Ni-charged beads, and lanes 7–9 represent negative controls with proteins pulled down with DRIL1 bound to Ni-charged beads. (B) Mapping the PKR interaction domain within hDUS2. hDUS2 and its deletion mutant Δ307 were in vitro translated. Hexahistidine-tagged, pure recombinant PKR protein was incubated with 5 μl of the reticulocyte lysates before pull-down with Ni-charged affinity resin. As a negative control, his-tagged DRIL1 protein that does not interact with hDUS2 was used in lanes 5 and 6. Lanes 1 and 2 represent total proteins from the translation mix (20% of input in lanes 4–9), lanes 3 and 4 represent proteins pulled down with hexahistidine-tagged pure recombinant PKR bound to Ni-charged beads, lanes 5 and 6 represent negative controls with proteins pulled down with his-tagged pure DRIL1 bound to Ni-charged beads.
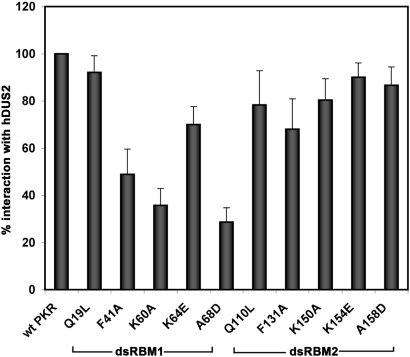
To further test the contribution of the two dsRBMs within DRBD of PKR in mediating interaction with hDUS2, we tested the ability of several PKR point mutants within the two motifs (52) to interact with pure recombinant hDUS2 using the in vitro interaction assay. The results are shown in Figure 4. As seen in the bar graph, F41A, K60A and A68D mutants within dsRBM1 showed severely reduced interaction with hDUS2. A mutation of the same residues in dsRBM2 was less detrimental to the interaction with hDUS2. These results strongly indicate that the interaction of PKR with hDUS2 is primarily mediated by its dsRBM1 of PKR and that the residues F41, K60, and A68 serve an important function.
Figure 4.
PKR's interaction with hDUS2 is mediated by dsRBM1 motif within its DRBD. PKR and its point mutants were analyzed for interaction with hexahistidine tagged pure, recombinant hDUS2 by Ni-agarose pull-down assay. PKR and its point mutants were in vitro translated. Hexahistidine-tagged, pure recombinant hDUS2 protein was incubated with 5 μl of the reticulocyte lysates before pull-down with Ni-charged affinity resin. As a negative control, his-tagged DRIL1 protein that does not interact with PKR was immobilized on Ni-agarose beads. The proteins that associate with recombinant hDUS2 are also pulled down with the beads and were analyzed by SDS–PAGE followed by phosphorimager analysis. The percent interaction was calculated using quantification of radioactivity present in the PKR band in the total and bound samples as follows: % interaction = 100 × (amount of radioactivity in the bound sample/amount of radioactivity in the total sample). The percent interactions of the point mutants are expressed relative to that of the wild-type PKR with wild-type PKR value taken as 100%. The amount of radioactivity in the negative control (this was never above background in any of the samples) was subtracted from the amount of radioactivity in the bound sample before calculations. Error bars represent SE calculated based on values from three different experiments.
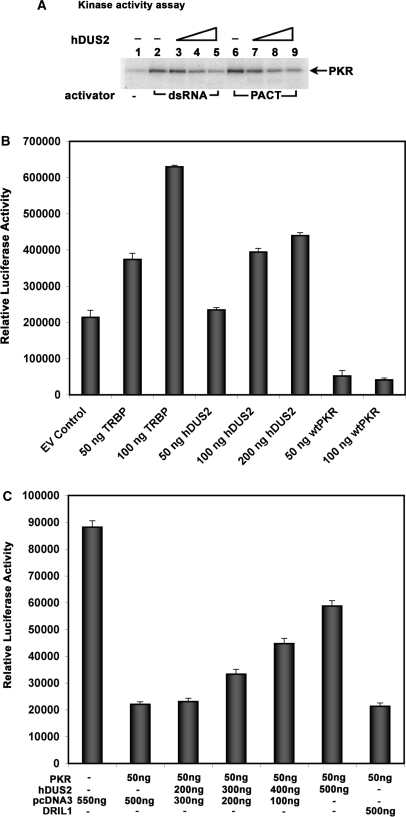
hDus2 inhibits PKR's kinase activity both in vitro and in vivo
In order to understand the functional consequence of hDUS2's interaction with PKR, we tested the effect of hDUS2 on PKR's kinase activity using the in vitro kinase activity assay. As seen in Figure 5A, recombinant hDUS2 inhibited the PKR kinase activity in a dose-dependent manner. hDUS2 was capable of inhibiting PKR activity when it was activated by dsRNA as well as PACT. These results demonstrate that hDUS2 acts as a PKR inhibitor. To test this further in vivo in mammalian cells, we performed the translation inhibition assay to check for PKR activation. The effect of a protein encoded by an expression construct on activation of PKR in mammalian cells can be measured by assaying the expression of a co-transfected reporter gene such as luciferase (12). Using the translation inhibition assay (46,55–58), we next determined the ability of the hDUS2 expression constructs to inhibit PKR in HeLa cells. As seen in Figure 5B, co-transfection with wild-type PKR reduced luciferase activity dramatically, whereas co-transfection with TRBP expression construct increased the reporter activity, due to inhibition of endogenous PKR activation. Co-transfection with hDUS2 expression construct also increased luciferase activity in a dose-dependent manner, indicating that hDUS2 inhibits the activation of PKR. These results further indicate that hDUS2 is not a very strong inhibitor of PKR since it did not increase the reporter activity to the same extent as the TRBP expression construct. At 100 ng DNA concentration, hDUS2 construct showed only about half as much increase in activity of the reporter than the TRBP construct. Thus, although hDUS2 does inhibit PKR activity, it does not act as efficiently as TRBP. These results indicate that to achieve an effective inhibition of PKR activity, hDUS2 may need to be expressed at high levels relative to PKR levels in mammalian cells. In order to test if hDUS2 overexpression can effectively inhibit activated PKR, we tested if co-transfected hDUS2 expression construct can relieve the translational repression of luciferase expression caused by active PKR. As seen in Figure 5C, co-transfection of the reporter plasmid with wild-type PKR expression construct reduced luciferase activity dramatically, whereas co-transfection with hDUS2 expression construct along with wt PKR increased the reporter activity in a dose-dependent manner. Co-transfection of PKR expression construct with an expression plasmid encoding DRIL1 protein, which has no effect on PKR activation showed no effect on the reporter gene activity thus demonstrating that the inhibitory effect of hDUS2 on PKR activation is specific. These results strongly indicate that hDUS2 is able to inhibit the PKR activity in mammalian cells.
Figure 5.
hDUS2 inhibits PKR activity in vitro and in vivo. (A) PKR kinase activity assay. PKR immunoprecipitated from HeLa cell extracts was activated by the addition of 0.1 μg/ml of poly(I): poly(C) (lanes 2–5) or 0.116 pmol of recombinant PACT (lanes 6–9). The effects of addition of increasing amounts of recombinant hDUS2 protein were assayed. Lane 1: no activator added, lanes 2 and 6: no hDUS2 added, lanes 3 and 7: 1.45 pmol (80 ng) hDUS2, lanes 3 and 8: 7.25 pmol (400 ng) hDUS2, lanes 4 and 9: 36.25 pmol (2 μg) hDUS2. The position of phosphorylated PKR is indicated by an arrow. (B) Translation inhibition assay. The translation inhibition assay was performed in HeLa cells grown in six-well plates. The reporter pGL2C (200 ng) was co-transfected using Effectene reagent with indicated amounts of the empty vector control or expression constructs for TRBP, hDUS2 or PKR. The amount of DNA in each well was kept constant at 600 ng by addition of required amounts of empty vector pCDNA3.1(–). At 24 h after transfection, the cells were treated with 100 U/ml interferon-β and 24 h after the treatment the luciferase activity was measured in the cell extracts and normalized to the total protein in the extract. All expression constructs were in pCDNA3.1(–). (C) hDUS2 overexpression relieves the translation inhibition caused by activated PKR. The translation inhibition assay was performed in HeLa cells grown in six-well plates. The reporter pGL2C (50 ng) was co-transfected using Effectene reagent with indicated amounts of the empty vector pCDNA3.1(–), expression constructs for PKR, hDUS2 and DRIL1. The amount of DNA in each well was kept constant by addition of required amounts of empty vector pCDNA3.1(–). At 24 h after transfection, the cells were treated with 100 U/ml interferon-β. Twenty-four hours after the treatment the luciferase activity was measured in the cell extracts and normalized to the total protein in the extract. All expression constructs were in pCDNA3.1(–).
Functional consequence of hDus2-mediated inhibition of PKR activity
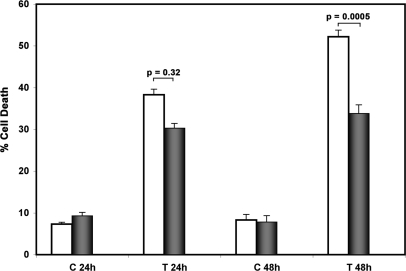
The inhibitory effect of hDUS2 being weaker than TRBP (Figure 5B), we tested the biological significance of it. Since PKR is known to be involved in stress signal-induced apoptosis, we tested if overexpression of hDUS2 can inhibit apoptosis induced by tunicamycin, an antibiotic that causes endoplasmic reticulum (ER) stress due to accumulation of unfolded proteins in the ER (59). PKR is involved in inducing apoptosis in response to ER stress (60,61). As seen in Figure 6, treatment of HT1080 cells with tunicamycin induced cell death in about 38% of cells transfected with the empty vector at 24 h, which further increased to about 52% at 48 h. In cells transfected with hDUS2 expression construct, tunicamycin induced cell death in about 30% cells at 24 h, which showed only an extremely marginal increase to about 34% at 48 h. The transfected samples that were not treated with tunicamycin did not show significant cell death. Thus, at 48 h after treatment with tunicamycin, there is a significant inhibition of cell death by overexpression of hDUS2 indicating that hDUS2 is able to inhibit PKR activation and the consequent apoptosis. These results establish that inhibition of PKR activity by hDUS2 may play an important role in regulating stress-induced apoptosis.
Figure 6.
hDUS2 overexpression inhibits stress-induced apoptosis. HT1080 cells were grown to 50% confluency on coverslips in six-well plates and transfected with 500 ng of pCDNA3.1(–) + 100 ng of pEGFPC1 (white bars) or 500 ng of hDUS2/pCDNA3.1(–) + 100 ng of pEGFPC1 (black bars) using the Effectene reagent. The cells were observed for GFP fluorescence 24 h after transfection using an inverted fluorescence microscope. At this point, they were treated with 0.1 μg/ml of tunicamycin. The morphology of cells was monitored at regular time intervals. At 24 and 48 h after treatment, the cells were fixed and mounted in Vectashield with DAPI nuclear stain. At least 300 fluorescent (GFP-positive) cells were counted as live or dead based on their morphology and nuclear condensation indicated by intense DAPI fluorescence. The experiment was repeated three times and the data is represented as averages of these values. The assays were counted in blinded manner to ascertain the validity of observed differences. The percentage of apoptotic cells were calculated using the formula, % apoptosis = (fluorescent dead cells with intense DAPI fluorescence/total fluorescent cells) × 100. The P-values, calculated using statistical analysis, are as indicated above the bars.
DISCUSSION
PKR activity is regulated very tightly in cells and several cellular proteins have been described as inhibitors of PKR (47). One of the best-studied proteins is p58IPK, which is a member of the tetratricopeptide repeat family (62–64). P58IPK inhibits PKR activity by interacting directly with it and by preventing its dimerization (65). Influenza virus evades the host antiviral response partially by recruiting p58IPK to repress PKR-induced eIF2α phosphorylation (66). TRBP is a cellular protein that was identified by its ability to bind human immunodeficiency virus type 1 (HIV-1) TAR RNA (67). TRBP inhibits PKR activity by sequestration of its activator dsRNA, and also by direct interaction with PKR (38,58). Recent data suggests that in astrocytes, low TRBP levels support and innate HIV-1 resistance via PKR activation (68). Thus, TRBP may play a major role in viral expression by regulating PKR activation. NPM is an abundant nucleolar phosphoprotein implicated in ribosome biogenesis (69). NPM interacts with PKR and inhibits eIF2α phosphorylation and PKR-mediated apoptosis (70). NPM has also been implicated in mediating antiviral activity of the tumor suppressor ARF thus linking the Arf/mdm2/p53 pathway to PKR (71). The heat shock proteins Hsp70 and Hsp90 have also been shown to bind to PKR and inhibit its activation (48,72). In addition to these cellular inhibitors of PKR, numerous viral proteins and RNAs have been established to act as PKR inhibitors during viral infection thereby evading the host cell's innate antiviral response (8,49).
In this article, we describe hDUS2 as a novel inhibitor of PKR activity. hDUS2 was reported recently to have 39% homology to Dus2 enzyme of Saccharomyces cerevisiae and to possess tRNA–DUS activity (50). It was also shown to exist in a complex with EPRS, a glutamy-prolyl tRNA synthetase and to enhance general translational efficiency in mammalian cells. hDUS2 was identified as a PACT-interacting protein in our yeast two-hybrid screen using PACT as a bait. Further characterization of the interaction between PACT and hDUS2 (data not shown) indicated that this interaction was mediated by the single copy of dsRBM present in hDUS2. Our work on PKR and PACT has demonstrated that PKR and PACT interact with each other via the dsRBM motifs (45). The amino-terminal two copies of dsRBM present in PACT are essential for mediating the high-affinity interaction with PKR via the two dsRBMs present in PKR. We reasoned that hDUS2 may also show interaction with PKR via the two dsRBMs. Originally identified as a PACT-interacting protein, hDUS2 did indeed interact with PKR equally well, both in vivo (in mammalian and yeast cells) and in vitro. Although there are two copies of dsRBM present in PKR, our results indicate that hDUS2 interacts with PKR mainly via the amino-terminal copy of the motif. Thus, the two dsRBMs do not contribute equally to PKR's interaction with hDUS2 and the first or the amino-terminal motif is responsible for mediating the interaction. Within this motif, mutations at position 41 (phenylalanine), 60 (lysine) and 68 (alanine) affected the interaction between PKR and hDUS2 adversely, thereby identifying these residues as essential for the interaction. Mutations of the same residues in motif 2 did not show much effect on the PKR–hDUS2 interaction. In addition to this, the interaction between hDUS2 and PKR is by direct protein–protein interaction and not likely to be mediated via dsRNA since mutations F131A, K150A, K154E and A158D that are known to destroy PKR's dsRNA binding (26,52) have very little adverse effect on PKR–hDUS2 interaction. Although hDUS2 carries one copy of the conserved dsRBM, it does not interact with dsRNA with high affinity. This may be attributed to the lack of a crucial conserved lysine at position 58 of the conserved motif (Figure 1E). This lysine has been shown to be essential for the high-affinity interaction with dsRNA (52). hDUS2 carries an aspartic acid residue at this position, which may contribute to its lack of high-affinity dsRNA binding.
Recombinant hDUS2 protein showed a marked inhibition of PKR's kinase activity in vitro. The inhibition was effective when PKR was activated either by addition of dsRNA or recombinant PACT. However, hDUS2 was effective in inhibiting PACT-induced PKR activation more efficiently than dsRNA-induced PKR activation since even the lowest amount of hDUS2 (80 ng) showed a marked reduction in PKR autophosphorylation when activated by PACT. This may be due to the ability of hDUS2 to interact both with PKR and PACT. hDUS2's interaction with PACT may inhibit PACT's interaction with PKR in addition to inhibition by directly interacting with PKR, thus resulting in a more efficient inhibition. The biological significance of this if any is currently unknown. hDUS2 also showed inhibition of PKR in mammalian cells as seen in the translation inhibition assay. However, hDUS2 did not inhibit PKR activation as efficiently as PKR's well-studied inhibitor TRBP (Figure 5B). Since this is not due to lower level of expression of hDUS2 as compared to TRBP (data not shown), we conclude that hDUS2 is not a strong inhibitor of PKR in vivo.
However, since the results shown in Figure 5C indicate that when overexpressed, hDUS2 can inhibit activated PKR it can be speculated that hDUS2 may regulate PKR activity under conditions (or in cells) where its expression is elevated. Thus, hDUS2's inhibition of PKR may be of biological relevance under conditions where hDUS2 expression is upregulated in mammalian cells. In this context, it has been recently reported that hDUS2 is overexpressed in human nonsmall cell lung carcinomas (NSCLC). It was observed that upregulation of hDUS2 was a relatively common feature of pulmonary carcinogenesis and that selective suppression of hDUS2 by SiRNA showed reduced growth rate in NSCLC cell lines (50). Inhibition of PKR activity in NIH 3T3 cells by expression of a trans-dominant negative PKR mutant has been shown to lead to cell transformation and tumor formation in nude mice (4,5). Overexpression of PKR inhibitors p58IPK and TRBP also results in tumorigenesis (73). It remains to be tested if overexpression of hDUS2 also can lead to cell transformation. In addition, it would also be interesting to examine if hDUS2-mediated inhibition of PKR is involved in tumorigenesis in cases of NSCLC. Since our results demonstrate that hDUS2 can inhibit apoptosis, overexpression of hDUS2 in NSCLC may offer a growth advantage to cancer cells.
hDUS2 has been shown to enhance the translation rate in mammalian cells possibly by enhancing the tRNA modification activity of EPRS complex (50). In view of the results presented here, it is also possible that hDUS2 may enhance the rate of translation by inhibition of eIF2α phosphorylation brought about by activated PKR. This may be of biological significance in cancer progression since enhanced PKR expression is reported in some types of cancers. In melanomas and colorectal cancers, PKR overexpression can be correlated with enhanced PKR activity (74). However, in breast cancer cells, PKR overexpression does not correlate with increased PKR activity (17,75). It is thought that an inhibitor of PKR may be expressed at high levels in such cancers, leading to an inhibition of PKR activity. In future, it would be of interest to examine if hDUS2 may be involved in PKR inhibition in these types of cancers.
ACKNOWLEDGEMENTS
Funding has been provided by National Institutes of Health (HL63359 to R.C.P.) and American Heart Association (0555503U to R.C.P.). The Open Access publication charges for this article were waived.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sen GC, Ransohoff RM. Interferon-induced antiviral actions and their regulation. Adv. Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 4.Meurs EF, Galabru J, Barber GN, Katze MG, Hovanessian AG. Tumor suppressor function of the interferon-induced double-stranded RNA-activated protein kinase. Proc. Natl Acad. Sci. USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 6.Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc. Natl Acad. Sci. USA. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hovanessian AG. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J. Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 8.Katze MG. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 1995;3:75–78. doi: 10.1016/s0966-842x(00)88880-0. [DOI] [PubMed] [Google Scholar]
- 9.Katze MG. Interferon, PKR, virology, and genomics: what is past and what is next in the new millennium? J. Interferon Cytokine Res. 2002;22:283–286. doi: 10.1089/107999002753675695. [DOI] [PubMed] [Google Scholar]
- 10.Hovanessian AG, Galabru J. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur. J. Biochem. 1987;167:467–473. doi: 10.1111/j.1432-1033.1987.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 11.Patel RC, Handy I, Patel CV. Contribution of double-stranded RNA-activated protein kinase toward antiproliferative actions of heparin on vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2002;22:1439–1444. doi: 10.1161/01.atv.0000028817.20351.fe. [DOI] [PubMed] [Google Scholar]
- 12.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Yang M, May WS. RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 14.Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 15.Colthurst DR, Campbell DG, Proud CG. Structure and regulation of eukaryotic initiation factor eIF-2. Sequence of the site in the alpha subunit phosphorylated by the haem-controlled repressor and by the double-stranded RNA-activated inhibitor. Eur. J. Biochem. 1987;166:357–363. doi: 10.1111/j.1432-1033.1987.tb13523.x. [DOI] [PubMed] [Google Scholar]
- 16.Samuel CE. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 17.Jagus R, Joshi B, Barber GN. PKR, apoptosis and cancer. Int. J. Biochem. Cell Biol. 1999;31:123–138. doi: 10.1016/s1357-2725(98)00136-8. [DOI] [PubMed] [Google Scholar]
- 18.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 19.Williams BRG. The role of the dsRNA-activated kinase, PKR, in signal transduction. Semin. Virol. 1995;6:191–202. [Google Scholar]
- 20.Kumar A, Yang YL, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, et al. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramana CV, Grammatikakis N, Chernov M, Nguyen H, Goh KC, Williams BR, Stark GR. Regulation of c-myc expression by IFN-gamma through Stat1-dependent and -independent pathways. EMBO J. 2000;19:263–272. doi: 10.1093/emboj/19.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Judware R, Petryshyn R. Partial characterization of a cellular factor that regulates the double-stranded RNA-dependent eIF-2 alpha kinase in 3T3-F442A fibroblasts. Mol. Cell Biol. 1991;11:3259–3267. doi: 10.1128/mcb.11.6.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzberg S, Mandelboim M, Zalcberg M, Shainberg A, Mandelbaum M. Interruption of myogenesis by transforming growth factor beta 1 or EGTA inhibits expression and activity of the myogenic-associated (2′–5′) oligoadenylate synthetase and PKR. Exp. Cell Res. 1995;219:223–232. doi: 10.1006/excr.1995.1222. [DOI] [PubMed] [Google Scholar]
- 24.Katze MG, Wambach M, Wong ML, Garfinkel M, Meurs E, Chong K, Williams BR, Hovanessian AG, Barber GN. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol. Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel RC, Sen GC. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J. Biol. Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 26.Green SR, Mathews MB. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 27.McCormack SJ, Thomis DC, Samuel CE. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology. 1992;188:47–56. doi: 10.1016/0042-6822(92)90733-6. [DOI] [PubMed] [Google Scholar]
- 28.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romano PR, Green SR, Barber GN, Mathews MB, Hinnebusch AG. Structural requirements for double-stranded RNA binding, dimerization, and activation of the human eIF-2 alpha kinase DAI in Saccharomyces cerevisiae. Mol. Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierro-Monti I, Mathews MB. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 32.Carpick BW, Graziano V, Schneider D, Maitra RK, Lee X, Williams BR. Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 33.Nanduri S, Rahman F, Williams BR, Qin J. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano PR, Garcia-Barrio MT, Zhang X, Wang Q, Taylor DR, Zhang F, Herring C, Mathews MB, Qin J, et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol. Cell Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor DR, Lee SB, Romano PR, Marshak DR, Hinnebusch AG, Esteban M, Mathews MB. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol. Cell Biol. 1996;16:6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Romano PR, Nagamura-Inoue T, Tian B, Dever TE, Mathews MB, Ozato K, Hinnebusch AG. Binding of double-stranded RNA to protein kinase PKR is required for dimerization and promotes critical autophosphorylation events in the activation loop. J. Biol. Chem. 2001;276:24946–24958. doi: 10.1074/jbc.M102108200. [DOI] [PubMed] [Google Scholar]
- 37.Patel RC, Stanton P, McMillan NM, Williams BR, Sen GC. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl Acad. Sci. USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cosentino GP, Venkatesan S, Serluca FC, Green SR, Mathews MB, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S, Kaufman RJ. Double-stranded (ds) RNA binding and not dimerization correlates with the activation of the dsRNA-dependent protein kinase (PKR) J. Biol. Chem. 1996;271:1756–1763. doi: 10.1074/jbc.271.3.1756. [DOI] [PubMed] [Google Scholar]
- 40.Patel RC, Sen GC. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol. Cell Biol. 1998;18:7009–7019. doi: 10.1128/mcb.18.12.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel RC, Stanton P, Sen GC. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J. Biol. Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 42.Fasciano S, Hutchins B, Handy I, Patel RC. Identification of the heparin-binding domains of the interferon-induced protein kinase, PKR. FEBS J. 2005;272:1425–1439. doi: 10.1111/j.1742-4658.2005.04575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fasciano S, Patel RC, Handy I, Patel CV. Regulation of vascular smooth muscle proliferation by heparin: inhibition of cyclin-dependent kinase 2 activity by p27(kip1) J. Biol. Chem. 2005;280:15682–15689. doi: 10.1074/jbc.M411458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters GA, Li S, Sen GC. Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J. Biol. Chem. 2006;281:35129–35136. doi: 10.1074/jbc.M607714200. [DOI] [PubMed] [Google Scholar]
- 45.Huang X, Hutchins B, Patel RC. The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR) Biochem. J. 2002;366:175–186. doi: 10.1042/BJ20020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters GA, Hartmann R, Qin J, Sen GC. Modular structure of PACT: distinct domains for binding and activating PKR. Mol. Cell Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Kato T, Daigo Y, Hayama S, Ishikawa N, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y. A novel human tRNA-dihydrouridine synthase involved in pulmonary carcinogenesis. Cancer Res. 2005;65:5638–5646. doi: 10.1158/0008-5472.CAN-05-0600. [DOI] [PubMed] [Google Scholar]
- 51.Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 52.Patel RC, Stanton P, Sen GC. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J. Biol. Chem. 1996;271:25657–25663. doi: 10.1074/jbc.271.41.25657. [DOI] [PubMed] [Google Scholar]
- 53.Yang M, May WS, Ito T. JAZ requires the double-stranded RNA-binding zinc finger motifs for nuclear localization. J. Biol. Chem. 1999;274:27399–27406. doi: 10.1074/jbc.274.39.27399. [DOI] [PubMed] [Google Scholar]
- 54.Green SR, Manche L, Mathews MB. Two functionally distinct RNA-binding motifs in the regulatory domain of the protein kinase DAI. Mol. Cell Biol. 1995;15:358–364. doi: 10.1128/mcb.15.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaufman RJ, Murtha P. Translational control mediated by eucaryotic initiation factor-2 is restricted to specific mRNAs in transfected cells. Mol. Cell Biol. 1987;7:1568–1571. doi: 10.1128/mcb.7.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies MV, Furtado M, Hershey JW, Thimmappaya B, Kaufman RJ. Complementation of adenovirus virus-associated RNA I gene deletion by expression of a mutant eukaryotic translation initiation factor. Proc. Natl Acad. Sci. USA. 1989;86:9163–9167. doi: 10.1073/pnas.86.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaufman RJ, Davies MV, Pathak VK, Hershey JW. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell Biol. 1989;9:946–958. doi: 10.1128/mcb.9.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, et al. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc. Natl Acad. Sci. USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onuki R, Bando Y, Suyama E, Katayama T, Kawasaki H, Baba T, Tohyama M, Taira K. An RNA-dependent protein kinase is involved in tunicamycin-induced apoptosis and Alzheimer's disease. EMBO J. 2004;23:959–968. doi: 10.1038/sj.emboj.7600049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee ES, Yoon CH, Kim YS, Bae YS. The double-strand RNA-dependent protein kinase PKR plays a significant role in a sustained ER stress-induced apoptosis. FEBS Lett. 2007;581:4325–4332. doi: 10.1016/j.febslet.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Ito M, Onuki R, Bando Y, Tohyama M, Sugiyama Y. Phosphorylated PKR contributes the induction of GRP94 under ER stress. Biochem. Biophys. Res.Commun. 2007;360:615–620. doi: 10.1016/j.bbrc.2007.06.087. [DOI] [PubMed] [Google Scholar]
- 62.Lee TG, Tomita J, Hovanessian AG, Katze MG. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68 000 from influenza virus-infected cells. Proc. Natl Acad. Sci. USA. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee TG, Tomita J, Hovanessian AG, Katze MG. Characterization and regulation of the 58 000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J. Biol. Chem. 1992;267:14238–14243. [PubMed] [Google Scholar]
- 64.Lee TG, Tang N, Thompson S, Miller J, Katze MG. The 58 000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol. Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan SL, Gale MJ, Jr, Katze MG. Double-stranded RNA-independent dimerization of interferon-induced protein kinase PKR and inhibition of dimerization by the cellular P58IPK inhibitor. Mol. Cell Biol. 1998;18:2431–2443. doi: 10.1128/mcb.18.5.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodman AG, Smith JA, Balachandran S, Perwitasari O, Proll SC, Thomas MJ, Korth MJ, Barber GN, Schiff LA, et al. The cellular protein P58IPK regulates influenza virus mRNA translation and replication through a PKR-mediated mechanism. J. Virol. 2007;81:2221–2230. doi: 10.1128/JVI.02151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gatignol A, Buckler WA, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 68.Ong CL, Thorpe JC, Gorry PR, Bannwarth S, Jaworowski A, Howard JL, Chung S, Campbell S, Christensen HS, et al. Low TRBP levels support an innate human immunodeficiency virus type 1 resistance in astrocytes by enhancing the PKR antiviral response. J. Virol. 2005;79:12763–12772. doi: 10.1128/JVI.79.20.12763-12772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim MJ, Wang XW. Nucleophosmin and human cancer. Cancer Detect. Prev. 2006;30:481–490. doi: 10.1016/j.cdp.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pang Q, Christianson TA, Koretsky T, Carlson H, David L, Keeble W, Faulkner GR, Speckhart A, Bagby GC. Nucleophosmin interacts with and inhibits the catalytic function of eukaryotic initiation factor 2 kinase PKR. J. Biol. Chem. 2003;278:41709–41717. doi: 10.1074/jbc.M301392200. [DOI] [PubMed] [Google Scholar]
- 71.Garcia MA, Collado M, Munoz-Fontela C, Matheu A, Marcos-Villar L, Arroyo J, Esteban M, Serrano M, Rivas C. Antiviral action of the tumor suppressor ARF. EMBO J. 2006;25:4284–4292. doi: 10.1038/sj.emboj.7601302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donze O, Abbas-Terki T, Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barber GN, Thompson S, Lee TG, Strom T, Jagus R, Darveau A, Katze MG. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc. Natl Acad. Sci. USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim SH, Gunnery S, Choe JK, Mathews MB. Neoplastic progression in melanoma and colon cancer is associated with increased expression and activity of the interferon-inducible protein kinase, PKR. Oncogene. 2002;21:8741–8748. doi: 10.1038/sj.onc.1205987. [DOI] [PubMed] [Google Scholar]
- 75.Savinova O, Joshi B, Jagus R. Abnormal levels and minimal activity of the dsRNA-activated protein kinase, PKR, in breast carcinoma cells. Int. J. Biochem. Cell Biol. 1999;31:175–189. doi: 10.1016/s1357-2725(98)00140-x. [DOI] [PubMed] [Google Scholar]