Abstract
A ribonuclease, RNase T-tat, specifically designed to hydrolyze the TAR RNA of HIV-1 virus has been engineered. The protein was made by domain swapping the TAT peptide at the loop 3 position of ribonuclease T1. The RNase T-tat maintains a guanine-specific RNA hydrolytic activity, and characteristically displayed a specific affinity for the TAR RNA of HIV-1. In the in vitro and in vivo assays, the RNase T-tat preferentially inhibited the expression of TAR-bearing mRNA through cis-TAR targeting, suggesting that RNase T-tat may be potentially useful for the disruption of the initial stage of the transcription process of HIV-1 virus.
INTRODUCTION
Recently, protein engineering has aimed at designing and producing novel proteins with desirable functions and this has emerged into a new stage due to significant developments in three-dimensional structure analysis combined with advanced knowledge of important cellular events. The design of the protein's properties that surpass the performance of native proteins by protein engineering has the potential to create protein drug and this has become a trend in pharmaceutical industry. The pre-requisite of this is to know where and how the engineering acts on the protein. For RNA virus related diseases, the utilization of an RNase- or ribozyme-based strategy aims for the destruction of the viral RNA (1,2). In the case of HIV-1 virus, the transcription of viral RNA is heavily dependent upon the formation of an RNP complex that results from the interaction between an early TAR (activation-responsive region) RNA transcript and the viral TAT (trans-activator of transcription) protein (3–6). A consequence of the formation of the RNP complex is the prompt recruitment of host cell transcription factors allowing efficient viral transcription (5,6). Thus, one way to interfere with the proliferation of HIV-1 virus in infected cells would lie in the destruction of the early TAR RNA scaffold. Based on this notion, the ribonuclease activity of ribonuclease T1 (RNase T1) was the choice for this purpose. This was made possible because the structure of RNase T1 is well understood and has proven to be a useful model for the study of protein folding and stability (7–10). More importantly, a recent study has indicated that the peripheral loop L3 of RNase T1 can be swapped with a specific RNA-binding motif without changing the base-specificity of RNase T1 but the change results in the gain of a double-stranded specificity (11). It was therefore our intention to use domain swapping to make a new RNase, denoted as RNase T-tat, which acts against TAR transcription of HIV-1 virus. RNase T-tat consists of the Tat peptide sequence of RKKRRQRRR from the TAT protein at the L3 position of the RNase T1. Based on the specific cis-acting recognition of TAR RNA by the Tat peptide (3–6,12,13), theoretically, the ribonuclease activity of RNase Tat is expected to inactivate the transcription of HIV-1 virus on gaining cell entry (5,6). With this expectation, RNase T-tat may be able to play a role to restraining the transcription of HIV-1 virus.
MATERIALS AND METHODS
Construction of the RNase T-tat gene
The gene coding for RNase T-tat was cloned by the polymerase chain reaction (PCR) using pGEM(T)/RNase T1 as the template by two primers, a C-primer (5′-NNNNNGGATCCCCAGGTGCGGACCGTGTGGTGTTC-3′) and a N primer (5′-NNNNNGGATCCTCTTCTTCTTTGTCTTCTTTTTTTTCTTCCAGAGCTCGAATCGGCCACTCATA-3′). The BamHI restriction sites are underlined. The N primer carried an insert site coding the Tat-peptide (27 bases in capital alphabet) plus 26 bases (in italics) that encode for the 3′ end of β2 strand of RNase T1. The PCR fragment (∼3.5 kb) was digested by restriction enzyme DpnI to eliminate the endogenous pGEM(T)/RNaseT1 plasmid and further digested with restriction enzyme BamHI to allow ligation to create the pGEM/T-tat plasmid. The cytosolic expression vector pET28a/T-tat was obtained using the same PCR strategy. The coding sequence of the RNase T-tat gene was verified by DNA sequencing. The pET28a/T-tat plasmid was propagated in Escherichia coli strain JM109, and protein was purified through Ni-chromatography. Similarly, the RNase T-tat gene was also cloned into a eukaryotic CMV-driven plasmid using the same strategy.
The procedure for the construction of pET28a/T-tatH95S was essentially the same as described earlier, except that pGEM(T)/RNase T1H92S was used as the template for PCR amplification. The mutant protein was purified by Ni-chromatography. A eukaryotic expression vector, pCMV/TAR-EGFP was also constructed using the same strategy. The vector carries a TAR RNA sequence (57-mer) joined to the NH2-terminal of the coding sequence of EGFP driven by a CMV promoter.
Molecular modeling
The tertiary structure of RNase T-tat was simulated on the basis of the crystal structure of RNase T1 (Brookhaven PDB entry iRNT). Using energy minimization and different conditions of constraints, a RNase T-tat structure was generated. All calculations were performed on an SGE Origin 2000 computer using the program DESCOVEER as implemented in the package Insight II (Molecular Simulation, San Diego, CA).
RNA-impregnated polyacrylamide gel electrophoresis
Purified recombinant RNase T-tat was analyzed electrophoretically in a gel contained 2.5 mg/ml of large fragments of ribosomal RNA. The presence of RNA fragments in the gel did not interfere with protein separation. After electrophoresis, the gel was rematured by incubation in buffer containing 10 mM Tri–HCl, pH 7.4 and 25% 2-propanol (to extract SDS), and this was followed by four washes with incubation buffer without 2-propanol. The gel was then stained with toluidine blue O (2% solution in H2O). A negative staining effect (white in color) against a blue background (the stained RNA) represented the ribonucleolytic activity of protein present on the gel.
The determination of the base specificity was carried out using the same principle except that that the gel contained mono-ribonucleotide polymer (poly U, Poly A and Poly G, or poly C) instead of RNA fragments.
RNA gel-mobility shift and Millipore assay
A 57-mer oligo RNA that mimics the structure of the TAR RNA was synthesized using phage T7 RNA polymerase (Promega Co. USA) and synthetic DNA oligomers as templates. The oligomers used had the sequence carried a complementary T7 promoter and the nucleotide sequence of TAR RNA. They were annealed at 90°C for 3 min followed by cooling on ice. The transcription reaction was carried out at 37°C for 1 h under conditions as described previously (11). At the end of transcription, the transcript was heated to 90°C for 2 min in buffer and renatured by gradual overnight cooling to 4°C, which allowed the formation of a secondary structure that imitated the structure of TAR RNA. The 57-mer TAR RNA was 3′ end-labeled with cytidine 3′,5′-[α-32P]bisphosphate as previously described (11).
The gel mobility shift assay was carried out in a 10 μl reaction mixture containing 53 ρM radioactive [32P]-57 mer TAR with various amounts of RNase T-tat or RNase T-tat H95S mutant protein in binding buffer (20 mM Tris–HCl, pH 8.0; 100 mM KCl; and 10 mM DTT, 1 mM EDTA and 10% glycerol) at 4°C or 25°C, for 15 min and analyzing using an 8% native polyacrylamide gel. The presence of radioactive RNA and RNP was detected using a phosphoimager.
The Millipore assay was carried out as follow: [32P]-TAR RNA or [32P]-5S rRNA was incubated with increasing amounts of RNase T-tat H95S protein in binding buffer for 15 min at 25°C. The reaction mixture was filtered through a nitrocellulose membrane (Millipore, type HA, 0.45 μm) that had been soaked in binding buffer; filtration was with a vacuum of 55 Torr. The radioactivity retained on nitrocellulose membrane was determined.
Cell entry assay
The ability of the RNase T-tat molecule to enter the cell was verified by both a protease-elimination assay and by direct microscopic fluorescent examination. The protease-elimination assay was carried by incubating embryonic kidney 293T cells with [35S]-RNase T-tat (0.5 μg, prepared by in vivo labeling) with or without chloroquine in PBS for 30 min at 25°C. At 6 h and 24 h post-incubation, the same numbers of treated and non-treated cells were taken and incubated with trypsin (5 μg/ml, 30 min at 25°C). After thoroughly washing cells with PBS, the trypsinized cells were then lysed with denaturation solution containing 1% SDS and analyzed on a SDS-containing polyacrylamide gel. The presence of a radioactive protein band was visualized by a phosphoimager.
The direct microscopic fluorescent examination was carried out as follows. Texas red-conjugated RNase T-tat (TR-RNase T-tat), RNase T1 (TR-RNase T1) and bovine serum albumin (TR-BSA) were prepared as previously described (14). These fluorescent proteins in PBS were separately applied to 293T cells that had been seeded on a 15 × 15 mm coverslip, for 30 min at 25°C. After removal of the excess fluorescent protein by extensively washing, growth was continued in fresh medium. At 6 h post-incubation, the cells were examined under a confocal fluorescence microscope and also by phase contrast microscopy.
The in vitro activity of RNase T-tat
The preferential inhibitory effect of RNase T-tat on in vitro translation of TAR-EGFP mRNA was carried out using two mRNAs (luciferase and TAR-EGFP mRNA; 5 μg each) in rabbit reticulocyte lysate containing [35S]methionine in the presence (0.2 μg) or the absence of RNase T-tat. The reaction was carried out for 60 min at 25°C and analyzed by SDS-containing polyacrylamide gel. The translated products were visualized using a phophoimager. Luciferase mRNA was purchased commercially. The TAR-EGFP mRNA was synthesized by the RiboMax procedure (Promega Co. USA) using the constructed vector (pCMV/TAR-EGFP) as the template.
Detection of the in vivo activity of RNase T-tat by a nanoparticulate single cell activity assay
A hepatoma cell line containing a stable gene coding for either TAR-EGFP or EGFP (kindly provided by Dr J. S. Su, Genome Center, National Yang-Ming University, Taiwan) was used to directly observe the in vivo activity of RNase T-tat. This was carried out by our currently developed nanoparticulate single cell activity assay (patent pending). In brief, nano-magnetic beads (NMB) (TANBead, USPIO-101; purchased from Taiwan Nanotech Inc. Taoyuan, Taiwan) were coated with polylysine (PLL, MW 4000; Sigma-Aldrich Co, USA) using EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride) conjugation. The polylysine-coated NMBs were magnetically separated from un-reacted EDC, and were subjected to labeling with Alexa fluor 555 (Molecular Probe, Eugene, USA) according to manufacture's suggested procedure. The fluorescent polylysine-coated NMB were coupled with an excess amount of plasmid pCMV/RNase T-tat under constant stirring. Uncoupled DNA plasmid was again removed by magnetic procedure. The NMB/plasmid complex was delivered into the hypatoma cells (with an excess number of NMB complexes to enhance cell entry) using electroporation. The electroporation took place in serum free DNEM medium in less than one second. Immediately after electroporation, the free NMB complexes were separated from cells by the magnetic removal procedure. Treated cells were then seeded on a coverslip for further growth. Under different fluorescent excitations, the transfected cells gave constant green fluorescence because of the expression of EGFP or TAR-EGFP, and the NMB showed up as a red spot. Thus, cells impregnated with a red fluorescent NMB indicated these cells had been successfully transfected with plasmid DNA containing the RNase T-tat gene. The fluorescent intensities in cells with and without internalized NMB were quantitatively measured. The measurement was carried out both for hepatoma cells containing TAR-EGFP and for cells containing EGFP.
RESULTS
Characterization of RNase T-tat
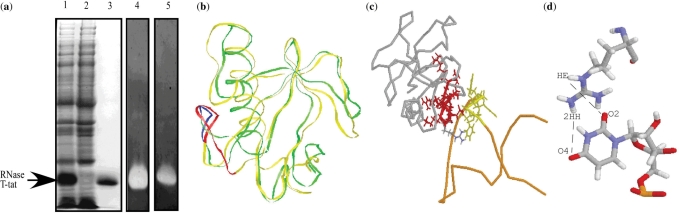
In this study, we have constructed a gene coding for the RNase T-tat by genetic manipulation. The RNase T-tat consists of the Tat peptide sequence of RKKRRQRRR, which was inserted at loop position of RNase T1. The gene was expressed in E.coli cells and the recombinant RNase T-tat was subsequently purified (Figure 1a). The recombinant protein RNase T-tat displays the same guanyl base-specificity as RNase T1 (7,8), which was detected by RNA-impregnated polyacrylamide gel assay (Figure 1a). This indicates that insertion of Tat peptide does not jeopardize the original ribonuclease activity and these results are similar to the reported case for RNase Tα (11).
Figure 1.
Characterization of RNase T-tat. (a) The expression, purification and ribonuclease activity of RNase T-tat. Lanes 1–3 are Coomassie staining of a 15% SDS-polyacrylamide gel analyzing the expression of RNase T-tat gene with (lane 1) and without (lane 2) IPTG induction and the purified recombinant protein RNase T-tat (lane 3). Lanes 4 and 5 are the RNA-impregnated gel electrophoresis analysis of the ribonuclease activity of recombinant RNase T-tat against naked RNA (lane 4) and poly G (lane 5). The gels were visualized using toluidine blue O staining of RNA. Negative results were obtained for poly A- and poly U-impregnated gels. (b) The simulated structure of RNase T-tat (yellow ribbon with the TAT peptide in red) superimposed on the structure of RNase T1 (green ribbon whose L3 loop is blue in color). (c) Molecular docking of RNase T-tat with TAR RNA. Both protein (gray) and RNA (orange) present in backbone mode except for the contact region, which is highlighted with a wire-structure in red color for the amino acid residues and yellow color for the UCU bulge. (d) Atomic detail of interaction between the Arg70 of the RNase T-tat and U33 of TAR RNA. The distances indicated (underlined) affect docking and the measurements were made after docking.
As an initial step, we have inspected the structure of the engineered RNase T-tat and its recognition of the TAR RNA by molecular modeling. The simulated structure of RNase T-tat has the basic β-pleated sheet conformation with a folded Tat peptide free on the surface of the protein molecule. The β-pleated sheet can be superimposed on the structure of RNase T1 (Figure 1b). In molecular docking, the Tat domain fitted the major groove of the TAR RNA with closed contact to the UCU bulge (Figure 1c), which is very like that found in previous studies of Tat-TAR interaction (4–6,12,13). When the intermolecular distances are measured (Figure 1d), they are close to the reported geometric values observed previously (13). This information supports the feasibility of the proposed RNase T-tat as a molecule that can recognize TAR RNA.
Specific binding of RNase T-tat to TAR RNA
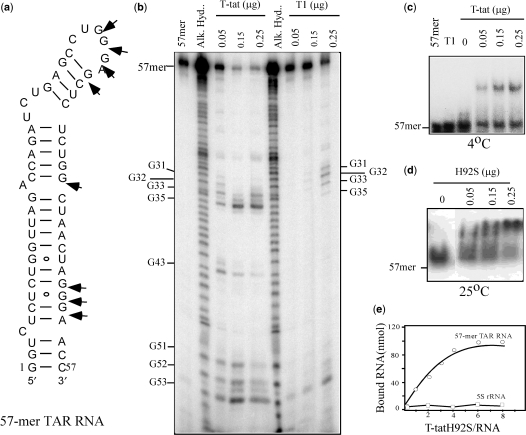
Experimentally, we tested the recognition TAR RNA by RNase T-tat using a gel-mobility shift assay using a synthetic 57-mer oligo TAR RNA (Figure 2a) as substrate. The results showed that RNase T-tat has ribonucleolytic activity and therefore was not surprising to find that the TAR RNA was cleaved by RNase T-tat at 25°C (Figure 2b). The cleavage occurred at both single stranded (G31, G32 and G33), and double stranded (G35, G43, G51, G52 and G53) guanine bases, which is different from the original RNase T1 single stranded-specificity (7) (Figure 2b).
Figure 2.
Recognition between RNase T-tat and a 57-mer TAR RNA. (a) The structure of 57-mer oligo TAR RNA. Arrows are the cleavage sites caused by RNase T-tat. (b) The hydrolysis of TAR RNA by RNase T-tat. The reaction mixture contained 3′end labeled 57-mer TAR RNA and various amounts of RNase T-tat. The analysis was on a 15% sequence polyacrylamide gel containing 8 M urea. Partial alkaline hydrolysis (Alk. Hyd.) was carried out according to a standard protocol. (c) Binding of RNase T-tat to the 57-mer TAR RNA at 4°C. The assay was analyzed using 8% native polyacrylamide gel. Lane T1 contains was 2 μg of RNase T1. (d) Binding of RNase T-tat mutant H95S to the 57-mer TAR RNA at 25°C. The radioactive RNA and RNP in (b), (c) and (d) were detected using a phosphoimager. (e) Millipore binding assay. Both binding of mutant RNase T-tat H95S to [32P]-TAR RNA and human [32P]-5S rRNA were carried out in parallel.
In order to avoid ribonuclease activity at normal temperature, the recognition between RNase T-tat and the TAR RNA was determined at 4°C. The result of the gel-mobility shift assay showed an up-shifted RNP complex (Figure 2c). The formation of this RNP is specific because in a parallel experiment, RNase T1 was unable to form such an up-shifted complex at 4°C (Figure 2c). In the effort to determine the affinity between RNase T-tat and TAR RNA, we created a defective mutant, RNase T-tatH95S (His95 mutated to serine), and used this in the kinetic binding assay. The corresponding mutation at this His residue in RNase T1 (His92) is known to reduce RNA hydrolysis by RNase T1 to a minimum but to maintain the protein's structural integrity (8,15,16). Accordingly, the gel-mobility shift assay was carried out again with RNase T-tatH95S at 25°C. The results showed that the RNase T-tatH95S up-shifted the TAR RNA, but produced no sign of RNA digestion (Figure 2d). Separately, a Millipore filtration assay was adapted to determine a saturation-binding curve (Figure 2e), and the apparent association constant for the interaction of RNase T-tatH95S with TAR RNA was determined by constructing a double reciprocal plot of the data. This yielded a Ka′ value of 1.8 × 107 M−1. The affinity suggests that RNase T-tat may prefer to recognize TAR RNA rather than any other RNA molecule.
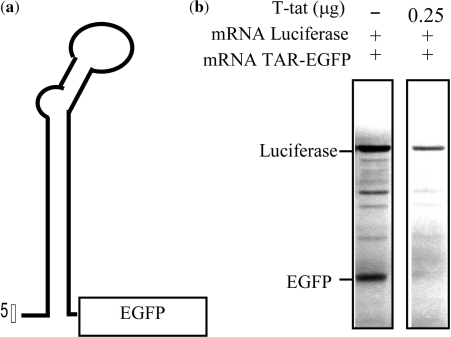
To test such preference, we followed up with a duo-mRNA translation assay containing two mRNAs, a luciferase mRNA and a TAR-carrying EGFP mRNA in the rabbit reticulocyte translation system with or without RNase T-tat. The TAR-carrying EGFP has a 57-mer TAR RNA sequence joined to the NH2-terminal end of the coding sequence of EGFP (Figure 3a). The results of the in vitro translation showed that both proteins were equally expressed in the absence of RNase T-tat (Figure 3b). However, in the presence of RNase T-tat, the production of luciferase protein was slightly reduced, but that of EGFP had totally been eliminated (Figure 3 b). Under the same conditions, the non-TAR-carrying mRNA (luciferase) was affected to a very much lesser extent (Figure 3b).
Figure 3.
The in vitro activity of RNase T-tat on the expression of TAR-carrying mRNA. (a) The schematic presentation of the structure of TAR-carrying EGFP (TAR-EGFP) that was used for the in vitro inhibitory translation assay. (b) The in vitro inhibitory translation assay. The translation of TAR-carrying EGFP mRNA and luciferase mRNA was carried out in the rabbit reticulocyte system for 45 min with or without RNase T-tat and analyzed using 15% SDS-polyacrylamide gel electrophoresis and a phosphoimager.
Cell entry of RNase T-tat
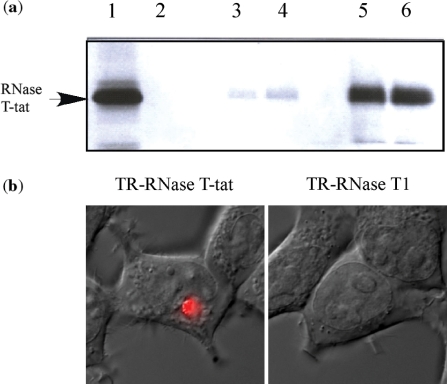
The Tat protein or a Tat-peptide conjugated protein has an ability to enter cells (17–22) or living mice (23) and this is well documented. Based on this, we have examined whether or not the Tat peptide within the structure of RNase T-tat was able to direct RNase T-tat into cells. This was carried out using a protease-elimination assay with radioactive [35S]-RNase T-tat and fluorescent microscopic examination. The results of elimination assay clearly showed that RNase T-tat was able to enter 293T cells (Figure 4a). Similarly, using fluorescent microscopy, fluorescent Texas red-conjugated RNase T-tat (TR-RNase T-tat) was located inside of the treated 293T cells after a short external exposure (Figure 4b), whereas Texas-red conjugated RNase T1 (TR-RNase T1) and Texas-red conjugated-BSA (TR-BSA) showed no sign of any entry (Figure 4b). It is of worth noting that the efficiency of cell entry as measured by the internalized fluorescent RNase T-tat was unexpectedly low with a yield of ∼20% compared with values in reported studies where the Tat peptide itself was used to mediate cell entry (24,25).
Figure 4.
Cell entry of RNase T-tat. (a) The protease elimination assay. Lane 1, [35S]-RNase T-tat alone; lane 2, trypsinization of [35S]-RNase T-tat; lanes 3 and 4, cell lysate 6 h post-treatment with or without chloroquine, respectively; lanes 5 and 6, cell lysate of 24 h post-treatment with or without cholorquine, respectively. The radioactive protein was detected by a phosphoimager using a SDS-polyacrylamide gel electrophoresis. (b) The cellular localization of the fluorescent RNase T-tat. Texas red-conjugated RNase T-tat (TR-RNase T-tat) and RNase T1 (TR-RNase T1) were separately applied to 293T cells as described in text. The presenting pictures were combining images that observed under a confocal fluorescence microscopy and phase contrast microscopy.
The in vivo activity of RNase T-tat
There remains a question of whether the described preferential targeting of TAR-carrying mRNA is able to cause in vivo inhibition under physiological conditions inside the cell. To test such physiological relevance, we have treated hepatoma cells that had carried a stable TAR-EGFP gene with different dosage of RNase T-tat, and measured the influence of RNase T-tat on the expression of EGFP. Using this approach (data not shown), it was difficult to see any difference caused by the action of RNase T-tat on the TAR-EGFP cells compared with EGFP cells. A similar non-significant outcome was also observed when the expression of EGFP mRNA was measured by northern blotting of both treated cells. Such observations could be attributed to the low percentage cell entry of RNase T-tat as described earlier. Interestingly, it was observed that the level of GAPDH mRNA in both treated cells was unaffected regardless of the presence or absence of RNase T-tat.
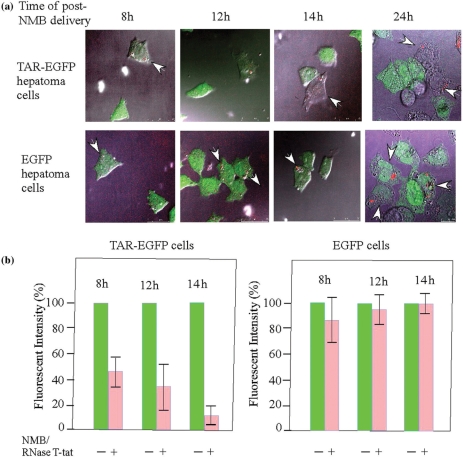
In an effort to demonstration that RNase T-tat has a specific in vivo activity on TAR-associated RNA, we carried out a single cell activity assay. The principle of this assay is the use of a fluorescent NMB as a gene-carrier and an indicator. Briefly, the NMB carried plasmid containing the CMV-driven RNase T-tat gene and this was mechanically delivered into the cells. The activity of RNase T-tat was followed in an individual cell impregnated with the fluorescent NMB (Figure 5a). The precise effect of expressed RNase T-tat on the expression of GFP was registered by measuring the GFP fluorescence intensity in single NMB-impregnated cells. This showed that the fluorescent intensity was gradually diminished in NMB-impregnated cell containing the stable TAR-EGFP gene with time after NMB delivery increased (Figures 5a and b). In parallel, little or no change in fluorescent intensity was found for NMB-impregnated cells that contained the stable EGFP gene (Figures 5a and b).
Figure 5.
The suppression of the expression of TAR-carrying EGFP by RNase T-tat as detected at single cell level. (a) The observations of the suppression of the expression of EGFP (shown in green) by internalized NMB (red) that carried RNase T-tat gene measured at different time intervals post-NMB delivery. The presenting photos are the merged images derived from results of phase contrast microscopic and fluorescent microscopic examination. Arrows showed cells that have impregnated with NMB(s). (b) The quantification of the effects of RNase T-tat as measured by the fluorescent intensity. The fluorescent intensity in a cell that contains no NMB was used as a 100% control for EGFP expression. Each bar represents an average measurement of 10 or more cells.
DISCUSSION
Utilizing an RNase- or ribozyme-based strategy against RNA related diseases has been suggested (1,2). Examples of fusing a RNA-binding domain to aid recognition and cleaved of HIV-RNA (26), the use of a ribozyme (27) or other approaches (25) to target the HIV-1 have been proposed in order to inactivate the machinery of HIV-1 transcription. Herein, we have successfully created an engineered ribonuclease, which potentially targets the initial stage of the transcription process of the HIV-1 virus. The action of RNase T-tat was first demonstrated by showing that RNase T-tat selectively recognized and hydrolyzed a synthetic 57-mer TAR RNA substrate. It is interesting that the engineering RNase T-tat not only displayed single stranded guanine base ribonuclease activity, but was also capable of cleaving guanine bases at the helix region around the bulge UCU of 57-mer TAR RNA. This expansion of the ribonuclease activity to double stranded cleavage can be attributed to conformational changes (4,6) in the helix region after the bulge UCU of the TAR RNA contacts the TAT-peptide of the RNase T-tat. Nevertheless, this is not a problem as the broadened specificity makes certain that RNase T-tat will effectively hydrolyze the TAR RNA. The observation that RNase T-tat selectively hydrolyzed the TAR RNA-associated mRNA in the in vitro duo-mRNA translation reaction clearly supports the protein's destructive ability.
In essence, by design, the protein transduction property of the Tat peptide (17,18,21) has been incorporated into the initial engineering blueprint of RNase T-tat. Indeed, positive results derived from both the protease-elimination assay and the fluorescent examination clearly confirmed the transductive nature of the Tat peptide in RNase T-tat as originally envisaged, except that a much lower efficiency of cell transduction was observed. Interestingly, we have observed that once RNase T-tat has entered a cell, it can easily reach the nucleus. Thus, despite the low efficiency of cell entry, the results show promise with respect to the action of RNase T-tat because the trans-activation of HIV-1 takes place in the nucleus (22), which is vital to final aim of disabling the proliferation of HIV-1.
In this study, the selective targeting of RNase T-tat to TAR-related RNA was first demonstrated by the in vitro duo-mRNA translation experiment, and this was followed up with a novel single cell activity assay system, which showed that internalized RNase T-tat molecules carried the same physiological relevance inside a cell. These results provide strong evidence that RNase T-tat is able to selectively prevent the expression of the TAR-associated EGFP, implying that RNase T-tat shows preferential recognition of the TAR-carrying mRNA (TAR-EGFP) through a cis-acting affinity and consequently hydrolyzes the RNA. It further suggests that RNase T-tat may be potentially useful for the disruption of TAR RNA of HIV-1 virus.
The significance of this study is providing a good example of the development of a RNA-oriented protein drug that can be used against a RNA virus. In addition, the technological platform of the single cell activity assay system demonstrates the ability to analyze gene action at a single cell level. The system is not only an excellent approach to the verification of gene activity, but also can be adapted to the screening potential drugs against HIV-1 virus, including chemicals and other types of biological materials.
ACKNOWLEDGEMENTS
This work was supported in part by National Science Council, ROC grants of nsc-91-2311-B010-008 and nsc92-2311-B-010-004. Special thanks to Professor R. Kirby, for his kind help in the preparation of this manuscript. Funding to pay the Open Access publication charges for this article was provided by Grant from the Ministry of Education, Aim for the Top University Plan, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Singwi S, Ramezani A, Ding SF, Joshi S. Targeted RNase: a feasibility study for use in HIV gene therapy. Gene Ther. 1999;6:913–921. doi: 10.1038/sj.gt.3300884. [DOI] [PubMed] [Google Scholar]
- 2.Singwi S, Joshi S. Potential nuclease-based strategies for HIV gene therapy. Front. Biosci. 2000;5:559–579. doi: 10.2741/singwi. [DOI] [PubMed] [Google Scholar]
- 3.Long KS, Crothers DM. Interaction of human immunodeficiency virus type 1 Tat-derived peptides with TAR RNA. Biochemistry. 1995;34:8885–8895. doi: 10.1021/bi00027a041. [DOI] [PubMed] [Google Scholar]
- 4.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J. Mol. Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 5.Verhoef K, Tijms M, Berkhout B. Optimal Tat-mediated activation of the HIV-1 LTR promoter requires a full-length TAR RNA hairpin. Nucleic Acids Res. 1997;25:496–502. doi: 10.1093/nar/25.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter S, Ping Y-H, Rana TM. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl Acad. Sci. USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sevcik J, Sanishvili RG, Pavlovsky AG, Polyakov KM. Comparison of active sites of some microbial ribonucleases: structural basis for guanylic specificity. Trends Biochem. Sci. 1990;15:158–162. doi: 10.1016/0968-0004(90)90217-y. [DOI] [PubMed] [Google Scholar]
- 8.Steyaert J. A decade of protein engineering on ribonuclease T1: atomic dissection of the enzyme-substrate interactions. Eur.J. Biochem. 1997;247:1–11. doi: 10.1111/j.1432-1033.1997.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- 9.Pace CN. Conformational stability of globular proteins. Trends Biochem. Sci. 1990;15:14–17. doi: 10.1016/0968-0004(90)90124-t. [DOI] [PubMed] [Google Scholar]
- 10.Pace CN, Heinemann U, Hahn U, Saenger W. Ribonuclease T1: structure, function, and stability. Angew. Chem. Int. Ed. Engl. 1991;30:343–360. [Google Scholar]
- 11.Chen D-T, Lin A. Domain swapping in ribonuclease T1 allows the acquisition of double stranded activity. Protein Eng. 2002;15:997–1003. doi: 10.1093/protein/15.12.997. [DOI] [PubMed] [Google Scholar]
- 12.Weeks KM, Crothers DM. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 13.Puglisi JD, Chen L, Blanchard S, Frankel AD. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 14.Wu WC, Liu HW, Lin A. Human ribosomal protein L7 displays an ER binding property and is involved in ribosome-ER association. FEBS Lett. 2007;581:651–657. doi: 10.1016/j.febslet.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Shirley BA, Stanssens P, Steyaert J, Pace CN. Conformational stability and activity of ribonuclease T1 and mutants. J. Biol. Chem. 1989;264:11621–11625. [PubMed] [Google Scholar]
- 16.Steyaert J, Wyns L. Functional interactions among the His40, Glu58 and His92 catalysts of ribonucelease T1 as studied by double and triple mutants. J. Mol. Biol. 1993;229:770–781. doi: 10.1006/jmbi.1993.1078. [DOI] [PubMed] [Google Scholar]
- 17.Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 18.Becker-Hapak M, McAllister SS, Dowdy SF. TAT-mediated protein transduction into mammalian cells. Methods. 2000;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- 19.Mann DA, Frankel AD. Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 1991;10:1733–1739. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teter SA, Klionsky DJ. How to get a folded protein across a membrane. Trends Cell Biol. 1999;9:428–431. doi: 10.1016/s0962-8924(99)01652-9. [DOI] [PubMed] [Google Scholar]
- 21.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl Acad. Sci. USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vives E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- 23.Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 24.Tachikawa K, Schroder O, Frey G, Briggs SP, Sera T. Regulation of the endogenous VEGF-A gene by exogenous designed regulatory proteins. Proc. Natl Acad. Sci. 2004;101:15225–15230. doi: 10.1073/pnas.0406473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner JJ, Arzumanov AA, Gait MJ. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melekhovets YF, Joshi S. Fusion with an RNA binding domain to confer target RNA specificity to an RNase: design and engineering of Tat-RNase H that specifically recognizes and cleaves HIV-1 RNA in vitro. Nucleic Acids Res. 1996;24:1908–1912. doi: 10.1093/nar/24.10.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barmlage B, Luzi E, Eckstein F. HIV-1 LTR as a target for synthetic ribozyme-mediated inhibition of gene expression: site selection and inhibition in cell culture. Nucleic Acids Res. 2000;28:4059–4067. doi: 10.1093/nar/28.21.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]