Abstract
The GIY-YIG nuclease domain was originally identified in homing endonucleases and enzymes involved in DNA repair and recombination. Many of the GIY-YIG family enzymes are functional as monomers. We show here that the Cfr42I restriction endonuclease which belongs to the GIY-YIG family and recognizes the symmetric sequence 5′-CCGC/GG-3′ (‘/’ indicates the cleavage site) is a tetramer in solution. Moreover, biochemical and kinetic studies provided here demonstrate that the Cfr42I tetramer is catalytically active only upon simultaneous binding of two copies of its recognition sequence. In that respect Cfr42I resembles the homotetrameric Type IIF restriction enzymes that belong to the distinct PD-(E/D)XK nuclease superfamily. Unlike the PD-(E/D)XK enzymes, the GIY-YIG nuclease Cfr42I accommodates an extremely wide selection of metal-ion cofactors, including Mg2+, Mn2+, Co2+, Zn2+, Ni2+, Cu2+ and Ca2+. To our knowledge, Cfr42I is the first tetrameric GIY-YIG family enzyme. Similar structural arrangement and phenotypes displayed by restriction enzymes of the PD-(E/D)XK and GIY-YIG nuclease families point to the functional significance of tetramerization.
INTRODUCTION
The Type II restriction endonucleases (REases) recognize short DNA sequences and cut phosphodiester bond at fixed position within or close to their recognition sites (1) [variable cuts, however, have been reported recently (2)]. Four families of REases have been described to date based on the presence of conserved amino acids residues: PD-(E/D)XK, PLD, HNH and GIY-YIG (3–5). PabI restriction enzyme that shows a novel nuclease fold according to the recently published crystal structure (6) may become a founding member of yet another nuclease family.
Among the REase families, the enzymes of PD-(E/D)XK clan are best characterized both structurally and with respect to the mechanism of DNA cleavage. Enzymes belonging to the latter family are Mg2+-dependent endonucleases that use acidic residues from the conserved PD-(E/D)XK motif to coordinate Mg2+-ions, which are necessary cofactor for catalysis to occur. Many PD-(E/D)XK family enzymes, like EcoRI or EcoRV, are homodimers that interact with palindromic DNA sequences and contain two distinct sites each responsible for catalyzing cleavage in one DNA strand (1). A number of PD-(E/D)XK family REases (e.g. SfiI, Cfr10I, NgoMIV and Bse634I) are arranged as tetramers that simultaneously bind two recognition sites and cleave concertedly four DNA strands (7–11). MspI, HinP1I, BcnI and MvaI restriction enzymes, however, act as monomers and use a single active site to cleave both DNA strands (12–15).
In contrast to the Mg2+-dependent endonucleases of PD-(E/D)XK family, the BfiI and BmrI restriction enzymes (3,16) do not require metal ions for catalysis and belong to the phospholipase D (PLD) superfamily that includes diverse enzymes that act on the phosphodiester bonds in various contexts (17). BfiI is arranged as a dimer and requires two recognition sites for DNA cleavage (18,19), however it uses a single active site to cut sequentially both DNA strands and employs a covalent DNA intermediate in catalysis (20,21).
Comparative amino acid analysis and mutational data show that KpnI, MnlI and Eco31I REases (22–24) are members of the HNH nuclease family that is a part of a wider group of enzymes called ββα-Me endonucleases, which also includes the His-Cys box enzymes (25). The HNH module is found in diverse proteins, including bacterial colicins E7 and E9 and homing endonucleases (26,27). Catalytic activity of MnlI REase is supported by a wide range of divalent metal ions, including Mg2+, Mn2+, Ca2+, Co2+, Zn2+ and Ni2+ (28). Interestingly, in the presence of Mg2+ ions, the KpnI REase displays promiscuous DNA cleavage activity that is suppressed by Ca2+ that abolishes DNA cleavage by the PD-(E/D)XK REases (29).
Protein sequence analysis predicted that Eco29kI, MraI and NgoMIII REases specific for the 5′-CCGC/GG-3′ sequence are members of the GIY-YIG nuclease family (4). Proteins belonging to this family share a well-defined ∼100 aa nucleolytic domain that harbours in the N-terminal part conserved triplets ‘GIY’ and ‘YIG’ (30). GIY-YIG nuclease domain is typically embedded within large modular proteins of diverse functions including homing endonucleases, transposases, DNA repair enzymes and proteins involved in maintenance of genome stability (31). Three-dimensional structures of GIY-YIG nuclease domain of homing endonuclease I-TevI and the catalytic domain of UvrC protein (32,33) revealed a single active site consisting of conserved arginine, tyrosine and glutamic acid residues. Based on crystallographic data, a reaction mechanism for the GIY-YIG nucleases has been proposed (33); however, it yet needs to be confirmed by kinetic and biochemical studies that are hindered by complex multi-domain organization of GIY-YIG family proteins and elaborate requirements for catalysis.
Due to the simple structural organization, REases of GIY-YIG family are a suitable model system to study the mechanistic and structural features of GIY-YIG nuclease domain. Indeed, Eco29kI REase that belongs to the GIY-YIG family (5) is a monomer in solution (34). Modelling studies suggest that in contrast to other GIY-YIG proteins that show modular architecture, Eco29kI is a single domain protein (4,5). The stoichiometry of DNA binding and mechanism of double-stranded DNA cleavage by Eco29kI yet has to be determined.
Restriction endonuclease Cfr42I (http://rebase.neb.com/rebase/rebase.html) like Eco29kI recognizes the palindromic sequence 5′-CCGC/GG-3′ and shares 32% overall sequence identity with Eco29kI (Figure 1). Moreover, all amino acid residues from the predicted active site of Eco29kI (5) are conserved in Cfr42I indicating that it also belongs to the GIY-YIG superfamily. We show here that Cfr42I restriction enzyme is functional as a tetramer and requires binding of two DNA copies for its optimal activity. We also demonstrate that Cfr42I is functional in the presence of a wide range of metal-ion cofactors providing novel insight into the mechanism of the GIY-YIG family enzymes.
Figure 1.
Alignment of Cfr42I and Eco29kI sequences. Identical amino acid residues (32%) are in black boxes, similar residues (32%) are shaded grey. Asterisks (*) mark amino acid residues Y49, R104, H108, E142 and N154 of Eco29kI that are critical for its catalytic activity and presumably form the active site of Eco29kI (5).
MATERIALS AND METHODS
Cloning, expression and purification of Cfr42I
Plasmid vector carrying the 8.4 kb chromosome fragment of Citrobacter freundii RFL42 strain carrying genes of the Cfr42I R-M system was kindly provided by Fermentas UAB (Vilnius, Lithuania). R.Cfr42I (REase) and M.Cfr42I (MTase) genes were cloned into the pET21b(+) and pACYC184 vectors, respectively. The Cfr42I protein was expressed in the BL21 (DE3) Escherichia coli strain that carries an additional laqIr gene in the pVH1 plasmid (35) and was purified by subsequent chromatography on Heparin Sepharose, Q-Sepharose (GE Healthcare) and phosphocellulose P11 (Whatman) columns using linear NaCl gradients for protein elution. Fractions containing Cfr42I REase activity were pooled, dialyzed against storage buffer [10 mM Tris-HCl (pH 7.4 at 25°C), 0.2 M KCl, 1 mM EDTA, 1 mM DTT, 50% (v/v) glycerol] and stored at −20°C. The protein was >90% pure as judged by SDS-PAGE.
Concentrations of Cfr42I were determined by absorbance at 280 nm using an extinction coefficient of 132 960 M−1 cm−1 for the tetramer. Enzyme concentrations are expressed in terms of tetramer if not stated otherwise.
Analytical ultracentrifugation
Equilibrium sedimentation experiments were done with an AN50Ti rotor in a Beckman–Coulter model XL-A analytical ultracentrifuge equipped with UV absorption detection. Six-channel centrepieces were used at 11 000 and 18 000 rpm, 20°C. Samples were spun until no change in absorbance profiles could be observed for at least 12 h at which time equilibrium was assumed to have been reached. Molar masses were evaluated from an average of the concentration gradients observed in these last 12 h as described (10).
Oligonucleotide substrates
All double-stranded oligonucleotide substrates used in this study are given in Table 1. The Cfr42I recognition sequences are underlined. ‘P’ denotes the 5′-terminal phosphate, and ‘bio-’ is the 5′-terminal biotin modification. All oligonucleotides were purchased from Metabion (Martinsried, Germany).
Table 1.
Oligonucleotide substrates
| Duplex | Sequence | Specification |
|---|---|---|
| 30/30 | 5′-AGACCCACGCCACCGCGGTGGAGATTACGG-3′ | 30 bp cognate substrate for Cfr42I (recognition sequence underlined) |
| 3′-TCTGGGTGCGGTGGCGCCACCTCTAATGCC-5′ | ||
| 16/16 | 5′-CGCCACCGCGGTGGCG-3′ | 16 bp cognate substrate for Cfr42I |
| 3′-GCGGTGGCGCCACCGC-5′ | ||
| 30-30 Bio-30-30 | 5′-AGACCCACGCCACCGCGGTGGAGATTACGG\ 3′-TCTGGGTGCGGTGGCGCCACCTCTAATGCC/ | 30 bp cognate hairpin substrate. Bio-30-30 has the same sequence, as shown, but with a biotin tag at the 5′ end |
| 30/(14p_16) | 5′-AGACCCACGCCACC-GCGGTGGAGATTACGG-3′ 3′–TCTGGGTGCGGTGGp CGCCACCTCTAATGCC-5′ | Duplex made from 3 oligos to give a nicked DNA akin to the Cfr42I product on one strand of 30/30; the nucleotide adjacent to the nick is 5′-phosphorylated, to mimic the terminus left by Cfr42I |
| 16/14p | 5′-AGACCCACGCCACC-GC pGGTGGAGATTACGG-3′ 3′–TCTGGGTGCGGTGGp CG-CCACCTCTAATGCC-5′ | Mixture of two 3′-tailed duplexes akin to the final Cfr42I product on the 30/30 substrate. 5′-termini of both duplexes are phosphorylated to mimic the Cfr42I products. |
| NS | 5′-CAGCACAGTTCAGCAGCCCAGTGCTACGCT-3′ 3′-GTCGTGTCAAGTCGTCGGGTCACGATGCGA-5′ | 30 bp non-cognate duplex used in gel-shift and immobilized DNA cleavage experiments |
| Bio-NS | 5′-AGACCCACGCTCACCGGTGAGAGATTACGT\ 3′-TCTGGGTGCGAGTGGCCACTCTCTAATGAT/ | 30 bp non-cognate hairpin substrate with a biotin tag at the 5′ end, used in DNA pull-down experiments |
Some duplexes were made by annealing two oligodeoxyribonucleotides with complementary sequences (30/30, 16/16, NS). Hairpin duplexes (30-30, bio-30-30 and Bio-NS) were made by annealing of the self-complementary oligonucleotides. Oligoduplex substrate containing a nick [30/(14p_16)], was made of three oligonucleotides: one 30 nt, one 16 nt and one 14 nt, while product DNA was made of four nucleotides: two 16 nt and two 14 nt. The 14 nt strand of the nicked and product duplexes was 5′-phosphorylated with ATP and T4 polynucleotide kinase to mimic the termini left by Cfr42I.
Radioactive labels were introduced either at the 5′ or at the 3′-ends of individual DNA strands prior to the annealing with unlabelled strands as described in (20).
Reactions with oligonucleotide substrates and different metal-ion cofactors
Reactions were typically carried out by preincubation of 50 nM of Cfr42I with 20 nM labelled oligoduplex in the Reaction Buffer (50 mM Tris-HCl pH 7.75 at 25°C, 100 mM NaCl, 0.1 mg/ml BSA and 0.01–0.1 mM EDTA) at 25°C. Reactions were initiated by addition of divalent metal ions (MgCl2, MnCl2, CoCl2, CaCl2, ZnCl2, NiCl2 and CuSO4) to a final concentration of 0.1, 1.0 or 10 mM. To equalize the ionic strength in all experiments, cleavage reactions with 0.1–1.0 mM cofactor concentrations were supplemented with extra 26–30 mM of NaCl.
Rapid DNA hydrolysis reactions with Mg2+, Mn2+ and Co2+ were studied in a quench-flow device (KinTek Corporation, Austin, TX), which was used to mix equal volumes of metal-ion and enzyme-DNA solutions (16 μl each). After the requisite time delay, the reactions were quenched with 2.0 M HCl. Immediately after quenching samples were neutralized by adding 3.5 M Tris and 3% SDS, mixed with loading dye solution (95% v/v formamide, 0.01% bromphenol blue, 25 mM EDTA, pH 8.0) and subjected to denaturing gel electrophoresis followed by phosphorimager detection as described in (20). Slower reactions with Zn2+, Ni2+, Ca2+ and Cu2+ where performed by manual mixing of metal cofactor and enzyme-DNA solutions. Samples were collected at timed intervals, quenched by mixing with loading dye solution and analysed as mentioned above.
Analysis of oligonucleotide hairpin cleavage
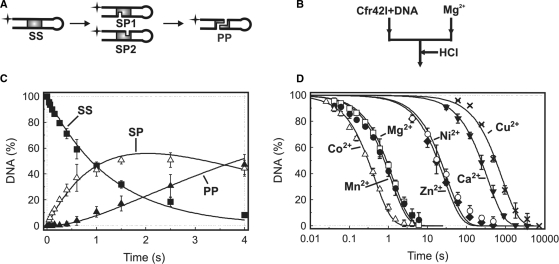
Cfr42I reaction on the 5′-radiolabelled 30 bp oligonucleotide hairpin 30-30 (Table 1) yields two shorter-labelled products of 44 and 16 nt (Figure 7A) that can be resolved by denaturing PAGE. Quantification of intact substrate and shorter products allows to evaluate amounts of intact substrate SS, nicked reaction intermediate SP and the final reaction product with a double-strand break PP as described in (11). The hairpin DNA cleavage data were described by the consecutive reaction scheme (1) as described in (36).
Figure 7.
Kinetic analysis of DNA cleavage by Cfr42I. (A) Schematic representation of the 30-30 hairpin DNA cleavage by restriction endonuclease Cfr42I. SS is the intact hairpin substrate, SP1 and SP2 are nicked duplexes cut at top or bottom DNA strands, respectively, PP is a final reaction product cleaved at both strands. (B) Setup of the quench-flow DNA cleavage experiments. A quench-flow device was used to mix equal volumes of preincubated enzyme-DNA and metal cofactor solutions. The reactions were quenched at timed intervals with 2.0 M HCl. (C) Hairpin DNA hydrolysis in the presence of Mg2+ ions. Final reaction mixtures at 25°C contained 20 nM cognate hairpin duplex (30-30, Table 1) and Cfr42I (50 nM of tetramer) in 50 mM Tris-HCl (pH 7.75 at 25°C), 100 mM NaCl, 0.1 mg/ml BSA and 10 mM MgCl2. After quenching the samples were analysed as described in ‘Materials and Methods’ to determine the amounts of the following DNA forms: intact hairpin substrate SS (filled squares), nicked duplex SP (open triangles) and final reaction product PP (filled triangles). All data points are presented as mean values from ≥3 repetitions ± 1SD. Continuous lines are the fit of Equation (1) to experimental data. The best fit gave k1(Mg2+) = 0.78 ± 0.05 s−1 and k2(Mg2+) = 0.29 ± 0.05 s−1. (D) DNA nicking by Cfr42I in the reactions with 10 mM of Mg2+ (open squares), 10 mM Mn2+ (filled circles), 10 mM Co2+ (open triangles), 10 mM Ca2+ (filled inverted triangles), 1 mM Cu2+ (crosses), 0.1 mM Zn2+ (filled diamonds) and 0.1 mM Ni2+ (open circles). Continuous lines are the fits of Equation (1) to the intact substrate SS depletion data. Determined rate constants are summarized in Table 2.
Reactions with immobilized oligonucleotide substrates
Streptavidin-coated magnetic beads (Promega, Madison, WI, US, binding capacity 1 nM/mg) were added to 80 μl bio-30-30 DNA (1 nM) in sodium citrate buffer (75 mM NaCl/7.5 mM sodium citrate, pH 7.2) to a final concentration of 0.12 mg/ml, to give ≈10 pmol biotin-binding sites on the beads for 0.08 pmol biotinylated oligonucleotide. After 5 min at 25°C, the beads were washed twice with sodium citrate buffer and resuspended in the Reaction Buffer with 10 mM MgCl2 before adding Cfr42I to a final concentration of 25 nM. In oligonucleotide activation experiments, the reactions were also supplemented with 50 nM of non-biotinylated oligonucleotides 30/30, NS, 30/(14p_16) and 16/14p (Table 1). Sample collection and analysis were as described in (36).
Biotin pull-down assay
The reactions at 25°C contained 10 nM of cognate biotinylated duplex bio-30-30, 10 nM of radiolabelled cognate duplex 30/30 and 0–250 nM Cfr42I enzyme in the Reaction Buffer supplemented with 0.1 mM EDTA. After 5 min at 25°C, 5 μl of a suspension of 0.5 mg/ml streptavidin-coated magnetic beads (binding capacity 1 nM/mg) were added. All further procedures were as described in (19).
DNA-binding studies
DNA binding by Cfr42I was analysed by the gel mobility-shift assay, using 30 bp 33P-labelled non-specific duplex NS and unmodified cognate duplex 30/30 (Table 1). Different amounts of protein in 20 μl of the Binding Buffer (30 mM MES, 30 mM His (pH 6.5 at 25°C), 0.1 mM EDTA, 0.1 mg/ml BSA and 10 (v/v) glycerol) were incubated with cognate or non-cognate oligoduplexes (1 nM) for 15 min at room temperature. Free DNA and protein-DNA complexes were resolved by native PAGE. The electrophoresis buffer was identical to the Binding Buffer except that it lacked BSA and glycerol. In protein-DNA complex composition analysis Cfr42I (12.5 nM of tetramer) was added to 30 bp or 16 bp 33P-labelled specific DNA duplexes (30/30 and 16/16, Table 1) alone or to the duplex mixtures in ratios varying from 9 : 1 to 1 : 9, keeping the total duplex concentration fixed at 20 nM.
Plasmid substrates
Plasmids pBRCFR-1 (one Cfr42I site) was derived from plasmid vector pBR322 (no Cfr42I sites) by inserting oligoduplex 30/30 (Table 1) through the NdeI site. Plasmid pBRCFR-2 (two Cfr42I sites) was derived from pBRCFR-1 by inserting a second copy of the 30/30 oligonucleotide through the Eco32I site. The supercoiled form of the substrates required for kinetic studies was purified by electrophoresis through agarose. Reaction mixtures typically contained 2.5 nM supercoiled substrate and 50 nM Cfr42I tetramer in the Reaction Buffer at 25°C. For the oligonucleotide activation experiments, the cognate and non-cognate duplexes 30/30 and NS (Table 1) were also added to a final concentration of 50 nM. DNA cleavage was initiated by mixing MgCl2 solution with the solution of preincubated enzyme and DNA. Reactions with the one-site plasmid were performed by manual mixing, experiments with the two-site plasmid employed the quench-flow equipment (see above). In all cases the reactions were quenched with 6 M guanidinium chloride, DNA was precipitated and analysed as described (19).
RESULTS
Cfr42I REase is a homotetramer
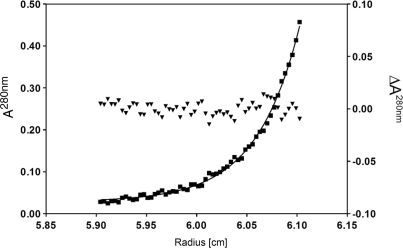
Most of the GIY-YIG family enzymes including Eco29kI REase are monomers (31,34). Surprisingly, sedimentation equilibrium analysis gives a molar mass of 99 kg/mol for Cfr42I (Figure 2). Since molar mass of a Cfr42I tetramer calculated from the amino acid sequence is 97.3 kg/mol, this shows the protein to be a tetramer. The residues of the least squares fit in Figure 2 show no indication of larger aggregates or smaller oligo/monomers. While tetrameric architecture is unusual for the GIY-YIG family enzymes, a number of PD-(E/D)XK family REases [Cfr10I, Bse634I, NgoMIV and SfiI (9,10,37,38)] are arranged as tetramers that simultaneously bind two recognition sites and cleave concertedly four DNA strands. Therefore, we have analysed if Cfr42I is able to bind simultaneously two DNA molecules forming a synaptic complex.
Figure 2.
Analytical ultracentrifugation of Cfr42I. Sedimentation diffusion equilibrium analysis of 3.2 µM Cfr42I (monomer) was performed at 18 000 rpm and 20°C in 0.01 M Tris-HCl pH 7.4, 0.2 M KCl, 0.1 mM EDTA. Filled squares denote the measured absorption (A280nm, left ordinate). The solid line shows the best least squares fit to the data for a single species with a molar mass of 99 ± 3 kg/mol (cf. (10)). Filled triangles denote the residues of that fit (ΔA280nm, right ordinate).
Cfr42 binds two DNA molecules according to the gel mobility shift assay
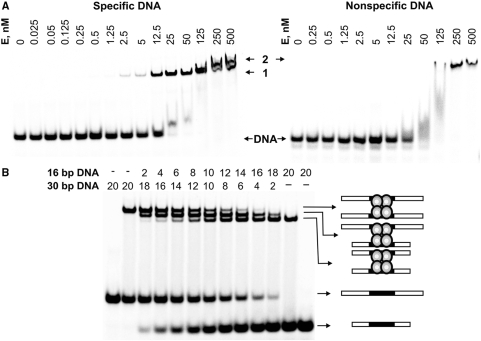
Gel mobility shift assay using 33P-labelled specific (30/30) and non-specific (NS) oligoduplexes (Table 1) revealed that Cfr42I shows different binding patterns to DNA duplexes containing and lacking the recognition site (Figure 3A). Indeed, Cfr42I binding to the cognate 30/30 oligoduplex yielded a single DNA-protein complex at low protein concentrations (band ‘1’ in Figure 3A) that was further converted into the second complex of lower electrophoretic mobility at protein concentrations >250 nM (band ‘2’ in Figure 3A). The NS oligoduplex formed only the latter slowly migrating complex, suggesting that band ‘2’ observed in the experiments with cognate DNA at highest protein concentrations may be due to the non-specific Cfr42I–DNA interactions (Figure 3A). Alternatively, the slowly migrating complex formed at high enzyme to DNA ratio can be the Cfr42I tetramer bound to one DNA duplex similarly to SfiI and BfiI REases (19,39). Hence, Cfr42I discriminates between cognate and non-cognate DNA even in the absence of divalent metal ions. This distinguishes Cfr42I from many PD-(E/D)XK REases that form specific complexes only in the presence of metal cofactor (40–43).
Figure 3.
DNA binding by Cfr42I endonuclease. (A) DNA-binding analysis by gel mobility-shift assay. The reactions contained 1 nM of the 33P-labelled specific oligoduplex 30/30 or the non-specific oligonucleotide NS (see Table 1 for sequence details), and the protein at concentrations (in terms of nM tetramer) as indicated above each lane. After 15 min at room temperature, the samples were subjected to PAGE for 2 h and analysed as described in ‘Materials and Methods’. (B) Analysis of the Cfr42I-DNA complex. Cfr42I (12.5 nM of tetramer) was added to the 30/30 or 16/16 33P-labelled specific DNA duplexes (Table 1) alone or to the duplex mixtures in ratios varying from 9:1 to 1:9 keeping the total DNA concentration fixed at 20 nM. Concentrations of both duplexes in the reactions are indicated above each line. The extreme left and right gel lanes contained no protein. After 15 min at room temperature, the samples were subjected to PAGE for 3 h and analysed as described in ‘Materials and Methods’. The cartoons illustrate protein-DNA complexes that correspond to each band.
To determine the number of DNA molecules in the specific complex ‘1’, we performed gel mobility-shift experiments with two cognate oligoduplexes of different length (Figure 3B). This approach was used previously to determine DNA-binding stoichiometry of the tetrameric PD-(E/D)XK REases SfiI and Bse634I (36,39). Cfr42I protein was incubated with 16 bp and 30 bp radiolabelled oligoduplexes (16/16 and 30/30, respectively; Table 1) either individually or mixed together at varying ratios but at fixed total DNA concentration of 20 nM. To avoid formation of the second slowly migrating complex (Figure 3A), the protein tetramer concentration was fixed at 12.5 nM. The mixtures of 16 bp and 30 bp duplexes and Cfr42I produced three different protein-DNA complexes (Figure 3B), two of which migrated as the individual complexes with 16 bp or 30 bp oligoduplexes, while the third one had an intermediate mobility. Formation of the intermediate complex can be explained only if the tetrameric Cfr42I REase simultaneously binds one 16 bp and one 30 bp cognate DNA molecules. Thus, individual complexes with 16 bp and 30 bp duplexes should also contain two copies of cognate DNA bound per protein molecule. Taken together, gel mobility shift experiments demonstrate that the Cfr42I tetramer binds two copies of cognate DNA.
Pull-down assay shows formation of the synaptic complex
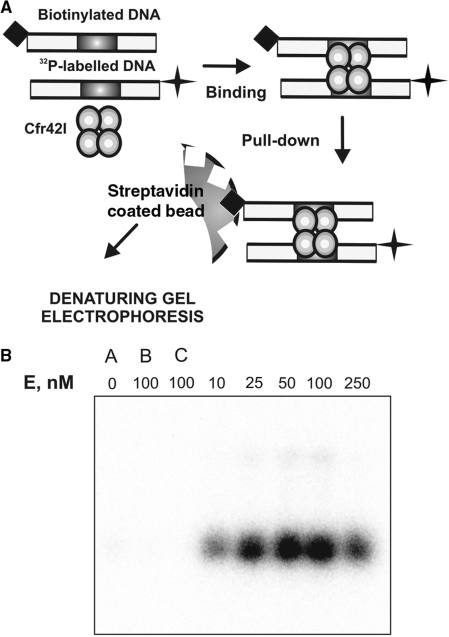
Interaction of Cfr42I with two DNA molecules was independently analysed by the DNA pull-down assay (19,44,45). The experimental setup of DNA pull-down assay is presented in Figure 4A. An oligoduplex containing a single recognition site for Cfr42I and a biotin tag at its 5′-end (bio-30-30, Table 1), was mixed with another cognate oligoduplex carrying a 32P radiolabel and incubated with various amounts of Cfr42I REase. The Cfr42I-DNA complexes containing biotinylated oligonucleotide were captured on streptavidin-coated magnetic beads. DNA recovered from the beads was then subjected to denaturing PAGE and the gels were analysed by phosphorescence, to quantify the 32P-labelled DNA pulled down together with the biotinylated DNA (Figure 4A and ‘Materials and Methods’). Amount of captured radiolabelled DNA is proportional to the concentration of synaptic complexes formed between Cfr42I, one biotinylated and one radiolabelled oligonucleotides. At all Cfr42I concentrations tested (10–250 nM of tetramer), significant amount of radiolabelled DNA was recovered from the beads (Figure 4B). The level of recovery increased with increasing concentrations of Cfr42I, rising to a maximum at 50–100 nM of protein and then declining at 250 nM concentration. It is likely that decreased recovery of the 32P-labelled DNA at highest Cfr42I concentrations tested is due to unfavourable enzyme to DNA ratio. At high Cfr42I concentrations, the synaptic complexes between one tetramer molecule and two DNAs are replaced by complexes with one tetramer molecule bound to a single DNA. Similar concentration dependence of the yield of synaptic complexes was reported previously for the BfiI REase (19). The high yield of synaptic complex recovered at the enzyme excess over the DNA (50–100 nM Cfr42I tetramer : 20 nM DNA) suggests that Cfr42I binds two copies of cognate DNA cooperatively.
Figure 4.
DNA synapsis by Cfr42I. (A) Schematic representation of the biotin pull-down assay. The reactions contained equimolar amounts of two specific 30 bp duplexes (10 nM each) with various amounts of Cfr42I. The first of the duplexes carried a biotin tag and the other was radiolabelled with 32P. After 5 min preincubation of enzyme and DNA, streptavidin-coated magnetic beads were added that adsorbed the biotin-tagged DNA. The beads were harvested and the amount of radiolabelled DNA associated with the beads was measured after denaturing PAGE. The radiolabelled DNA is pulled down with the beads only if Cfr42I forms complexes with two DNA molecules. (B) Results of the pull-down assay. Cfr42I concentrations were as indicated above each lane. The gel lanes marked ‘A’, ‘B’ and ‘C’ are control pull-down experiments performed in the absence of protein (lane ‘A’), in the presence of non-specific biotinylated DNA (lane ‘B’) or in the absence of biotinylated DNA (lane ‘C’).
Control experiments with no protein, no biotinylated DNA and with non-specific biotinylated DNA (Figure 4B, lanes ‘A’, ‘B’ and ‘C’, respectively) retrieved only trace amounts of labelled DNA, indicating that 32P-DNA recovered in the experiments described above was solely due to synaptic complexes formed between Cfr42I and two copies of cognate DNA. Thus, DNA pull-down experiments provide direct physical evidence for incorporation of two cognate DNA molecules into a synaptic complex with one Cfr42I molecule.
Synaptic Cfr42I complex with two DNA sites is functionally significant
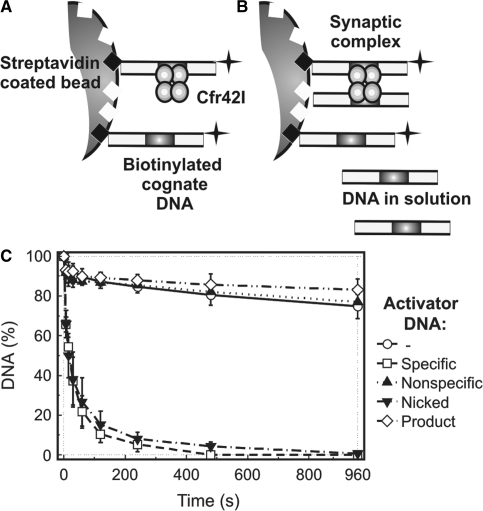
Type IIF restriction enzymes that belong to the PD-(E/D)XK family show optimal catalytic activity upon interaction with two copies of cognate DNA and display only low residual level of activity when bound to a lone copy of the recognition site (7,36,46,47). In order to find out if tetrameric Cfr42I REase that belongs to the GIY-YIG family requires synapsis of two DNA sites for its optimal catalytic activity, we employed immobilized oligonucleotide DNA cleavage assay (11,36,47). Experimental strategy of this method is depicted in Figure 5A and B. First, a 30 bp cognate hairpin duplex bio-30-30 (Table 1) that carries a centrally located Cfr42I site, a 32P-label at the 3′-terminus and a 5′-terminal biotin tag was immobilized on the surface of streptavidin-coated beads at a low density (see ‘Materials and Methods’). This prevents simultaneous binding of two DNA duplexes to a single Cfr42I molecule, therefore kinetic studies with surface-bound substrates reveal the catalytic activity of enzyme acting on a lone copy of cognate DNA (Figure 5A). Alternatively, addition of non-biotinylated DNA into the reaction mixture in trans may trigger formation of synaptic complexes between the enzyme, the immobilized DNA and the DNA in solution (Figure 5B) thereby stimulating cleavage of the surface-bound substrate.
Figure 5.
Immobilized DNA cleavage by Cfr42I. Cartoons in (A) and (B) depict the experimental strategy (36). (A) Oligonucleotide duplex bio-30-30 (Table 1), which carries a biotin at the 5′-end and a 32P label at the 3′ end, is immobilized on the streptavidin-coated magnetic beads at low density. These reaction conditions prevent formation of synaptic complexes between tetrameric REase and two DNA molecules and thus reveal catalytic activity of enzyme bound to a single DNA site. (B) Alternatively, addition of non-biotinylated oligonucleotide duplex (activator DNA) into the reaction mixture enables formation of synaptic complexes between the enzyme, the immobilized DNA and the activator DNA in solution. (C) Experimental results. Upon addition of 25 nM of tetrameric Cfr42I, cleavage of the immobilized hairpin duplex (∼1 nM) was monitored by removing samples at timed intervals (open circles) and analysing them as described in ‘Materials and Methods’. Cleavage of immobilized substrate was also performed in the presence of various non-biotinylated activator DNAs: 50 nM non-specific duplex NS (filled triangles), 50 nM cognate duplex 30/30 (open squares), 50 nM product DNA 16/14p (open diamonds) and 50 nM cognate nicked duplex 30/(14p_16) (inverted filled triangles) (see Table 1 for oligonucleotide sequences). All data points are presented as mean values from ≥3 independent experiments ±1SD.
Immobilized DNA hydrolysis experiments (Figure 5C) revealed that both in the absence of non-biotinylated DNA or in the presence of either non-specific duplex NS or product DNA 16/14p (Table 1), the Cfr42I reaction rates are extremely low (reaction half-time for immobilized DNA cleavage >4000 s). Thus, Cfr42I displays only trace catalytic activity when bound to a single DNA copy and does not form catalytically competent synaptic complexes between cognate immobilized DNA and non-specific or product DNA in solution (Figure 5C). On the other hand, specific oligoduplexes 30/30 (intact) and 30/(14p_16) (nicked) dramatically accelerate cleavage of the surface-coupled oligonucleotide (reaction half-time for immobilized DNA cleavage ∼20 s). Thus, Cfr42I forms catalytically competent synaptic complexes either between two intact specific duplexes or between one intact substrate and one nicked reaction intermediate (Figure 5C). In conclusion, experiments with immobilized DNA clearly demonstrate functional importance of the synaptic complex formed between the Cfr42I tetramer and the two copies of cognate DNA.
Functional significance of the synaptic complex formed by Cfr42I was also confirmed in a set of experiments using supercoiled plasmid substrates bearing one and two copies of the Cfr42I recognition site (Figure 6). It was shown previously that many REases that require two recognition sites for the optimal activity cleave the two-site plasmid much more rapidly than a single-site plasmid because synaptic complexes between two sites in cis are much more stable than in trans complexes between separate single-site plasmids (7,9,10,19). Indeed, Cfr42I cleaved the supercoiled form of the two-site plasmid substrate >400-fold faster than the single-site substrate (Figure 6A and B). Moreover, cleavage of the one-site plasmid by Cfr42I was significantly enhanced by addition of cognate oligoduplex into the reaction mix (data not shown), presumably due to formation of mixed synaptic complexes between the enzyme, the one-site plasmid and the oligoduplex.
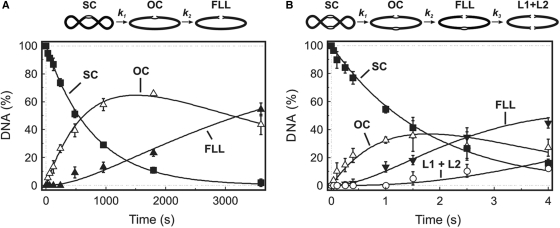
Figure 6.
Cfr42I reactions on plasmids with one or two recognition sites. The reactions were performed by mixing solution of MgCl2 (final concentration 10 mM) with the preincubated mixture of enzyme and plasmid DNA (final concentrations 50 nM enzyme tetramer and 2.5 nM DNA). The plasmids were pBRCFR-1 (one Cfr42I site) for (A), and pBRCFR-2 (two Cfr42I sites) for (B). Samples were quenched with 6 M guanidinium chloride and analysed as described in ‘Materials and Methods’ to determine the amounts of the following forms of the DNA: supercoiled DNA (SC), filled squares; open-circular DNA (OC), open triangles; linear DNA cut at one Cfr42I site (FLL), filled triangles; and, only in (B), linear DNA cut at both Cfr42I sites (L1 + L2), open circles. Cartoons above the graphs depict the two-step and three-step consecutive reaction schemes that were used to quantify the one-site and two-site plasmid DNA cleavage data as described (19). The solid lines in panel (A) show the best least squares fit of the two-step reaction scheme to the one-site plasmid data that gave k1 = 0.0013 ± 0.0001 s−1 and k2 = 0.0004 ± 0.0001 s−1. The solid lines in panel (B) show the best fit of the three-step scheme to the two-site plasmid data that gave k1 = 0.56 ± 0.02 s−1, k2 = 0.56 ± 0.03 s−1 and k3 = 0.17 ± 0.02 s−1.
Kinetic analysis of DNA hydrolysis by restriction endonuclease Cfr42I
In order to quantitatively describe the Cfr42I DNA hydrolysis reaction and to determine the reaction rates with different metal-ion cofactors, we performed transient kinetics experiments with limiting substrate and excess enzyme concentrations. To monitor the amounts of intact DNA duplex, nicked intermediate and the final reaction product cut at both strands, we employed the 30 bp hairpin DNA duplex 30-30 (Table 1), assembled by self-annealing of a 60 nt synthetic oligonucleotide. The Cfr42I recognition sequence 5′-CCGCGG-3′ was embedded in the centre of the hairpin and was symmetrically flanked by identical 3 bp DNA sequences. The 5′-end of the oligonucleotide was used for radioactive labelling. Cleavage of the 5′-radiolabelled 60 nt DNA hairpin by Cfr42I (Figure 7A) generates 16 and 44 nt radiolabelled products that can be separated by denaturing PAGE. Quantification of intact hairpin DNA and the cleavage products allows direct monitoring of concentrations of the substrate (SS), the nicked intermediate (SP) and the final reaction product (PP) as described in ‘Materials and Methods’.
The DNA hydrolysis reactions were initiated by adding metal-ion cofactor solution to the pre-incubated mixture of Cfr42I and DNA at concentrations (50 nM Cfr42I tetramer and 20 nM oligoduplex) favouring synaptic complex formation (Figure 4B). Addition of the metal ion to the pre-formed Cfr42I-DNA complex triggers DNA cleavage and reaction rates measured in such experimental setup most likely correspond to the chemical step. Due to high reaction rates, oligonucleotide cleavage experiments with Mg2+ were performed in a quench-flow device that was used to mix equal volumes of metal-ion and Cfr42I-DNA solutions. The reactions at timed intervals were quenched with 2.0 M HCl, neutralized and analysed as described in ‘Materials and Methods’.
The time course of hairpin DNA cleavage in the presence of 10 mM Mg2+ shows (Figure 7B) that Cfr42I forms a significant amount of nicked DNA intermediate. Thus, Cfr42I similarly to many PD-(E/D)XK family REases, cleaves double-stranded DNA substrate sequentially, by first converting oligoduplex into a nicked reaction intermediate and only then cleaving the second DNA strand (19,36,48–50). Such DNA hydrolysis reactions obey a simple Equation (1):
| (1) |
where SS, SP and PP are intact substrate, nicked intermediate and final product resulting from a double-strand break, k1 is the rate constant for cleavage of the first DNA strand (rate constant for DNA nicking) and k2 is the rate constant for cleavage of the second DNA strand. Fitting Equation (1) to experimental data in Figure 7B gave rate constants k1(Mg2+) = 0.78 ± 0.05 s−1 and k2(Mg2+) = 0.29 ± 0.05 s−1.
Sequential reaction pathway suggested for the hairpin DNA cleavage by Cfr42I is supported by plasmid DNA cleavage experiments (Figure 6A and B). Indeed, Cfr42I first cuts one strand of the supercoiled plasmid to generate nicked intermediate that is further converted to the linear reaction products. While the nicking and linearization rates of the single site plasmid are low, corresponding rates for the two-site plasmid (k1 = 0.56 ± 0.02 s−1, k2 = 0.56 ± 0.03 s−1, Figure 6B) are in reasonable quantitative agreement with the oligonucleotide cleavage data.
DNA cleavage with different divalent metal-ion cofactors
All Type II REases except BfiI (3) require divalent metal-ions as a cofactor for DNA cleavage. Restriction enzymes of the PD-(E/D)XK superfamily display optimal activity with Mg2+ that in some cases can be substituted with Mn2+ (or Fe2+, Co2+, Ni2+, Zn2+, Cd2+, depending on the enzyme), but not with Ca2+ (51). REases of the HNH family also depend on metal ions but may use a broad range of divalent cations including Ca2+. For example, MnlI is active with Mg2+, Mn2+, Ca2+, Zn2+, Ni2+ and Co2+ (28). Metal-ion requirements for catalysis by GIY-YIG family enzymes were not studied before. Therefore, we monitored DNA cleavage by Cfr42I in the presence of various metal ions (Ca2+, Co2+, Zn2+, Cu2+ and Ni2+) (see, ‘Materials and Methods’ for the details). It turned out, that Cfr42I cleaved DNA with all divalent metal ions tested. In all cases the DNA cleavage reactions followed the consecutive reaction scheme (1). DNA cleavage profiles and rate constants determined with different cofactors are summarized in Figure 7C and Table 2, respectively.
Table 2.
DNA cleavage by Cfr42I with different metal-ion cofactors
| Metal-ion cofactor | k1, s−1a | k2, s−1b |
|---|---|---|
| Mg2+ (10 mM) | 0.78 ± 0.05 | 0.29 ± 0.05 |
| Co2+ (10 mM) | 2.40 ± 0.04 | 0.37 ± 0.03 |
| Mn2+ (10 mM) | 0.95 ± 0.06 | 0.17 ± 0.01 |
| Ca2+ (10 mM) | 0.0027 ± 0.0002 | 0.0020 ± 0.0003 |
| Cu2+ (1 mM) | 0.0010 ± 0.0001 | 0.0003 ± 0.0001 |
| Zn2+ (0.1 mM) | 0.036 ± 0.001 | 0.017 ± 0.002 |
| Ni2+ (0.1 mM) | 0.026 ± 0.002 | 0.010 ± 0.002 |
Highest rates are achieved at 10 mM concentrations of Co2+, Mn2+ and Mg2+. Decreasing of Co2+, Mn2+ and Mg2+ concentrations to 1 mM resulted in a 2–5-fold drop in the reaction rates (data not shown), indicating that equilibrium dissociation constants for these cofactors in the active site of Cfr42I are >1 mM. DNA cleavage in the presence of 10 mM Ca2+ was 300-fold slower in comparison to the reaction in the buffer supplemented with 10 mM Mg2+ (Table 2 and Figure 7C). Decrease of the Ca2+ concentration to 1 mM decreased the reaction rate by 5-fold (data not shown). Zn2+, Cu2+ and Ni2+ at 10 mM concentrations completely abolished the catalytic activity of Cfr42I, however, the enzyme showed significant activity at 0.1–1.0 mM concentrations of these metal ions. The reaction rates determined at 0.1 mM of Zn2+ and Ni2+ are ∼30-fold lower compared to the reaction with 10 mM Mg2+, while reaction with 1 mM Cu2+ is comparable to the 10 mM Ca2+ reaction (Table 2 and Figure 7C). In conclusion, our data provide the first experimental evidence that GIY-YIG family nucleases are rather promiscuous regarding their divalent metal-ion cofactor requirement.
DISCUSSION
Cfr42I REase––a novel member of GIY-YIG family
Cfr42I recognizes symmetrical sequence 5′-CCGC/GG-3′ and cleaves it yielding 2 nt 3′-overhangs. Protein sequence analysis reveals that Cfr42I like Eco29kI REase is a member of the GIY-YIG family (Figure 1). Proteins belonging to the GIY-YIG family share a well-defined ∼100 aa nucleolytic domain (30) that is embedded into a different structural contexts to generate proteins with diverse functions. Many of the GIY-YIG family enzymes are monomers built of separate modules (31). For example, GIY-YIG homing endonucleases consist of the N-terminal nuclease domain connected to variable C-terminal DNA-binding domains (32,52,53). Unlike homing endonucleases of the GIY-YIG family, the Eco29kI restriction enzyme is a single-domain protein that has the conserved GIY-YIG nuclease core and presumably uses loop insertions and terminal extensions instead of the separate DNA-binding domain to interact with the target site 5′-CCGC/GG-3′ (5). Protein sequence similarities (Figure 1) suggest the similar structure for the Cfr42I REase.
Tetrameric architecture of Cfr42I REase
Surprisingly, in solution Cfr42I is a homotetramer, which interacts simultaneously with two copies of specific DNA (Figures 3B and 4). To our knowledge, this is the first tetrameric GIY-YIG family enzyme: all previously characterized family members were monomers (31). Moreover, the Eco29kI REase that shares ∼30% of identical residues with Cfr42I and recognizes the same DNA sequence 5′-CCGC/GG-3′ is a monomer in solution (34). A monomeric architecture, however, causes a problem for the REase that has to interact with a symmetrical recognition site and make a double-strand break using a single active site.
The PD-(E/D)XK superfamily REases solved this problem using two different strategies. First, monomeric Type IIP REases use a single active site to cut both strands sequentially (54). Monomeric homing endonucleases of the GIY-YIG family may also employ the same strategy (55,56). Second, many Type IIP REases interact with symmetrical target sites as homodimers. Each monomer within a dimer binds to a half of the target site and uses a single active site within each monomer for catalyzing cleavage in one DNA strand. Moreover, some PD-(E/D)XK superfamily REases belonging to Type IIF are arranged as dimers of dimers (9,10,37,38). Each primary dimer within the tetramer is responsible for binding and cleavage of one cognate DNA molecule.
We propose that Cfr42I may have a similar arrangement. In this case Cfr42I monomer is responsible for recognition and cleavage of one half-site of the symmetric recognition site 5′-CCGC/GG-3′. Four subunits of the enzyme, therefore, can bind and introduce double-strand breaks into two cognate DNA molecules. The Eco29kI, unlike Cfr42I, has been reported to be a monomer in solution (34) raising a question that related REases Cfr42I and Eco29kI may use different strategies to cleave homologous recognition sites. One cannot exclude, however, that Eco29kI may change its oligomeric state upon DNA binding. Indeed, it has been reported that PD-(E/D)XK family REase SgrAI binds to the recognition site as a dimer and transiently makes a tetramer for DNA cleavage (57). On the other hand, it has been demonstrated that a single amino acid replacement can disrupt the tetramer structure. Indeed, W228A mutation transforms the Bse634I tetramer into a functional dimer (36), while R222A mutation converts Bse634I tetramer into monomers (11). Keeping in mind these data, the difference between oligomeric states of Cfr42I and Eco29kI seems not surprising.
Functional importance of the tetrameric structure
Biochemical experiments with plasmid and oligonucleotide substrates (Figures 5 and 6) clearly demonstrate the functional importance of the synaptic complex between the Cfr42I tetramer and two copies of cognate DNA. Indeed, Cfr42I displays only residual catalytic activity on a single copy of cognate DNA and requires two DNA copies for maximum activity (Figures 5 and 6). In that respect Cfr42I is virtually indistinguishable from the previously characterized PD-(E/D)XK family REases Bse634I, NgoMIV and SfiI that are homotetramers and require binding of two DNAs for the optimal catalytic activity (7,9,36). It would be interesting to see if Cfr42I tetramer has a similar structural arrangement and employ similar mechanism of communication proposed for the Bse634I and SfiI restriction enzymes (11,36,47).
The functional and structural similarity of the GIY-YIG family REase Cfr42I to the tetrameric PD-(E/D)XK family enzymes of the Type IIF subtype has important implications for our understanding of REase evolution and functions. Our studies demonstrate that despite of different evolutionary origins of the PD-(E/D)XK and GIY-YIG REases, some members of these families are homotetramers and require simultaneous binding of two cognate DNAs for catalytic activity. Similar structures and phenotypes displayed by REases belonging to the different nuclease families’ points to the functional significance of tetramerization, which is still poorly understood. It was suggested that tetramerization may be obligatory to stabilize the functional dimer of some REases like Bse634I (36) or contribute to the REase specificity due to the requirement to bind two recognition sites simultaneously (39). Both hypotheses, however, have their own limitations and may prompt new models to account for the evolutionary selection of tetrameric REases interacting with two copies of cognate DNA.
GIY-YIG family nuclease Cfr42I is active with a wide range of metal-ion cofactors
Crystal structures of catalytic GIY-YIG domains of UvrC and I-TevI nucleases revealed a single divalent cation (Mg2+ or Mn2+) coordinated by a single glutamate residue (E76 in case of UvrC from Thermotoga maritima, E75 in case of I-TevI) and a cluster of well-ordered water molecules bound to the metal ion (32,33). The metal-ion cofactor is presumably not directly involved in generation and orientation of the attacking water nucleophile but helps to stabilize the negative charge formed at the phosphorane transition state (33). Interestingly, the relative positions of active site residues R27, E75 and Y17 of I-TevI homing endonuclease, which belongs to the GIY-YIG family, are quite similar to conserved active-site residues of the His-Cys box enzyme I-PpoI (R61, N119 and H98, respectively) that is a member of the ββα-Me family (58). In addition, the position of bound Mn2+ in I-TevI is nearly identical to the site of the same bound metal ion in I-PpoI ternary complexes. The ββα-Me family enzymes show catalytic activity with a wide range of metal ions (22,28).
We demonstrate that GIY-YIG nuclease Cfr42I hydrolyzes DNA using a wide range of divalent metal-ion cofactors (Mg2+, Mn2+, Co2+, Zn2+, Ni2+, Ca2+ and Cu2+) (Figure 7C and Table 2), indicating that active site of Cfr42I is extremely tolerant to the size and coordination requirements of the metal ions. This observation is in agreement with the loose coordination of metal ions observed in the I-TevI and UvrC structures and the proposed catalytic role of the metal ion as Lewis acid (33). This similarity implies that the GIY-YIG nuclease domain might utilize a similar DNA-strand cleavage mechanism as the His-Cys box enzymes. However, a structure of a GIY-YIG domain bound to DNA is still required to confirm the reaction mechanism (33). Due to relatively simple structural organization, high solubility and simple requirements for DNA binding, Cfr42I is a promising target for crystallographic studies.
ACKNOWLEDGEMENTS
We are grateful to Fermentas UAB for the Cfr42I R-M system clone. We thank S. Klimašauskas for the opportunity to use the quench-flow equipment. Research in the VS laboratory was supported by an intramural grant from the Institute of Biotechnology and FP6 Marie Curie Research Training network ‘DNA enzymes’. The Cfr42I sequence has been deposited in GenBank under the accession number EU262735. Funding to pay the Open Access publication charges for this article was provided by FP6 Marie Curie Research Training project “DNA enzymes”.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 2.Sukackaite R, Lagunavicius A, Stankevicius K, Urbanke C, Venclovas C, Siksnys V. Restriction endonuclease BpuJI specific for the 5'-CCCGT sequence is related to the archaeal Holliday junction resolvase family. Nucleic Acids Res. 2007;35:2377–2389. doi: 10.1093/nar/gkm164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sapranauskas R, Sasnauskas G, Lagunavicius A, Vilkaitis G, Lubys A, Siksnys V. Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J. Biol. Chem. 2000;275:30878–30885. doi: 10.1074/jbc.M003350200. [DOI] [PubMed] [Google Scholar]
- 4.Bujnicki JM, Radlinska M, Rychlewski L. Polyphyletic evolution of type II restriction enzymes revisited: two independent sources of second-hand folds revealed. Trends Biochem. Sci. 2001;26:9–11. doi: 10.1016/s0968-0004(00)01690-x. [DOI] [PubMed] [Google Scholar]
- 5.Ibryashkina EM, Zakharova MV, Baskunov VB, Bogdanova ES, Nagornykh MO, Den'mukhamedov MM, Melnik BS, Kolinski A, Gront D, et al. Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct. Biol. 2007;7:48. doi: 10.1186/1472-6807-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyazono K, Watanabe M, Kosinski J, Ishikawa K, Kamo M, Sawasaki T, Nagata K, Bujnicki JM, Endo Y, et al. Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 2007;35:1908–1918. doi: 10.1093/nar/gkm091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wentzell LM, Nobbs TJ, Halford SE. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 8.Embleton ML, Siksnys V, Halford SE. DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol. 2001;311:503–514. doi: 10.1006/jmbi.2001.4892. [DOI] [PubMed] [Google Scholar]
- 9.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 10.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 11.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Allosteric communication network in the tetrameric restriction endonuclease Bse634I. J. Mol. Biol. 2006;363:800–812. doi: 10.1016/j.jmb.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 12.Xu QS, Kucera RB, Roberts RJ, Guo HC. An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site. Structure. 2004;12:1741–1747. doi: 10.1016/j.str.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Horton JR, Maunus R, Wilson GG, Roberts RJ, Cheng X. Structure of HinP1I endonuclease reveals a striking similarity to the monomeric restriction enzyme MspI. Nucleic Acids Res. 2005;33:1892–1901. doi: 10.1093/nar/gki337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokolowska M, Kaus-Drobek M, Czapinska H, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007;369:722–734. doi: 10.1016/j.jmb.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Kaus-Drobek M, Czapinska H, Sokolowska M, Tamulaitis G, Szczepanowski RH, Urbanke C, Siksnys V, Bochtler M. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007;35:2035–2046. doi: 10.1093/nar/gkm064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SH, Bao Y, Ciszak E, Laget S, Xu SY. Catalytic domain of restriction endonuclease BmrI as a cleavage module for engineering endonucleases with novel substrate specificities. Nucleic Acids Res. 2007;35:6238–6248. doi: 10.1093/nar/gkm665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting CP, Kerr ID. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagunavicius A, Sasnauskas G, Halford SE, Siksnys V. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003;326:1051–1064. doi: 10.1016/s0022-2836(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 20.Sasnauskas G, Halford SE, Siksnys V. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl. Acad. Sci. U.S.A. 2003;100:6410–6415. doi: 10.1073/pnas.1131003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasnauskas G, Connolly BA, Halford SE, Siksnys V. Site-specific DNA transesterification catalyzed by a restriction enzyme. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2115–2120. doi: 10.1073/pnas.0608689104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 2004;32:6129–6135. doi: 10.1093/nar/gkh951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriukiene E, Lubiene J, Lagunavicius A, Lubys A. MnlI––The member of H-N-H subtype of Type IIS restriction endonucleases. Biochim. Biophys. Acta. 2005;1751:194–204. doi: 10.1016/j.bbapap.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Jakubauskas A, Giedriene J, Bujnicki JM, Janulaitis A. Identification of a single HNH active site in type IIS restriction endonuclease Eco31I. J. Mol. Biol. 2007;370:157–169. doi: 10.1016/j.jmb.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 26.Pommer AJ, Cal S, Keeble AH, Walker D, Evans SJ, Kuhlmann UC, Cooper A, Connolly BA, Hemmings AM, et al. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 2001;314:735–749. doi: 10.1006/jmbi.2001.5189. [DOI] [PubMed] [Google Scholar]
- 27.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J. Mol. Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 28.Kriukiene E. Domain organization and metal ion requirement of the Type IIS restriction endonuclease MnlI. FEBS Lett. 2006;580:6115–6122. doi: 10.1016/j.febslet.2006.09.075. [DOI] [PubMed] [Google Scholar]
- 29.Chandrashekaran S, Saravanan M, Radha DR, Nagaraja V. Ca(2+)-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J. Biol. Chem. 2004;279:49736–49740. doi: 10.1074/jbc.M409483200. [DOI] [PubMed] [Google Scholar]
- 30.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat. Struct. Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 33.Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten, et al. Structural insights into the first incision reaction during nucleotide excision repair. Embo. J. 2005;24:885–894. doi: 10.1038/sj.emboj.7600568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertzev AV, Kravetz AN, Mayorov SG, Zakharova MV, Solonin AS. Isolation of a strain overproducing endonuclease Eco29kI: purification and characterization of the homogeneous enzyme. Biochemistry (Mosc) 1997;62:732–741. [PubMed] [Google Scholar]
- 35.Kupper D, Reuter M, Mackeldanz P, Meisel A, Alves J, Schroeder C, Kruger DH. Hyperexpressed EcoRII renatured from inclusion bodies and native enzyme both exhibit essential cooperativity with two DNA sites. Protein Expr. Purif. 1995;6:1–9. doi: 10.1006/prep.1995.1001. [DOI] [PubMed] [Google Scholar]
- 36.Zaremba M, Sasnauskas G, Urbanke C, Siksnys V. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: oligomeric structure-stability-function correlations. J. Mol. Biol. 2005;348:459–478. doi: 10.1016/j.jmb.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 37.Grazulis S, Deibert M, Rimseliene R, Skirgaila R, Sasnauskas G, Lagunavicius A, Repin V, Urbanke C, Huber R, et al. Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence. Nucleic Acids Res. 2002;30:876–885. doi: 10.1093/nar/30.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanamee ES, Viadiu H, Kucera R, Dorner L, Picone S, Schildkraut I, Aggarwal AK. A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA. Embo. J. 2005;24:4198–4208. doi: 10.1038/sj.emboj.7600880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Embleton ML, Williams SA, Watson MA, Halford SE. Specificity from the synapsis of DNA elements by the SfiI endonuclease. J. Mol. Biol. 1999;289:785–797. doi: 10.1006/jmbi.1999.2822. [DOI] [PubMed] [Google Scholar]
- 40.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 41.Lagunavicius A, Grazulis S, Balciunaite E, Vainius D, Siksnys V. DNA binding specificity of MunI restriction endonuclease is controlled by pH and calcium ions: involvement of active site carboxylate residues. Biochemistry. 1997;36:11093–11099. doi: 10.1021/bi963126a. [DOI] [PubMed] [Google Scholar]
- 42.Skirgaila R, Siksnys V. Ca2+-ions stimulate DNA binding specificity of Cfr10I restriction enzyme. Biol. Chem. 1998;379:595–598. [PubMed] [Google Scholar]
- 43.Tamulaitis G, Solonin AS, Siksnys V. Alternative arrangements of catalytic residues at the active sites of restriction enzymes. FEBS Lett. 2002;518:17–22. doi: 10.1016/s0014-5793(02)02621-2. [DOI] [PubMed] [Google Scholar]
- 44.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 45.Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 46.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 47.Bellamy SR, Milsom SE, Kovacheva YS, Sessions RB, Halford SE. A switch in the mechanism of communication between the two DNA-binding sites in the SfiI restriction endonuclease. J. Mol. Biol. 2007;373:1169–1183. doi: 10.1016/j.jmb.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright DJ, Jack WE, Modrich P. The kinetic mechanism of EcoRI endonuclease. J. Biol. Chem. 1999;274:31896–31902. doi: 10.1074/jbc.274.45.31896. [DOI] [PubMed] [Google Scholar]
- 49.Erskine SG, Baldwin GS, Halford SE. Rapid-reaction analysis of plasmid DNA cleavage by the EcoRV restriction endonuclease. Biochemistry. 1997;36:7567–7576. doi: 10.1021/bi970155s. [DOI] [PubMed] [Google Scholar]
- 50.Sasnauskas G, Jeltsch A, Pingoud A, Siksnys V. Plasmid DNA cleavage by MunI restriction enzyme: single-turnover and steady-state kinetic analysis. Biochemistry. 1999;38:4028–4036. doi: 10.1021/bi982456n. [DOI] [PubMed] [Google Scholar]
- 51.Pingoud A, Jeltsch A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 52.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. Embo. J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horton JR, Zhang X, Maunus R, Yang Z, Wilson GG, Roberts RJ, Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. Embo. J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter JM, Friedrich NC, Kleinstiver B, Edgell DR. Strand-specific contacts and divalent metal ion regulate double-strand break formation by the GIY-YIG homing endonuclease I-BmoI. J. Mol. Biol. 2007;374:306–321. doi: 10.1016/j.jmb.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 57.Daniels LE, Wood KM, Scott DJ, Halford SE. Subunit assembly for DNA cleavage by restriction endonuclease SgrAI. J. Mol. Biol. 2003;327:579–591. doi: 10.1016/s0022-2836(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 58.Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ, Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat. Struct. Biol. 1999;6:1096–1099. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]