Abstract
Azidothymidine (AZT, zidovudine) is one of the few nucleoside inhibitors known to inhibit foamy virus replication. We have shown previously that up to four mutations in the reverse transcriptase gene of simian foamy virus from macaque (SFVmac) are necessary to confer high resistance against AZT. To characterize the mechanism of AZT resistance we expressed two recombinant reverse transcriptases of highly AZT-resistant SFVmac in Escherichia coli harboring three (K211I, S345T, E350K) or four mutations (K211I, I224T, S345T, E350K) in the reverse transcriptase gene. Our analyses show that the polymerization activity of these mutants is impaired. In contrast to the AZT-resistant reverse transcriptase of HIV-1, the AZT resistant enzymes of SFVmac reveal differences in their kinetic properties. The SFVmac enzymes exhibit lower specific activities on poly(rA)/oligo(dT) and higher KM-values for polymerization but no change in KD-values for DNA/DNA or RNA/DNA substrates. The AZT resistance of the mutant enzymes is based on the excision of the incorporated inhibitor in the presence of ATP. The additional amino acid change of the quadruple mutant appears to be important for regaining polymerization efficiency.
INTRODUCTION
Foamy viruses belong to the retroviridae but follow a replication pattern unique among retroviruses: (i) reverse transcription occurs before the virus leaves the host cell, (ii) the pol-gene is expressed from an separate mRNA and (iii) the viral protease is not cleaved off the Pol-polyprotein (1,2). Only the integrase is removed from Pol (3). Thus, the FV reverse transcriptase (PR–RT) harbors a protease, polymerase and RNase H domain.
Apart form the nucleoside inhibitor tenofovir, only azidothymidine (AZT, zidovudine) is known to inhibit FV reverse transcriptase in vivo in cell culture assays at concentrations as low as 5 μM (4–6). We have shown recently that four point mutations involving the amino acids 211 (K211I), 224 (I224T) 345 (S345T) and 350 (E350K) located in the PR–RT gene are involved in AZT resistance of SFVmac. The fully resistant SFVmac virus harboring all four mutations was able to replicate in the presence of 1 mM AZT (7). While AZT resistance in HIV-1 is based on the excision of incorporated AZT-monophosphate (AZTMP), AZT-resistant HIV-2 can distinguish between AZT-triphosphate (AZTTP) and TTP during incorporation (8–11).
For FVs, the resistance mechanism is not known. To elucidate the mechanism of AZT resistance we set out to express partially and fully AZT-resistant SFVmac PR–RTs harboring either three or all four AZT resistance mutations in Escherichia coli. We were able to show that the mechanism of AZT resistance in SFVmac PR–RTs is based on AZTMP excision from a terminated primer in the presence of ATP. Although the resistant PR–RTs are impaired in their polymerase activities, the faster excision of AZTMP in the presence of ATP confers high resistance against AZT. The I224 mutation appears to be primarily important for regaining polymerization activities for efficient viral replication.
MATERIALS AND METHODS
Cloning, expression and purification of PR–RTs
The wild-type PR–RT gene was cloned into the vector pET28c (Novagen, Germany) via PCR amplification and by using the restriction sites XhoI and NcoI. The expressed proteins contain a 6× His tag at the C-terminus. To avoid degradation of the PR–RT by autocatalytic activity of the PR, a mutant enzyme was constructed harboring an active site mutation in the PR region (D24A), which leads to an inactive PR. Thus the AZT-resistant mutants also contain the D24A mutation in the PR. The AZT-resistance mutations were created by site-directed mutagenesis according to the QuickChange kit from Stratagene (Heidelberg, Germany). The following potentially AZT-resistant PR–RT mutants were obtained:
mt3: (D24A) K211I, S345T, E350K
mt4: (D24A) K211I, I224T, S345T, E350K
The activities of the mutants were compared to wild-type PR–RT either without or with the PR D24A mutation (WT and WT*, respectively).
The corresponding plasmids were transformed into the E. coli strain Rosetta (DE3) (Novagen, Germany). Expression of the PR–RT genes was induced at an optical density of the culture of ca. 0.8–1.0 at 600 nm by the addition of 0.2 mM IPTG and incubated further over night at 25°C. The enzymes were purified via Ni-affinity chromatography (HisTrap, GE Healthcare, Munich, Germany), followed by chromatography over a heparin column (HiTrap heparin, GE Healthcare, Munich, Germany). The integrity of the proteins was verified by peptide mass fingerprints (Zentrale Bioanalytik, Zentrum für Molekulare Medizin, Köln, Germany). The purity of the proteins was >95% as judged by SDS–PAGE.
Quantitative polymerization assay
RNA-dependent DNA polymerase activity was quantitated on a poly(rA)/oligo(dT)15 substrate (0.2 U/ml) (Roche Diagnostics GmbH, Mannheim, Germany) in a standard assay (30 μl reaction volume) as described previously (12,13) with 150 μM TTP and 41.7 Ci/ml [3H]TTP (49.9 Ci/mmol; MP Biomedicals Inc., Irvine, CA, USA) in reaction buffer [50 mM Tris/HCl, pH 8.0, 80 mM KCl, 6 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.05% Triton X-100]. Samples were pre-incubated for 2 min at 37°C. The reaction was started by the addition of 12 nM of PR–RT. After 10 min, 7.5 μl aliquots were taken out and spotted on DEAE filter paper and treated as described (12,13). Under these conditions 1 U of enzyme activity catalyzes the incorporation of 1 nmol of TTP into poly(rA)/oligo(dT)15 in 10 min at 37°C. For the determination of KM and vmax values, reactions were performed with increasing concentrations of TTP of 25, 50, 75 or 125 μM. KM– and vmax–values were calculated by linear regression using Eadie–Hofstee plots.
5′-end labeling of primers
One hundred picomoles of M13 primer (5′-GTAAAACGACGGCCAGT) or P30 primer (GCTCTAATGGCGTCCCTGTTCGGGCGCCTC) (IBA, Göttingen, Germany) was labeled with 60 μCi γ[32P]-ATP with 2 U T4 polynucleotide kinase (New England Biolabs, Frankfurt, Germany) for 1 h at 37°C. After inactivation of the kinase for 20 min at 65°C the primer was purified via a MicroSpin column (GE Healthcare, Munich, Germany).
Chain termination assay
Chain termination assays were performed using single-stranded M13mp18 (Roche Diagnostics GmbH, Mannheim, Germany). The 5′ 32P-labeled M13 primer was hybridized to a 1.2-fold molar excess of the M13 DNA in a buffer containing 50 mM Tris/HCl, pH 8.0 and 80 mM KCl by heating to 95°C for 2 min, followed by a transfer to a heating block at 70°C and slow cooling to room temperature. Reaction mixtures contained 6 nM of primer/template substrate (P/T), 85 nM of PR–RT, 150 μM of each dNTP and increasing concentrations of AZTTP (GeneCraft GmbH, Lüdinghausen, Germany) in a total volume of 10 μl. After a pre-incubation time of 5 min, reactions were carried out for 10 min at 37°C in reaction buffer (see above). Reactions were stopped by adding 10 μl of urea loading buffer [1 mM EDTA, 0.1% xylene cyanole, 0.1% bromophenol blue, 8 M urea in 1 × TBE (Tris/Borate/EDTA)] and analyzed by denaturing gel electrophoresis (10% polyacrylamide, 7 M urea). The reaction products were visualized by autoradiography or phosphoimaging and quantitated by densitometry using a phosphoimaging device (FLA 3000, raytest, Straubenhardt, Germany).
Fluorescence anisotropy measurements
Fluorescence equilibrium titrations were performed to determine the dissociation constants (KD) for nucleic acid binding with a 24/40-mer DNA/DNA or DNA/RNA P/T substrate with the following sequences for the primer 5′-ATCACCAGGAGAGGGGAAAGCGGA and template 5′-DY647-CTAATTCCGCTTTCCCCTCTCCTGGTGATCCTTTCCATCC (biomers.net GmbH, Ulm, Germany). The RNA template sequence was identical, containing U instead of T. The templates harbored the fluorescent dye DY647 at their 5′ ends. Titrations were performed in fluorescence buffer (50 mM Tris/HCl, pH 8.0; 80 mM KCl, 10 mM EDTA, 0.5 mM DTT) in a total volume of 1 or 2 ml using a 10 × 4 mm quartz cuvette (Hellma GmbH, Mühlheim, Germany). The excitation wavelength was at 552 nm, and the emission intensity was measured at 573 nm. Slit widths were set at 4.9 and 5.0 nm for excitation and emission, respectively. All anisotropy measurements were performed at 25°C with 15 nM of fluorescently labeled P/T using an L-format Jobin-Yvon Horiba Fluoromax fluorimeter equipped with an automatic titration device (Hamilton). Following sample equilibration, at least six data points with an integration time of 1 s were collected for each titration point.
Data fitting
Data were fitted to a two-component binding equation to determine the equilibrium dissociation constant (KD) using standard software. The anisotropy was calculated from:
| 1 |
where A, Acomplex and ARNA represent the anisotropy values and fcomplex, fRNA the fractional intensities. The change in fluorescence intensity has to be taken into account, so that the fraction bound is given by
| 2 |
with
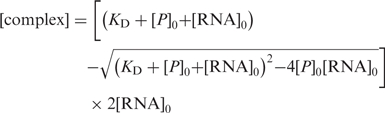 |
3 |
where A is the anisotropy, ARNA is the initial free anisotropy, Acomplex is the anisotropy of the protein–RNA complex and P0 and RNA0 represent the total protein and RNA concentrations, respectively. R is the ratio of intensities of the bound and free forms.
Termination of the radioactively labeled P/T with AZTTP
The [32P] end labeled P30 DNA primer was hybridized to a template deoxyoligonucleotide T50 (5′-GCTGTGGAAAATCTCATGCAGAGGCGCCCGAACAGGGACGCCATTACAGC) (IBA; Göttingen, Germany) as described for the M13 DNA and used for incorporation of AZTTP. P30/T50 measuring 100 nM were mixed with 100 μM AZTTP and 150 nM WT PR–RT in reaction buffer and incubated for 2 h at 37° C. After phenol extraction and ethanol precipitation, the P/T substrate was purified over two MicroSpin columns (GE Healthcare, Munich, Germany) to eliminate protein and excess AZTTP.
Excision assay
Ten nanomolar of the [32P] P30-AZTMP/T50 substrate were incubated with 20 nM PR–RT in a volume of 10 μl in reaction buffer for the times indicated. Either 150 μM of Na-pyrophosphate (PPi) or 5 mM of ATP was present in the mixture. Reactions were started by the addition of enzyme. Where stated, the samples were pre-incubated for 5 min with 0.02 U of pyrophosphatase (Sigma–Aldrich Chemie GmbH, Taufkirchen, Germany). When different concentrations of PR–RTs were tested, the reactions were stopped after 20 min. An equal volume of urea loading buffer was added and the products were analyzed as stated above on denaturing polyacrylamide urea gels.
Primer rescue
One hundred micromolar of dCTP, dGTP, TTP and ddATP was added to the samples with 5 mM ATP described above to allow for elongation by 4 nt once the AZTMP is excised. Samples were pre-incubated for 5 min with 0.02 U of pyrophosphatase before the reaction was started with 40 nM of PR–RT. Reactions were stopped after 10 min and treated further as described above.
RESULTS
We have shown previously that it is possible to generate AZT-resistant SFVmac in cell culture which is able to replicate in medium containing 1 mM AZT (7). Four mutations were necessary to confer high resistance to the virus: K211I, I224T, S345T and E350K. Since one published genomic sequence of wild-type SFVmac already harbors a threonine at position 224 of Pol and since several other primate FV Pol proteins also possess a threonine at position 224, the wild-type SFVmac might have a polymorphism at this site. Thus, we decided to analyze a triple mutant PR–RT lacking the I224T mutation [mt3; (D24A), K211I, S345T and E350K] as well as the quadruple mutant harboring I224T [mt4; (D24A), K211I, I224T, S345T and E350K]. To avoid autoprocessing of the PR domain, the enzymes also contained a D24A amino acid exchange in the active site of the PR. Our data below indicate that this mutation does not influence the polymerization activities of the mutants. The purified enzymes were used to determine kinetic parameters of polymerization and to analyze the AZT resistance mechanism.
Polymerization activities
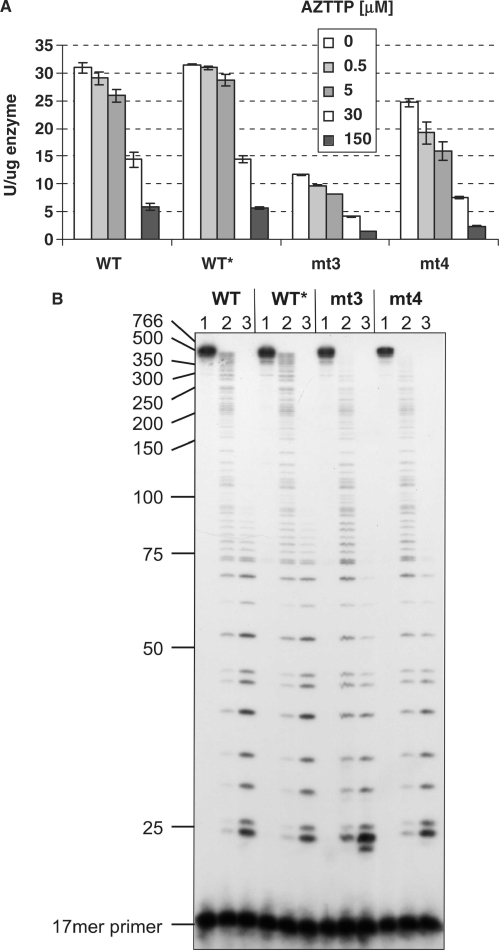
In order to characterize the AZT-resistant PR–RT enzymes we performed various polymerization assays. First, the specific activities of the enzymes were determined by observing the 3H-TTP incorporation into poly(rA)/oligo(dT)15 (Table 1). Our results indicate that the D24A mutation of the WT* does not interfere with polymerization activities. Furthermore, the activity of mt3 is reduced to ∼38% of WT activity, whereas the additional mutation I224T of mt4 helps this enzyme to regain activity (80% of WT). These effects are even more pronounced regarding the replication activity of the corresponding mutant viruses (7): the virus replication activity of the virus containing mt3 was severely reduced (8.6% of WT) whereas the virus containing mt4 displayed a replication activity similar to the WT virus (113% of WT).
Table 1.
Quantitative analysis of RNA-dependent DNA polymerase activities on a homopolymeric substrate
| Enzyme | U/μg protein *10 min |
|---|---|
| WT | 30.9 (± 0.9) |
| WT* | 31.5 (± 0.1) |
| mt3 | 11.6 (± 0.2) |
| mt4 | 24.6 (± 0.7) |
Activities are given in units per microgram of protein, where 1 U catalyzes the incorporation of 1 nmol TTP in poly(rA)/oligo(dT)15 in 10 min at 37°C.
Dissociation constants
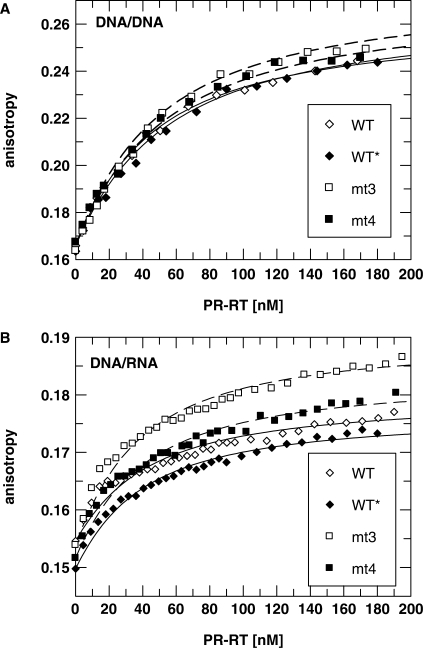
As shown above, polymerization activities of the two mutants are impaired. Since this might have an impact on AZT resistance, we wanted to analyze some kinetic parameters. To check if the reduced polymerization activity is due to changes in the affinity for nucleic acids, we determined the KD-values for nucleic acid binding. Measurements were performed using fluorescence anisotropy titrations with 24/40mer DNA/RNA or DNA/DNA P/T substrates harboring a fluorescent dye (DY647) at the 5′ end of the template strand (Table 2, Figure 1). The KD-values obtained show that there is no significant difference in substrate binding affinities of WT and mutant enzymes, implying that neither the D24A mutation nor the AZT-resistance mutations influence substrate binding. The affinity for the DNA/RNA substrate appears to be slightly higher than for DNA/DNA.
Table 2.
Parameters for P/T binding and the incorporation of dNTPs
| Enzyme | KD DNA/RNA (nM) | KD DNA/DNA (nM) | KM (μM) | Vmax (pmol/min) |
|---|---|---|---|---|
| WT | 32.4 (± 4.2) | 36.4 (± 2.4) | 40.1 (± 4.0) | 29.6 (± 1.7) |
| WT* | 30.4 (± 2.4) | 44.0 (± 3.7) | 40.3 (± 4.0) | 29.6 (± 1.3) |
| mt3 | 28.3 (± 2.7) | 39.5 (±3.0) | 103.0 (± 16.0) | 25.8 (± 2.5) |
| mt4 | 31.3 (± 3.2) | 42.4 (± 3.0) | 112.0 (± 4.0) | 30.1 (± 3.3) |
KD-values were obtained by using Equation (3) to fit a curve to the titration data (see ‘Materials and Methods section’). KM- and vmax-values were determined by Eadie–Hofstee plots.
Figure 1.
Determination of KD-values by fluorescence anisotropy measurements. Fifteen nanomolar of a fluorescently labeled DNA/DNA (A) or DNA/RNA (B) P/T substrate was titrated with different PR–RTs at 25°C. The curves show the best fit to Equation (3) (‘Materials and methods’ section) describing the binding equilibrium with KD-values shown in Table 2.
Determination of KM and vmax values
This analysis was performed using poly(rA)/oligo(dT)15 as a substrate (Table 2). Both AZT-resistant enzymes, mt3 and m4, reveal elevated KM-values as compared to the two WT proteins. However, the vmax value of mt4 is higher than that of mt3 and comparable to the WT proteins, indicating that mt4 is able to exhibit similar polymerization activities like the WT at saturating dNTP concentrations. This might explain the high virus replication activities observed with mt4 containing virus in cell culture assays (7).
Polymerization in the presence of AZTTP
Two mechanisms for AZT resistance have been described. HIV-2 RT controls the incorporation of the inhibitor nucleotide AZTTP (11), whereas for HIV-1 RT excision of the incorporated AZTMP has been recognized as the mechanism of resistance (8–10). Thus, we first analyzed the polymerization behavior of the enzymes in the presence of AZTTP to check for incorporation control. We performed polymerization assays on poly(rA)/oligo(dT)15 in the presence of increasing AZTTP concentrations up to 150 μM (Figure 2A). The TTP concentration was kept constant (150 μM) in all assays. Our data indicate that mt3 and mt4 do not exhibit AZT resistance in this assay.
Figure 2.
Polymerization activities in the presence of AZTTP. (A) Specific activities on 6 nM of poly(rA)/oligo(dT)15 with 12 nM of the various SFVmac PR–RTs, 150 μM TTP and 0, 0.5, 5.0, 30.0 or 150 μM AZTTP. The reaction was stopped after 10 min at 37°C. (B) Chain termination by AZTMP incorporation during DNA polymerization with M13 ssDNA (for conditions and analysis see ‘Materials and methods’ section). Either no AZTTP (lane 1), 5 μM (lane 2) or 50 μM (lane 3) of AZTTP was added to the PR–RTs. DNA size markers are indicated on the left.
We then used the heteropolymeric single-stranded M13 substrate with a 32P-endlabeled DNA-primer for polymerization in the absence of inhibitor or in the presence of 5 and 50 μM AZTTP and analyzed the polymerization products on denaturing polyacrylamide gels (Figure 2B). As already described for the homopolymeric substrate, all enzymes are sensitive to AZTTP addition in the M13 assay. This result is reminiscent of HIV-1 RT (14), where the AZT resistance was also not visible in steady-state polymerization assays or during pre-steady-state analyses and could only be detected with an AZTMP-terminated P/T substrate (8–10). Our results indicate that the resistance mechanism of SFVmac PR–RTs is not comparable to HIV-2 RT where discrimination between the inhibitor and TTP takes place during incorporation (11).
AZTMP excision form a terminated primer
For HIV-1 RT, it has been shown previously that AZTMP can be excised from an AZTMP-terminated P/T substrate in the presence of PPi or ATP (8–10). We thus tested these possibilities.
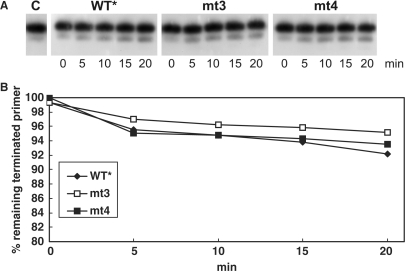
The 32P-endlabeled and AZTMP-terminated substrate P30-AZTMP/T50 was incubated with 150 μM Na-PPi or 5 mM ATP and PR–RT. Time course experiments were performed (Figures 3 and 4) and aliquots were analyzed on denaturing sequencing gels and quantified by densitometry. Our data indicate that in the presence of PPi the WT* PR–RT can excise AZTMP from the terminated primer with similar efficiency as mt3 or mt4 (Figure 3B). Obviously, the ability to perform the reverse reaction of nucleotide incorporation is an intrinsic property of RTs and might be used as a general proof reading function.
Figure 3.
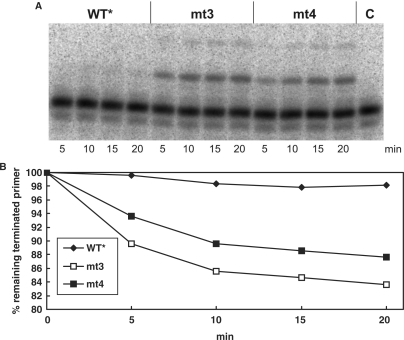
Time course of AZTMP removal in the presence of PPi. (A) One hundred micro molar NaPPi was present in a mix containing 10 nM of an AZTMP-terminated P/T P30-AZTMP/T50 that was labeled with 32P at the 5′ end of the primer. The reaction was started by the addition of 20 nM of the different PR–RTs and stopped at the time points indicated. Lane C, no enzyme added. (B) Quantification of pyrophosphorolytic removal of chain-terminating AZTMP was achieved by densitometry. The percentage of remaining terminated primer is shown.
Figure 4.
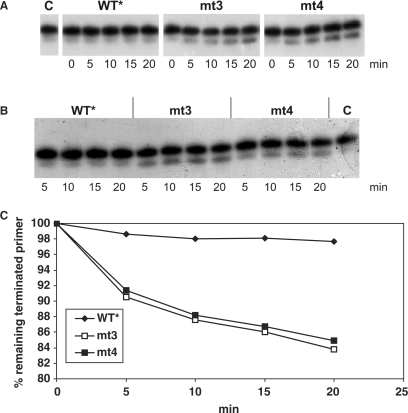
Time course of AZTMP removal in the presence of ATP. (A) Five millimolar ATP was present in a mix containing 10 nM of the AZTMP-terminated P/T P30-AZTMP/T50 that was labeled with 32P at the 5′ end of the primer. The reaction was started by the addition of 20 nM of the different PR–RTs and stopped at the time points indicated. (B) Addition of 0.02 U pyrophosphatase to the mixture described in (A) 5 min before the PR–RT enzymes were added. Lane C, no enzyme added. (C) Quantification of (B) by densitometry. The percentage of remaining terminated primer is shown.
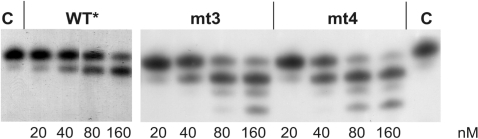
In contrast, when ATP is added (Figure 4), even after an incubation time of 20 min the WT* enzyme does not exhibit significant AZTMP removal activity, whereas mt3 and mt4 are able to excise AZTMP efficiently. To exclude an influence of PPi in the excision reaction, an additional assay was performed after pre-incubating the reaction mix with pyrophosphatase (Figure 4B). The results shown in Figure 4A and B look very similar, indicating that only insignificant amounts of PPi were present in the reactions. Quantification of the data of Figure 4B by densitometry (Figure 4C) demonstrates that the excision reactions of mt3 and mt4 are much faster than that of the WT*. However, they slow down when about 10% of the incorporated AZTMP is eliminated from the primer. This might be due to product inhibition by AZTp4A (15). These data clearly indicate that AZTMP excision in the presence of ATP is the valid mechanism for AZT resistance of SFVmac.
To substantiate our results, we performed the excision reactions with ATP using increasing concentrations of enzyme (Figure 5). The assays were also pre-incubated with pyrophosphatase. Again, our results demonstrate very clearly that in the presence of ATP the mutant enzymes are more efficient in AZTMP removal than the WT*. Furthermore, in the case of the mutants, large excess of enzyme leads to the excision of more than one nucleotide.
Figure 5.
AZTMP removal by ATP in the presence of pyrophosphatase using increasing PR–RT concentrations. Five millimolar ATP and 10 nM of P30-AZTMP/T50 were pre-incubated for 5 min at 37°C in reaction mix. The reaction was started by the addition of different concentrations of the various PR–RTs as indicated and stopped after 20 min. Lane C, no enzyme added.
Primer rescue
The data delineated above are further confirmed by testing the enzymes in the presence of dNTPs to allow for extension of the primer after AZTMP removal (Figure 6). The assay was performed in the presence of ATP and pyrophosphatase. Due to the addition of ddATP, elongation of the primer comes to a halt after the incorporation of 4 nt. Figure 6 shows that only mt3 or mt4 can rescue DNA synthesis, whereas the WT* enzyme is not able to extend the primer.
Figure 6.
ATP-dependent rescue of AZTMP terminated DNA synthesis. (A) Primer rescue reactions using 10 nM P30-AZTMP/T50 were performed with 5 mM ATP, 0.02 U pyrophosphatase and 100 μM dCTP, dGTP, TTP and ddATP. Lane C, no enzyme added. (B) Quantification of (A) by densitometry. The percentage of the remaining unextended primer is shown.
We thus conclude that AZT resistance of SFVmac is due to AZTMP removal by ATP. Furthermore, the AZTMP excision activity obtained with the triple mutant is comparable to that of mt4. This data indicates that the additional I224T change of mt4 is not important for the AZT-resistance mechanism but is necessary to improve the polymerization efficiency.
DISCUSSION
We have shown previously that SFVmac can gain resistance to the nucleoside inhibitor AZT (7). Here, we analyzed the corresponding mutated PR–RTs to elucidate the mechanism of AZT resistance. Our results obtained with purified SFVmac PR–RTs demonstrate that in the case of SFVmac the AZT-resistance mechanism is due to AZTMP removal in the presence of ATP. Remarkably, mt3 which exhibited severely impaired polymerization activities on homo- and heteropolymeric substrates (Table 1 and Figure 2) also shows higher AZTMP excision activities than the WT* enzyme when ATP is present in the reaction (Figures 4–6). Although mt3 and mt4 are also able to excise AZTMP in the presence of PPi (Figure 3) the WT* PR–RT exhibits similar efficiency in this reaction, indicating that this cannot be the mechanism of AZT resistance.
Interestingly, compared to the WT SFVmac PR–RTs, mt3 and mt4 exhibit differences in kinetic parameters. This is also noteworthy since the AZT-resistant HIV-1 RT did not differ from the WT HIV-1 RT in its kinetic parameters (14,16–18). The KM values of the mutant SFVmac PR–RTs are about 2.5-fold higher than those of the WT PR–RTs. While mt3 also shows a reduced value for vmax, the I224T mutation of mt4 is obviously responsible for an increase of vmax similar to that of the WT levels (Table 2), implying that if saturating dNTP concentrations are present in infected cells, reverse transcription will not be greatly impaired in SFVmac viruses harboring mt4. This result indicates that the mutation I224T is important for viral fitness since it can reconstitute the polymerization activity of mt4 in SFVmac-infected cells (7).
It has been demonstrated previously that the RTs of HIV-1 and HIV-2 use different mechanisms for AZT resistance. HIV-2 can discriminate between AZTTP and TTP during nucleotide incorporation (11). In contrast, although certain HIV-1 RT mutations confer a 100-fold decrease in the sensitivity to AZT in vivo (19,20), this effect could not be demonstrated in in vitro assays (14,16–18), indicating that HIV-1 RT is not able to discriminate between AZTTP and TTP. In fact, the mechanism appears to be due to a removal of the chain terminating AZTMP residue after it has been incorporated in the DNA chain. The mutations involved in the enhanced excision of AZTMP in HIV-1 RT are M41L, D67N, K70R, T215Y/F and K219Q/E (Figure 7) (8–10,21,22). Removal of the inhibitor was suggested to be accomplished by two mechanisms that use different substrates to carry out the reaction. AZTMP removal can take place either in the presence of PPi or ATP. The chemistry involved in pyrophosphorolysis and the ribonucleotide-dependent phosphorolysis reaction is similar. Removal of the chain-terminating AZT results from nucleophilic attack of a polyphosphate in the phosphodiester bond between the last but one nucleotide and the AZTMP. In case of PPi, this leads to removal of the 3′ AZTMP by creating AZTTP.
Figure 7.
Sequence alignment of the relevant regions from the HIV-1 and SFVmac RT domains. The amino acids conferring AZT resistance are indicated by filled gray boxes. The numbers represent the amino acid numbering in the HIV-1 RT and SFVmac PR–RT. Sequence alignment was performed with the program lalign (28).
There is evidence that the phosphate donor in the excision reaction of AZT-resistant HIV-1 RT is ATP, leading to an ATP-AZTMP dinucleotide-tetraphosphate (adenosine-3′azido,3′deoxthymidine-5′-5′-tetraposphate, AZTp4A). For HIV-1 RT it was concluded from biochemical and structural data that the exchange of T215 to an aromatic residue (T215F/Y) enhances binding of ATP, but not PPi, thus facilitating excision. The model suggests that in the AZT-resistant enzyme, the adenine moiety of the incoming ATP makes π–π interactions with the aromatic ring of the mutated amino acid (22–26).
Our results obtained with SFVmac PR–RT are especially interesting when comparing the AZT-resistance mutations of HIV-1 RT and SFVmac PR–RT since in the latter enzyme no mutation leading to an aromatic side chain is present. In addition, sequence alignments of the polymerase domains of HIV-1 and SFVmac reveal that the amino acid exchanges obtained with SFVmac are not the ones corresponding to the exchanges in HIV-1 RT (Figure 7). Furthermore, although the homology between PFV and SFVmac is around 90%, introduction of the SFVmac RT mutations into PFV did not result in AZT-resistant viruses (7).
These findings might indicate structural differences between HIV-1 and SFVmac RTs. This appears to be plausible since the RT domains of HIV-1 and SFVmac are phylogenetically rather distantly related (27). Thus, the interpretation of alignment data is also rather difficult. In addition, differences in the mechanism of ATP binding and/or ATP-mediated excision are possible. Structural analysis of WT and AZT-resistant SFVmac PR–RT are under way and will help to elucidate the differences between WT and mutant PR–RTs and also between HIV-1 and SFVmac.
ACKNOWLEDGEMENTS
We thank Philipp Weiglmeier for help with protein purifications and Prof. Paul Rösch for continuous support. The project was funded by the Deutsche Forschungsgemeinschaft DFG (Re627/7-1, Re627/8-1, SFB 479, Wo630/7-1), the Bavarian International Graduate School of Science (BIGSS) and the University of Bayreuth. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft DFG, grant Wo630/7-1.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rethwilm A. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 2003;277:1–26. doi: 10.1007/978-3-642-55701-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Linial M. Foamy viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Vol. 2, Lippincott Williams & Wilkins; 2007. pp. 2245–2262. [Google Scholar]
- 3.Pfrepper KI, Rackwitz HR, Schnölzer M, Heid H, Löchelt M, Flügel RM. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 1997;71:7305–7311. doi: 10.1128/jvi.71.10.7305-7311.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenblum LL, Patton G, Grigg AR, Frater AJ, Cain D, Erlwein O, Hill CL, Clarke JR, McClure MO. Differential susceptibility of retroviruses to nucleoside analogues. Antivir. Chem. Chemother. 2001;12:91–97. doi: 10.1177/095632020101200202. [DOI] [PubMed] [Google Scholar]
- 6.Lee CC, Ye F, Tarantal AF. Comparison of growth and differentiation of fetal and adult rhesus monkey mesenchymal stem cells. Stem Cells Dev. 2006;15:209–220. doi: 10.1089/scd.2006.15.209. [DOI] [PubMed] [Google Scholar]
- 7.Kretzschmar B, Nowrouzi A, Hartl MJ, Gärtner K, Wiktorowicz T, Herchenröder O, Kanzler S, Rudolph W, Mergia A, et al. AZT-resistant Foamy Virus. Virology. 2008;370:151–157. doi: 10.1016/j.virol.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 9.Meyer PR, Matsuura SE, So AG, Scott WA. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl Acad. Sci. USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 11.Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS. Pathog. 2006;2:e10. doi: 10.1371/journal.ppat.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacques PS, Wöhrl BM, Ottmann M, Darlix J-L, Le Grice SFJ. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 1994;269:26472–26478. [PubMed] [Google Scholar]
- 13.Werner S, Wöhrl BM. Soluble Rous Sarcoma Virus reverse transcriptases α, αβ and β purified from insect cells are processive DNA polymerases that lack an RNase H 3′ > 5′ directed processing activity. J. Biol. Chem. 1999;274:26329–26336. doi: 10.1074/jbc.274.37.26329. [DOI] [PubMed] [Google Scholar]
- 14.Krebs R, Immendörfer U, Thrall S, Wöhrl BM, Goody RS. Single-step kinetics of HIV-reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3TC. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 15.Dharmasena S, Pongracz Z, Arnold E, Sarafianos SG, Parniak MA. 3′-Azido-3′-deoxythymidine-(5′)-tetraphospho-(5′)-adenosine, the product of ATP-mediated excision of chain-terminating AZTMP, is a potent chain-terminating substrate for HIV-1 reverse transcriptase. Biochemistry. 2007;46:828–836. doi: 10.1021/bi061364s. [DOI] [PubMed] [Google Scholar]
- 16.Carroll SS, Geib J, Olsen DB, Stahlhut M, Shafer JA, Kuo LC. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry. 1994;33:2113–2120. doi: 10.1021/bi00174a018. [DOI] [PubMed] [Google Scholar]
- 17.Lacey SF, Reardon JE, Furfine ES, Kunkel TA, Bebenek K, Eckert KA, Kemp SD, Larder BA. Biochemical studies on the reverse transcriptase and RNase H activities from human immunodeficiency virus strains resistant to 3′-azido-3′-deoxythymidine. J. Biol. Chem. 1992;267:15789–15794. [PubMed] [Google Scholar]
- 18.Kerr SG, Anderson KS. RNA dependent DNA replication fidelity of HIV-1 reverse transcriptase: evidence of discrimination between DNA and RNA substrates. Biochemistry. 1997;36:14056–14063. doi: 10.1021/bi971385+. [DOI] [PubMed] [Google Scholar]
- 19.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high- level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 20.Larder BA, Kellam P, Kemp SD. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 21.Meyer PR, Matsuura SE, Schinazi RF, So AG, Scott WA. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 2000;44:3465–3472. doi: 10.1128/aac.44.12.3465-3472.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 2002;76:3248–3256. doi: 10.1128/JVI.76.7.3248-3256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 2002;76:9143–9151. doi: 10.1128/JVI.76.18.9143-9151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarafianos SG, Clark AD, Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, et al. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002;21:6614–6624. doi: 10.1093/emboj/cdf637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AJ, Meyer PR, Asthana D, Ashman MR, Scott WA. Intracellular substrates for the primer-unblocking reaction by human immunodeficiency virus type 1 reverse transcriptase: detection and quantitation in extracts from quiescent- and activated-lymphocyte subpopulations. Antimicrob. Agents Chemother. 2005;49:1761–1769. doi: 10.1128/AAC.49.5.1761-1769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt VM. Retroviral virions and genomes. In: Coffin J, Hughes SH, Varmus H, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 27–69. [PubMed] [Google Scholar]
- 28.Huang X, Miller W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. [Google Scholar]