Abstract
Insulin-like growth factor (IGF) activity is regulated by six high affinity binding proteins (IGFBPs) and possibly by some of the nine IGFBP-related proteins (IGFBP-rPs). To determine the phylogenetic relationship of this proposed gene superfamily, we conducted maximum likelihood (ML) and Bayesian inference analyses on a matrix of amino acid sequences from a diversity of vertebrate species. A single most likely phylogram, ML bootstrap, and Bayesian consensus tree of 10,000,000 generations revealed a monophyletic IGFBP lineage independent of the IGFBP-rPs. The IGFBPs segregated into three distinct clades: IGFBP-1, -3, and -6. Subsequent gene duplication events within the IGFBP-1 and -3 clades resulted in the production and divergence of IGFBP-2 and -4 within the IGFBP-1 clade and IGFBP-5 in the IGFBP-3 clade. By contrast, the IGFBP-rPs were distributed paraphyletically into two clades: IGFBP-rP1, 5, and 6 in one clade and the CCN family (IGFBP-rP2-4,7-9) in another. A recently identified IGFBP-3 homolog in rainbow trout localized to the IGFBP-2 subclade. Subsequence analysis identified a RGD motif common to IGFBP-2 orthologs, but did not identify the nuclear localization sequence present in IGFBP-3 and -5 homologs. The putative trout IGFBP-3 was 36-55% identical to different IGFBP-2 proteins, but only 24-27% identical to IGFBP-3 proteins. These results suggest that the IGFBPs and IGFBP-rPs are at best distantly related and that the limited similarities likely resulted from exon shuffling. They also suggest that rainbow trout, and possibly other salmonids, possess two IGFBP-2 paralogs as the putative trout IGFBP-3 is misannotated.
Introduction
The bioavailability of both insulin-like growth factor (IGF)-I and –II is mediated by six high affinity IGF binding proteins (IGFBP-1 to -6). This occurs locally at the cell and tissue level as well as systemically where the IGFBPs significantly enhance the circulating half-life of the IGFs (Hwa et al., 1999b). Several recent studies, however, suggest that some IGFBPs, in particular IGFBP-3 and -5, can modulate cellular activities without binding to either IGF (Duan and Xu, 2005; Firth and Baxter, 2002; Lee and Cohen, 2002; Oufattole et al., 2006). Despite minor differences, the IGFBPs share common amino and carboxy terminal domains, both of which facilitate IGF binding, although only the former domain is critical, and each IGFBP is well conserved in the different vertebrate classes. Indeed, conservation extends beyond primary sequence as all of the IGFBP genes characterized to date are similarly organized (Hwa et al., 1999b). By contrast, the IGFBP-related proteins (IGFBP-rP1 to 9) share very little overall homology with the IGFBPs, although they possess amino terminal domains similar to those of the IGFBPs and some are capable of IGF binding, albeit with considerably lower affinity (Kim et al., 1997). Their ability to influence IGF action, however, has not been convincingly demonstrated as most of their defined functions are either independent of IGF-binding or have not been thoroughly described (Hwa et al., 1999b; Perbal, 2004).
The cysteine rich amino terminal domains of most IGFBPs, excluding only IGFBP-6 homologs, and IGFBP-rPs contain a GCGCCXXC or “IGFBP” motif whose function is poorly understood (Hwa et al., 1999b). The IGFBPs additionally contain a thyroglobulin-type I motif within their carboxy terminal domains while the IGFBP-rPs contain many different motifs depending upon the specific protein. For instance, IGFBP-rP1 and -rP5 (a.k.a. IGFBP-7/Mac25 & L56, respectively) also possess Kazal-type serine proteinase inhibitor (both rP1 & 5), immunoglobulin-like (rP1) and serine protease (rP5) motifs (Hwa et al., 1999b). The CCN (CTGF; Cef10/Cyr61 & Nov) sub-family of IGFBP-rPs, which include IGFBP-rP2 (a.k.a. connective tissue growth factor, CTGF), -rP3 (NovH), -rP4 (Cyr61), -rP7 (WISP2), -rP8 (WISP1) and -rP9, also possess Von Willebrand factor type C repeats and thrombospondin type I repeats, none of which are found in any IGFBP (Rachfal and Brigstock, 2005). Despite the structural differences noted and the low level of overall homology, previous reviews have suggested a common ancestral link between the two protein families (Hwa et al., 1999a; Hwa et al., 1999b; Kelley et al., 2000). Kelley et al. (2000) suggested that the IGFBP-rPs arose from a thyroglobulin-like ancestor and that a primordial IGFBP subsequently diverged. Hwa et al. (1999a, 1999b) further suggested that the entire IGFBP/IGFBP-rP “superfamily” evolved from an ancestral “IGF binder” into two distinct protein families with different binding affinities for the IGFs: high affinity IGF binders (IGFBPs) and low affinity binders (IGFBP-rPs). However, such conclusions may be unsubstantiated in the absence of a rigorous phylogenetic analysis of the putative superfamily.
In order to determine the true phylogenetic relationship between these gene families, we conducted maximum likelihood (ML) and Bayesian inference analyses on 64 IGFBP and IGFBP-rP amino acid sequences representing most vertebrate classes. These computational methods generate a ML point estimate of the IGFBP phylogeny and two measures of branch support: ML bootstraps and Bayesian posterior probabilities of tree distributions from millions of generations (Huelsenbeck et al., 2002; Huelsenbeck and Ronquist, 2001; Huelsenbeck et al., 2001). Our results indicate that five independent gene duplication events are responsible for the divergence of the IGFBPs into three distinct subclades: the IGFBP-6 clade, the IGFBP-1 clade, which also contains IGFBP-2 and -4, and the IGFBP-3 clade, which also contains IGFBP-5. They also suggest that the two families (IGFBPs and IGFBP-rPs) may not necessarily share a common ancestor, but that the similar amino terminal domains may be a product of exon shuffling. The lack of clear phylogenetic and functional relationships between the IGFBPs and the IGFBP-rPs suggests that the current classification of these two largely unrelated groups as a “superfamily” should be revised.
Materials and Methods
Phylogenetic Analysis
A single matrix composed of homologous amino acid sequences from species in all vertebrate classes except Agnatha and Reptilia (no known homologs) was constructed using Vector NTI for the Macintosh. This included IGFBP sequences from bovine (Bos taurus), zebrafish (Danio rerio), chicken (Gallus gallus), human (Homo sapiens), little skate (Leucoraja erinacea), mouse (Mus musculus), striped bass (Morone saxatilis), sheep (Ovis aries), rainbow trout (Oncorhynchus mykiss) and African clawed frogs (Xenopus laevus & Xenopus tropicalus). Human and mouse myostatin (MSTN) sequences were also included as an unrelated outgroup. A list of the accession numbers for each IGFBP, IGFBP-rP and MSTN homolog or the matrix itself will be provided upon request.
Maximum likelihood (ML) and ML boootstrap analyses of the IGFBP matrix were performed using the proml and seqboot modules of PHYLIP 3.66 (Felsenstein 2007). ML phylogram reconstruction used the Jones-Taylor-Thornton probability model, constant rates, global rearrangements, and random sequence order. Bootstrap analyses were run on 100 replicate data matrices under the same conditions as above but without global rearrangements due to time constraints.
Bayesian inference analysis was performed on the matrix using MrBayes v.3.0 (Huelsenbeck and Ronquist, 2001). Ten million generations were performed with four chains (Markov Chain Monte Carlo) and a tree was saved every 100 generations. Priors included a mixed amino acid model allowing for optimization of the model during the analysis (Minin et al., 2003). Multiple analyses were started from different random locations within the tree space in order to test for the occurrence of stationarity, convergence and mixing within the ten million generations. Posterior probability distributions from separate replicates were compared for convergence to the same posterior probabilities across branches. Majority rule consensus trees of the 80,000 sampled during the Bayesian inference analyses yielded probabilities indicating monophyletic clades (Lewis, 2001). The trees from the MrBayes analysis were then loaded into PAUP* after discarding the trees generated within the first 2,000,000 generations. Thus, the sampled trees included only those obtained post “burnin” of the chain (Huelsenbeck and Ronquist, 2001) and after stationarity was established. Bayesian posterior probability (pp) values are therefore presented above branches when greater than 50% and as a consensus topology.
BLAST Analysis and Alignment of IGFBP-3 Orthologs
The amino terminal 150 amino acid sequence of human IGFBP-3 was compared to the zebrafish expressed sequence tag (EST) database using the basic local alignment tool for protein sequences (BLASTP). A multiple sequence alignment of human and zebrafish IGFBP-3 and -5 proteins as well as a putative rainbow trout IGFBP-3 homolog was constructed using Vector Align X and the BLOSUM 62 matrix (gap penalty 10, extension 0.05).
Results
Phylogenetic Analysis
The ML analysis of the IGFBP amino acid matrix resulted in one most likely tree (-lnL = 22742.11243; Fig. 1). ML bootstrap results suggest many strongly supported branches, particularly within IGFBP clades, but some lack of support for the exact relationships among the major clades. Posterior probability (PP) distributions from the Bayesian inference analysis resulted in congruent topologies to the ML analyses and similar support for branches when branches with PP values ≥ 95% are compared with those with bootstrap values ≥ 70% (Fig. 1).
Figure 1. Phylogenetic relationship of IGFBP and IGFBP-rP gene products.
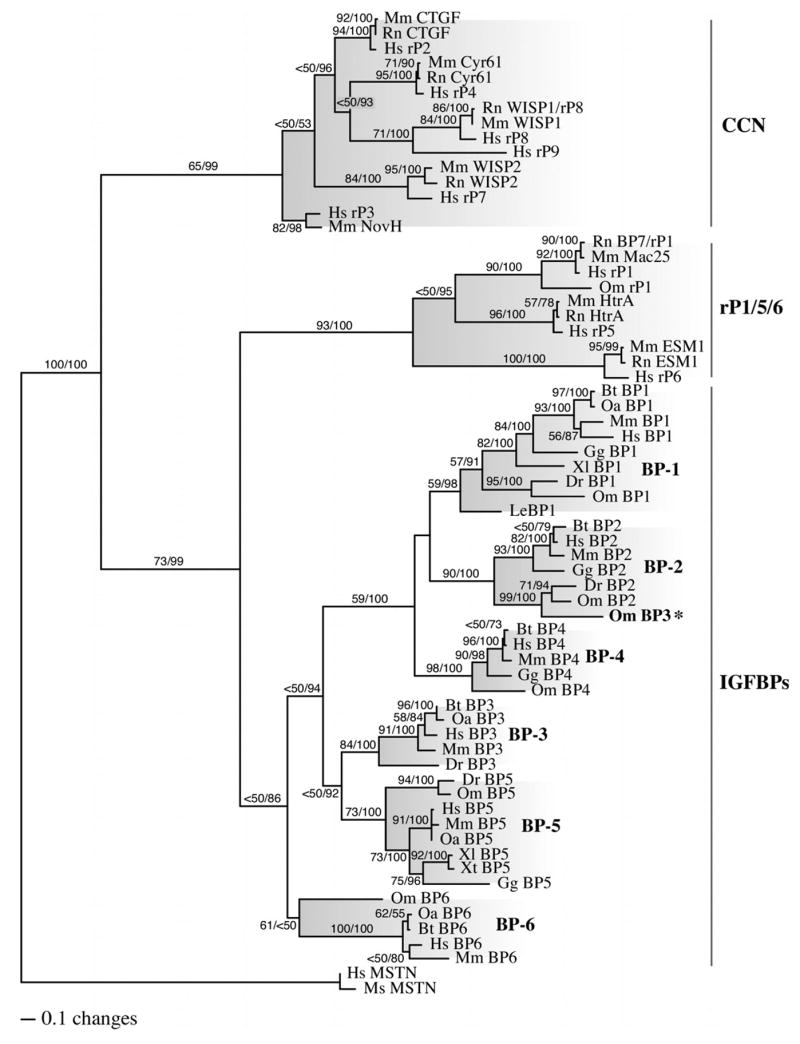
Maximum likelihood (ML), ML boootstrap, and Bayesian inference analyses were performed on a single matrix composed of homologous amino acid sequences from various vertebrate species using PHYLIP 3.66 and MrBayes v.3.0 (Bt, Bos taurus; Dr, Danio rerio; Gg, Gallus gallus; Hs, Homo sapiens; Le, Leucoraja erinacea; Mm, Mus musculus; Ms, Morone saxatilis; Oa, Ovis aries; Om, Oncorhynchus mykiss; Xl, Xenopus laevus; Xt, Xenopus tropicalus). A total of 100 ML bootstrap iterations and 10,000,000 Bayesian generations were performed (trees saved every 100 generations). Trees from the first 2,000,000 generations were discarded as burn-in to assure that stationarity was established and the remaining 80,000 post burn-in trees were used to construct the majority rule consensus tree shown with PAUP*. Two independent analyses were performed and produced identical results. ML bootstrap and Bayesian posterior probability values are shown above each branch (ML/Bay. PP) when greater than 50%. The major subclades are shaded and the monophyletic IGFBP group and the paraphyletic IGFBP-rP groups are labeled on the right. (BP, IGFBP; rP, IGFBP-rP; MSTN, myostatin; CCN proteins include CTGF, Cyr61, WISP, NovH & IGFBP-rP2-4,7-9; IGFBP-rP1,5,6 include Mac25, HtrA & ESM; * misannotated)
The phylogenetic hypothesis presented here of IGFBP/BP-rP superfamily diversification reveals a monophyletic distribution of all IGFBPs that was independent of the IGFBP-rPs (Fig. 1). The IGFBP family was divided into three distinct sister clades: the IGFBP-6 clade, the IGFBP-1 clade that also included IGFBP-2 and -4 and the IGFBP-3 clade that also included IGFBP-5. A total of five independent gene duplication events were therefore detected. The first created the IGFBP-1/-3 and -6 sister clades while the second created the IGFBP-1 and -3 clades. This was followed by two additional duplication events in the IGFBP-1 clade and one event in the IGFBP-3 clade (Fig. 1).
By contrast, the IGFBP-rPs are distributed paraphyletically into two clades: the IGFBP-rP1, 5, and 6 clade, and the CCN family (IGFBP-rP2, -rP3, -rP4, -rP7, -rP8 and –rP9). None of the IGFBP-rPs segregated within any of the IGFBP subclades or vice versa, suggesting the two “gene families” evolved independent from one another and that any relationship between the two families is distant and occurred prior to the appearance of protochordates. Indeed, individual IGFBP-rPs are only 13-19% identical to human IGFBP-3 overall as the limited homology is confined to small regions within the carboxy terminal domains commonly referred to as IGFBP motifs. Similar pair wise comparisons between representatives of the three different IGFBP-rP clades reveal very little homology between clades. For example, IGFBP-rP1 and -rP5 are 14.3-17.5% and 7.5-16.6% identical to the CCN family members, respectively. The same is true for IGFBP-rP6 (9.1-15.4%) and an unrelated zebrafish myostatin homolog (Kerr et al., 2005) (zfMSTN-2, 10.1-13.4%). By contrast, the different CCN proteins themselves are up to 48% identical.
Low stringency BLASTP analysis of the zebrafish EST database with the amino terminal domain of human IGFBP-3 (first 150 amino acids) retrieved all of the previously identified zebrafish IGFBPs (Duan et al., 1999; Li et al., 2005; Maures and Duan, 2002) and in addition, a tankyrase homolog (TANK1; GID, 68397005) that also contained a carboxy terminal IGFBP motif (Fig. 2, shaded residues 60-70). However, the TANK1 protein is only 10.9% identical to IGFBP-3 overall and the tankyrase family as a whole is functionally unrelated to either the IGFBPs or IGFBP-rPs. These data together suggest that the limited homology shared between all three protein families possibly occurred via exon shuffling and that other proteins may also possess similar domains and motifs.
Figure 2. Partial amino acid alignment of human (Hs) IGFBP-3 and a novel zebrafish (Dr) tankyrase homolog (TANK1).
BLASTP analysis of the zebrafish EST database using a 150 amino acid sequence from the amino terminus of human IGFBP-3 identified a novel tankyrase homolog (GenBank: XM_682318.1, GI:68397005) that shares motifs common to the IGFBPs and IGFBP-rPs. Alignment positions are numbered above the sequences whereas residue numbers within a specific sequence are indicated to the left and in parentheses. Conserved regions are shaded and residues necessary for IGF-binding (Buckway et al., 2001; Yan et al., 2004) are indicated with asterisks.
Identification of a Novel Rainbow Trout IGFBP-2 Paralog
Each IGFBP clade (Fig. 1) contained only orthologs for that specific IGFBP with one exception. A recently identified rainbow trout IGFBP-3 homolog segregated within the IGFBP-2 subclade rather than with the IGFBP-3 subclade, suggesting that this particular homolog is actually an IGFBP-2 paralog as another rainbow trout IGFBP-2 was also characterized (Kamangar et al., 2006). The true identity of the putative IGFBP-3 was therefore determined by comparative alignments and subsequence analysis.
Paired and multiple sequence alignments of the putative rainbow trout IGFBP-3 with different mammalian and fish orthologs of IGFBP-2, -3 and -5 indicate that the trout IGFBP-3 shares more identities with IGFBP-2 sequences than with those of IGFBP-3. Indeed, it is only 24% identical overall to human and zebrafish IGFBP-3 and 27% identical to IGFBP-5s (Fig. 3). By contrast, it is 36, 53 and 55% identical to human, zebrafish and rainbow trout IGFBP-2, respectively. Comparative subsequence analysis identified several motifs in the trout IGFBP-3 that are also found in IGFBP-2 orthologs including a RGD motif within the carboxy-terminal domains (Fig. 3). However, the nuclear localization sequence found in all IGFBP-3 and -5 orthologs is not present in the rainbow trout IGFBP-3. The low level of sequence homology with IGFBP-3 proteins, the high level with IGFBP-2s and the presence and absence of clade-specific motifs all complement the phylogenetic analysis and indicate that the putative rainbow trout IGFBP-3 is actually a second IGFBP-2 paralog rather than an IGFBP-3 ortholog. The putative rainbow trout IGFBP-4 sequence used in this study was obtained from a partial EST clone. Nevertheless, it segregated with other IGFBP-4 homologs. This suggests that unlike the misannotated IGFBP-3, the rainbow trout IGFBP-4 is indeed a true ortholog, although this can only be confirmed by isolating a complete clone.
Figure 3. Amino acid alignment of human (Hs), zebrafish (Dr) and rainbow trout (Om) IGFBP-3 homologs.
Individual sequences were aligned using Vector AlignX. Alignment positions are numbered above the sequences whereas residue numbers within a specific sequence are indicated to the left and in parentheses. The nuclear localization sequence conserved among all IGFBP-3 (BP3) and IGFBP-5 (BP5) proteins (KGRKR) and the integrin-binding RGD sequence commonly found in IGFBP-2 proteins are boxed. Conserved residues within the alignment are shaded and residues involved in IGF binding (Buckway et al., 2001; Yan et al., 2004) are highlighted with asterisks.
Discussion
The monophyletic distribution of the IGFBPs in contrast to the paraphyletic distribution of the IGFBP-rPs indicates that the IGFBPs are distinct, at best only distantly related to IGFBP-rPs and that members of the putative IGFBP-rP gene family may not be as closely related as originally presumed. In fact, the extreme low level of homology shared between the three different IGFBP-rP clades suggests that they are as dissimilar as comparisons between the IGFBPs and IGFBP-rPs. Previous attempts to define the evolutionary relationships between these families were based either on sequence similarities and associations with hox gene clusters (Kelley et al., 2000) or with a rudimentary and unrooted tree (Hwa et al., 1999b). Several differences were noted when the tree described by Hwa et al. (1999b) was compared to that reported herein (Fig. 1).
Hwa et al. (1999b) presented four models that could have explained the evolution of the proposed IGFBP superfamily: three based on the divergence of a primitive IGFBP, IGFBP-rP or common ancestral gene and one on the shuffling of an amino terminal module. The complete lack of any significant amino acid conservation among the IGFBPs and IGFBP-rPs outside of the IGFBP motifs in addition to the monophyletic distribution of IGFBPs strongly suggests that the similar amino terminal domains in both protein groups resulted from exon shuffling. This model cannot be definitively proved, although it was also favored by Hwa et al. and was based in part on the fact that the common domains are encoded by a single exon in every gene. Shared motifs are not necessarily indicative of exon shuffling, however, as they could have evolved convergently. The similarities within the amino terminal regions discussed extend beyond the GCGCCXXC IGFBP motif in some, but not all, of the IGFBP-rPs and possibly other proteins as well. This includes TANK1 whose amino terminal domain is far more similar to the IGFBPs than are comparable domains from any of the IGFBP-rPs (Fig. 2). In addition, several residues critical to IGF binding (Buckway et al., 2001; Yan et al., 2004) are also found in TANK1, although they are lacking in most of the IGFBP-rPs. It is unknown whether TANK1 can or does bind the IGFs. However, Yan et al. (2006) recently determined that the amino terminal domain of CCN3 (a.k.a. IGFBP-rP3 & NovH) cannot replace the similar domain of IGFBP-3 as the IGF affinity of the chimeric IGFBP-3 was significantly reduced to levels comparable of CCN3. This indicates that the IGFBP motif alone does not confer high-affinity binding as other amino terminal motifs/residues are required. Exon shuffling, therefore, seems likely responsible for the similarities noted, although it was followed by significant genetic divergence. The less likely alternative explanation, extreme functional divergence of a common ancestor, cannot be entirely excluded without further analysis, especially given the very early divergence of IGFBPs and IGFBP-rPs.
Kamangar et al. (2006) recently characterized several cDNA clones for all six rainbow trout IGFBP homologs and IGFBP-rP1. This includes a putative IGFBP-3 that segregated along with IGFBP-2 orthologs in our phylogenetic analysis. Subsequence analysis and sequence alignments (Fig. 3) also suggest that it is actually an additional IGFBP-2 paralog rather than the trout IGFBP-3. Duplicate genes are common in the bony fishes due to a genome wide duplication event that occurred early in their evolution (Amores et al., 1998; Postlethwait et al., 1998). An additional genome duplication event specifically within the salmonids (Hordvik, 1998) is responsible for up to four unique copies of some genes in some species including the rainbow trout (Brunelli et al., 2001; Garikipati et al., 2006; Kavsan et al., 1993; McKay et al., 2004). Thus, the existence of two IGFBP-2 paralogs in rainbow trout is not necessarily surprising. The possible lack of an IGFBP-3 ortholog in salmonids (none have been cloned to date) is highly unusual and suggests that, if true, compensatory mechanisms may have evolved specifically within these fishes. Indeed, 244984 ESTs (83,863 unique, see www.TIGR.org) have been sequenced to date from different rainbow trout tissues including the liver and no IGFBP-3 homologs have been identified (Rexroad et al., 2003).
Given the lack of a clear ancestral relationship between the IGFBPs and the IGFBP-rPs, and among the three different IGFBP-rP clades as well, it is questionable whether the “IGFBP superfamily” classification is justified as it does not reflect the phylogenetic relationships of the different protein families. The physiological significance of low affinity binding interactions between IGFBP-rPs and the IGFs remains equally questionable, especially in the presence of the IGFBPs which bind with much higher affinity, as other IGF-independent functions have already been well established for many IGFBP-rPs (Bleau et al., 2005; Perbal, 2004; Rachfal and Brigstock, 2005). The alternative IGFBP-rP nomenclature (WISP, NovH, CTGF, CCN etc.) may therefore be more appropriate in light of future revisions. Nevertheless, the IGFBP phylogenies described will help in better defining the evolution of this diverse gene family. Future studies, however, may require the identification of homologs from more basal groups and from invertebrates as the origin of IGFBPs have yet to be described.
Acknowledgments
This work was supported by grants from the United States Department of Agriculture (2004-34468-15199) and the National Institutes of Health (1R03AR051917) to Buel D. Rodgers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, Westerfield M, Ekker M, Postlethwait JH. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–4. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Perbal B. CCN proteins and cancer: two to tango. Front Biosci. 2005;10:998–1009. doi: 10.2741/1594. [DOI] [PubMed] [Google Scholar]
- Brunelli JP, Robison BD, Thorgaard GH. Ancient and recent duplications of the rainbow trout Wilms' tumor gene. Genome. 2001;44:455–62. [PubMed] [Google Scholar]
- Buckway CK, Wilson EM, Ahlsen M, Bang P, Oh Y, Rosenfeld RG. Mutation of three critical amino acids of the N-terminal domain of IGF-binding protein-3 essential for high affinity IGF binding. J Clin Endocrinol Metab. 2001;86:4943–50. doi: 10.1210/jcem.86.10.7936. [DOI] [PubMed] [Google Scholar]
- Duan C, Ding J, Li Q, Tsai W, Pozios K. Insulin-like growth factor binding protein 2 is a growth inhibitory protein conserved in zebrafish. Proc Natl Acad Sci U S A. 1999;96:15274–9. doi: 10.1073/pnas.96.26.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C, Xu Q. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen Comp Endocrinol. 2005;142:44–52. doi: 10.1016/j.ygcen.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 6.44. Department of Genetics; University of Washington, Seattle: 2007. Distributed by the author. [Google Scholar]
- Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–54. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- Garikipati D, Gahr SA, Rodgers BD. Identification, Characterization and Quantitative Expression Analysis of Rainbow Trout Myostatin-1a and -1b Genes. J Endocrinol. 2006 doi: 10.1677/joe.1.06866. in press. [DOI] [PubMed] [Google Scholar]
- Hordvik I. The impact of ancestral tetraploidy on antibody heterogeneity in salmonid fishes. Immunol Rev. 1998;166:153–7. doi: 10.1111/j.1600-065x.1998.tb01260.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Larget B, Miller RE, Ronquist F. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst Biol. 2002;51:673–88. doi: 10.1080/10635150290102366. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–4. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Hwa V, Oh Y, Rosenfeld RG. Insulin-like growth factor binding proteins: a proposed superfamily. Acta Paediatr Suppl. 1999a;88:37–45. doi: 10.1111/j.1651-2227.1999.tb14349.x. [DOI] [PubMed] [Google Scholar]
- Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev. 1999b;20:761–87. doi: 10.1210/edrv.20.6.0382. [DOI] [PubMed] [Google Scholar]
- Kamangar BB, Gabillard JC, Bobe J. Insulin-like growth factor-binding protein (IGFBP)-1, -2, -3, -4, -5, and -6 and IGFBP-related protein 1 during rainbow trout postvitellogenesis and oocyte maturation: molecular characterization, expression profiles, and hormonal regulation. Endocrinology. 2006;147:2399–410. doi: 10.1210/en.2005-1570. [DOI] [PubMed] [Google Scholar]
- Kavsan V, Koval A, Petrenko O, Roberts CT, Jr, LeRoith D. Two insulin genes are present in the salmon genome. Biochem Biophys Res Commun. 1993;191:1373–8. doi: 10.1006/bbrc.1993.1369. [DOI] [PubMed] [Google Scholar]
- Kelley KM, Desai P, Roth JT, Haigwood JT, Arope SA, Flores RM, Schmidt KE, Perez M, Nicholson GS, Song WW. Evolution of endocrine growth regulation: the insulin like growth factors (IGFs), their regulatory binding proteins (IGFBPs) and IGF receptors in fishes and other ectothermic vertebrates. In: Fingerman M, Nagabhushanam R, editors. Recent Advances in Marine Biotechnology. Science Publishers, Inc; Enfield: 2000. p. 292. [Google Scholar]
- Kerr T, Roalson EH, Rodgers BD. Phylogenetic analysis of the myostatin gene sub-family and the differential expression of a novel member in zebrafish. Evol Dev. 2005;7:390–400. doi: 10.1111/j.1525-142X.2005.05044.x. [DOI] [PubMed] [Google Scholar]
- Kim HS, Nagalla SR, Oh Y, Wilson E, Roberts CT, Jr, Rosenfeld RG. Identification of a family of low-affinity insulin-like growth factor binding proteins (IGFBPs): characterization of connective tissue growth factor as a member of the IGFBP superfamily. Proc Natl Acad Sci U S A. 1997;94:12981–6. doi: 10.1073/pnas.94.24.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Cohen P. Nuclear effects: unexpected intracellular actions of insulin-like growth factor binding protein-3. J Endocrinol. 2002;175:33–40. doi: 10.1677/joe.0.1750033. [DOI] [PubMed] [Google Scholar]
- Lewis PO. Phylogenetic systematics turns over a new leaf. Trends in Ecology & Evolution. 2001;16:30–37. doi: 10.1016/s0169-5347(00)02025-5. [DOI] [PubMed] [Google Scholar]
- Li Y, Xiang J, Duan C. Insulin-like growth factor-binding protein-3 plays an important role in regulating pharyngeal skeleton and inner ear formation and differentiation. J Biol Chem. 2005;280:3613–20. doi: 10.1074/jbc.M411479200. [DOI] [PubMed] [Google Scholar]
- Maures TJ, Duan C. Structure, developmental expression, and physiological regulation of zebrafish IGF binding protein-1. Endocrinology. 2002;143:2722–31. doi: 10.1210/endo.143.7.8905. [DOI] [PubMed] [Google Scholar]
- McKay SJ, Trautner J, Smith MJ, Koop BF, Devlin RH. Evolution of duplicated growth hormone genes in autotetraploid salmonid fishes. Genome. 2004;47:714–23. doi: 10.1139/g04-018. [DOI] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol. 2003;52:674–83. doi: 10.1080/10635150390235494. [DOI] [PubMed] [Google Scholar]
- Oufattole M, Lin SW, Liu B, Mascarenhas D, Cohen P, Rodgers BD. Ribonucleic acid polymerase II binding subunit 3 (Rpb3), a potential nuclear target of insulin-like growth factor binding protein-3. Endocrinology. 2006;147:2138–46. doi: 10.1210/en.2005-1269. [DOI] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–4. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, Goutel C, Fritz A, Kelsh R, Knapik E, Liao E, Paw B, Ransom D, Singer A, Thomson M, Abduljabbar TS, Yelick P, Beier D, Joly JS, Larhammar D, Rosa F, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–9. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Rachfal AW, Brigstock DR. Structural and functional properties of CCN proteins. Vitam Horm. 2005;70:69–103. doi: 10.1016/S0083-6729(05)70003-0. [DOI] [PubMed] [Google Scholar]
- Rexroad CE, 3rd, Lee Y, Keele JW, Karamycheva S, Brown G, Koop B, Gahr SA, Palti Y, Quackenbush J. Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res. 2003;102:347–54. doi: 10.1159/000075773. [DOI] [PubMed] [Google Scholar]
- Yan X, Baxter RC, Perbal B, Firth SM. The aminoterminal insulin-like growth factor (IGF) binding domain of IGF binding protein-3 cannot be functionally substituted by the structurally homologous domain of CCN3. Endocrinology. 2006;147(11):5268–74. doi: 10.1210/en.2005-1568. [DOI] [PubMed] [Google Scholar]
- Yan X, Forbes BE, McNeil KA, Baxter RC, Firth SM. Role of N- and C-terminal residues of insulin-like growth factor (IGF)-binding protein-3 in regulating IGF complex formation and receptor activation. J Biol Chem. 2004;279:53232–40. doi: 10.1074/jbc.M409345200. [DOI] [PubMed] [Google Scholar]




