Abstract
MSX1 has been considered a strong candidate for orofacial clefting, based on mouse expression studies and knockout models, as well as association and linkage studies in humans. MSX1 mutations are also causal for hereditary tooth agenesis. We tested the hypothesis that individuals with orofacial clefting with or without tooth agenesis have MSX1 coding mutations by screening 33 individuals with cleft lip with or without cleft palate (CL/P) and 19 individuals with both orofacial clefting and tooth agenesis. Although no MSX1 coding mutations were identified, the known 101C>G variant occurred more often in subjects with both CL/P and tooth agenesis (p = 0.0008), while the *6C-T variant was found more often in CL/P subjects (p = 0.001). Coding mutations in MSX1 are not the cause of orofacial clefting with or without tooth agenesis in this study population. However, the significant association of MSX1 with both phenotypes implies that MSX1 regulatory elements may be mutated.
Keywords: MSX1, cleft lip, cleft palate, tooth agenesis
INTRODUCTION
Isolated cleft lip with or without cleft palate (CL/P) is a common birth defect that affects about 1/700 births, depending on the population. Asian or Amerindian populations have the highest frequencies (1/500 or higher), Caucasian populations have intermediate frequencies (1/1000), and African populations have the lowest reported frequencies (1/2500) (Vanderas, 1987; Wyszynski et al., 1996; Mossey and Little, 2002).
CL/P presents with characteristics of a genetically complex trait. It has been suggested that from 3 to 14 genes, interacting multiplicatively, may be involved in the etiology of CL/P (Schliekelman and Slatkin, 2002). MSX1, a non-clustered homeobox gene, has been considered a strong candidate for clefting in humans, based on the biological evidence composed of expression studies (Robert et al., 1989) and a knockout mouse model (Satokata and Maas, 1994) as well as association studies (Lidral et al., 1998; Beaty et al., 2001; Fallin et al., 2003; Jugessur et al., 2003; Vieira et al., 2003), complete sequencing (Jezewski et al., 2003; Suzuki et al., 2004), and linkage studies (Moreno et al., 2004; Schultz et al., 2004) in humans.
We have identified a susceptibility locus for isolated CL/P in the 4p16 region in families from Ohio (Moreno et al., 2004). The positive marker D4S2366, located 4.63 cM proximal from the MSX1 gene, presented a LOD score of 1.53 under the parametric recessive linkage analysis. Because of the important role that MSX1 plays in the etiology of orofacial clefting, we performed a MSX1 mutation screen in individuals with isolated CL/P from the same Ohio population.
Furthermore, data from genetic studies are consistent with a contribution of MSX1 to CL/P and tooth agenesis (Slayton et al., 2003) and isolated tooth agenesis (Vieira et al., 2004). MSX1 mutations and rare variants have been previously described in individuals with CL/P and/or hereditary tooth agenesis (Vastardis et al., 1996; van den Boogaard et al., 2000; Jumlongras et al., 2001; Lidral and Reising, 2002; Jezewski et al., 2003; Suzuki et al., 2004; De Muynck et al., 2004). Also, Msx1-deficient mice have both cleft of the secondary palate and failure of tooth development (Satokata and Maas, 1994).
Tooth agenesis affects 1.6% to 9.6% of the Caucasian general population, excluding third molars (Graber, 1978). This malformation is found more frequently in children affected with CL/P than in the general population (Ranta, 1986; Shapira et al., 1999). The prevalence of hypodontia, both in the vicinity of the cleft and outside the cleft area, in the permanent dentition is significantly higher in children with cleft lip, cleft palate, or both, and the prevalence of hypodontia increases markedly with the severity of cleft.
We believe that the occurrence of both CL/P and tooth agenesis in some individuals is caused by the same genetic mutation, and that MSX1 is a very plausible candidate. Therefore, we hypothesized that mutations in MSX1 are causal for orofacial clefting with or without tooth agenesis.
MATERIALS & METHODS
Subjects
The study group consisted of 52 unrelated individuals with orofacial clefting recruited from the Children's Hospital in Columbus, OH, USA. The age range of these individuals was from 7 to 16 yrs old. The inclusion criterion was the diagnosis of orofacial clefting with or without congenital agenesis of at least one permanent tooth, not including third molars, as verified by radiographs and dental history. Instances of tooth agenesis adjacent to a cleft site were not included, because the absence of such teeth is likely the consequence of local developmental anomalies at the cleft site. To identify any syndromes or phenocopies, we examined individuals clinically and interviewed them using a clinical survey to gather information regarding medical history, family history, and gestational environmental exposures.
Thirty-three of the individuals had only isolated orofacial CL/P. They were included in this mutation search, however, because we have found suggestive linkage to the MSX1 region in this population. Nineteen of the individuals had both orofacial clefting and tooth agenesis, and they were included in this study to test the hypothesis that MSX1 is mutated in patients with both phenotypes. Six of these 19 had additional major anomalies or facial dysmorphology (Table 1).
Table 1.
Phenotypes of Individuals with Both Orofacial Clefting and Tooth Agenesis
| ID Number | Gender | Cleft Lipa |
Cleft Palateb |
Family History of Cleftingc |
Family History of Tooth Agenesisc |
Other Findings |
|---|---|---|---|---|---|---|
| 556-1 | F | B | H, S | − | 0 | |
| 585-1 | F | - | H, S | + | 0 | |
| A039* | M | R | H, S | − | − | Sub-aortic stenosis |
| A052* | M | - | S | − | − | |
| A054* | F | - | S | − | − | |
| A063* | M | B | H, S | 0 | 0 | |
| A075* | F | B | H, S | − | − | |
| A100* | F | - | SM | + | − | Facial dysmorphology |
| A107 | M | L | H, S | 0 | 0 | |
| A108 | M | B | H, S | 0 | 0 | |
| A109 | M | L | H, S | + | 0 | Short stature and facial dysmorphology |
| A111 | M | L | H, S | 0 | 0 | |
| A113 | M | B | H, S | − | − | |
| A115 | M | R | H, S | − | − | |
| A117 | M | L | H, S | + | + | |
| A118 | F | B | H, S | 0 | 0 | Ventricular septal defect |
| A119 | F | L | H, S | 0 | 0 | |
| A120 | M | B | H, S | 0 | 0 | Facial dysmorphology |
| A121 | F | - | SM | 0 | 0 | Facial dysmorphology |
Individuals also screened by SSCP for MSX1 mutations as reported by Lidral and Reising (2002).
B, bilateral cleft lip; R, right cleft lip; L, left cleft lip; -, no cleft lip.
H, cleft hard palate; S, cleft soft palate; SM, submucous cleft palate.
+, positive family history; −, negative family history; 0, unknown family history.
The study was approved by the institutional review boards at the Ohio State University and the University of Iowa, and written, informed consent was obtained from each person included in the study.
Direct Sequencing
DNA was extracted from whole blood or cheek swabs with the use of a commercial kit (Puragene, Gentra, Minneapolis, MN, USA). The entire coding region of the MSX1 gene was direct-sequenced in both directions. Four primer pairs were used to amplify overlapping regions of the 2 exons of the MSX1 gene (Appendix). Genomic DNA was amplified by PCR under the following conditions: 0.24μM each primer, 200μM dNTPs, 50 mM KCl, 10 mM Tris Cl, 1 or 1.5 mM MgCl2, 0.01% gelatin, 0.045 U Taq polymerase, 10% (v/v) DMSO, and 20 ng/μL DNA in a 30-μL reaction volume. Templates included either PCR products purified by QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) or QIAquick PCR Purification Kit (Qiagen). Cycle sequencing was performed in a 10-μL reaction with 1μL of ABI Big Dye Terminator sequencing reagent (version 1.1, Applied Biosystems, Foster City, CA, USA), 0.35μL of 20μM/L sequencing primer, 0.5μL DMSO, 1μL of 5X buffer, and 2.5 ng/100 base pair of DNA template. Following a denaturation step at 96°C for 30 sec, reactions were cycle-sequenced at 96°C for 10 sec, at TM (melting temperature) of the primer − 5°C for 5 sec, and 60°C for 4 min for 35 cycles.
Clean-up of amplicons was performed through the AMPure™ PCR Purification system (Agencourt, Beverly, MA, USA), with the use of Agencourt's solid-phase paramagnetic bead technology. Beads were washed with 85% ethanol to remove excess oligonucleotides, nucleotides, salts, and enzymes, and purified products were eluted from the magnetic beads with sterilized water and injected onto an Applied Biosystems 3700 sequencer.
Sequence Analysis
First-pass base-calling (Perkin Elmer) was performed with the ABI sequence software (version 2.1.2). Chromatograms were transferred to a Unix workstation (Sun Microsystems Inc., Mountain View, CA, USA), base-called with Phred (version 0.961028), assembled with Phrap (version 0.960731, scanned by PolyPhred (version 0.970312), and the results viewed with the Consed program (version 4.0) (Nickerson et al., 1997).
Case-Control Comparisons
The case-control comparisons in this study used individuals with orofacial clefting recruited from the Children's Hospital in Columbus, OH, and included in our mutation search. Thirty-three unrelated individuals with only isolated CL/P, and 19 unrelated individuals with both orofacial clefting and tooth agenesis were used in the analysis.
In this study, we used controls of the same ethnic group (Caucasian) from a population-based case-control study within the University of Iowa Craniofacial Anomalies Research Center (CARC), previously genotyped by Lidral et al. (1998). Using a pseudo-random number generator (Romitti et al., 1998), we selected controls from all Iowa live births (between 1 January 1987 and 31 December 1991) not reported to the Iowa Birth Defects Registry.
We also analyzed the group with both orofacial clefting and tooth agenesis, excluding six individuals with clefting, tooth agenesis, and additional major anomalies or facial dysmorphology (Table 1).
Data were analyzed by means of 2 x n contingency tables, which were evaluated by either the Pearson χ2 test or Fisher's exact test, when any of the cells had an expected frequency of ≤ 5. P values < 0.05 were considered statistically significant.
RESULTS
Phenotypes of the individuals who presented with both orofacial clefting and tooth agenesis are described in Tables 1 and 2. Most individuals were missing only 1 tooth outside of the cleft area, and the highest number of missing teeth in an individual was 8 (Fig.). Of the 33 individuals with isolated CL/P, 20 had a positive family history for CL/P. Among the 11 individuals with both orofacial clefting and tooth agenesis, and for whom the family history was known, only four presented a positive family history for CL/P. Among the eight individuals with both orofacial clefting and tooth agenesis, and for whom the family history was known, one presented a positive family history for tooth agenesis.
Table 2.
Summary of Congenitally Missing Teeth in Individuals with Both Orofacial Clefting and Tooth Agenesis
| Right | Left | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID Number | 8* | 7 | 6 | 5 | 4 | 3 | 2 | 1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 556-1 | Maxillary | ||||||||||||||||
| Mandibular | * | * | |||||||||||||||
| 585-1 | Maxillary | * | * | ||||||||||||||
| Mandibular | * | * | |||||||||||||||
| A039 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A052 | Maxillary | ||||||||||||||||
| Mandibular | * | * | |||||||||||||||
| A054 | Maxillary | ||||||||||||||||
| Mandibular | * | ||||||||||||||||
| A063 | Maxillary | ||||||||||||||||
| Mandibular | * | ||||||||||||||||
| A075 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A100 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A107 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A108 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A109 | Maxillary | * | * | ||||||||||||||
| Mandibular | * | * | |||||||||||||||
| A111 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A113 | Maxillary | ||||||||||||||||
| Mandibular | * | ||||||||||||||||
| A115 | Maxillary | * | * | ||||||||||||||
| Mandibular | * | * | |||||||||||||||
| A117 | Maxillary | * | |||||||||||||||
| Mandibular | |||||||||||||||||
| A118 | Maxillary | * | * | * | |||||||||||||
| Mandibular | |||||||||||||||||
| A119 | Maxillary | * | * | ||||||||||||||
| Mandibular | |||||||||||||||||
| A120 | Maxillary | * | * | * | * | ||||||||||||
| Mandibular | * | * | |||||||||||||||
| A121 | Maxillary | * | * | ||||||||||||||
| Mandibular | * | * | * | * | * | * | |||||||||||
1 = central incisor; 2 = lateral incisor; 3 = canine; 4 and 5 = first and second premolars, respectively; 6, 7, and 8 = first, second, and third molars, respectively.
Figure.
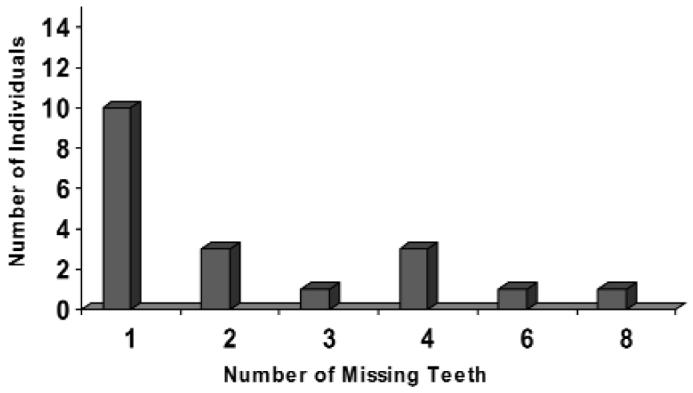
Number of missing teeth in individuals with orofacial clefting and tooth agenesis.
Neither new mutations nor new variants were found in the MSX1 coding regions of the 52 individuals. However, 4 known polymorphic variants were observed in the study population (Table 3). The *6C>T variant, 6 nucleotides 3′ of the stop codon, was observed more frequently in subjects with CL/P than in controls (p = 0.001). The exon 1 101C>G variant, causing an Ala34Gly substitution, was observed more frequently in subjects with both orofacial clefting and tooth agenesis than in controls (p = 0.0008) (Table 3). This difference was still significant after exclusion of the six individuals presenting additional major anomalies or facial dysmorphology (Table 3).
Table 3.
Association of Observed Polymorphism with Orofacial Clefting with or without Tooth Agenesis
| Nucleotide Positiona |
Nucleotide Variant |
Amino Acid Variant |
Case-Control Comparisons | Controlsd, N = 165 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Clefting (CL/P), n = 33* | Clefting and Tooth Agenesisb, n = 19 | Clefting and Tooth Agenesisc, n = 13 | |||||||
| Number of Variantse (%) | p value | Number of Variantse (%) | p value | Number of Variantse (%) | p value | ||||
| 5′UTR | |||||||||
| 434 | −36G>A | - | 4/66 ( 6.1%) | 0.22 | 0/38 ( 0%) | 0.11 | 0/38 ( 0%) | 0.11 | 20/330 ( 6.1%) |
| EXON 1 | |||||||||
| 570 | 101C>G | Ala34Gly | 12/66 (18.2%) | 0.14 | 16/38 (42.1%) | 0.0008 | 9/26 (34.6%) | 0.028 | 60/330 (18.2%) |
| 799 | 330C>T | Gly110Gly | 3/66 ( 4.5%) | 0.11 | 0/38 ( 0%) | 0.56 | 0/38 ( 0%) | 0.56 | 5/318 ( 1.6%) |
| 3′UTR | |||||||||
| 3695 | *6C>T | - | 30/66 (45.4%) | 0.001 | 4/38 (10.5%) | 0.014 | 2/26 ( 7.7%) | 0.017 | 66/248 (26.6%) |
n = number of individuals included in the analysis.
Positions of variants are referred to by the nucleotide position within the GenBank entry, AF426432.
Analysis included 19 individuals with clefting and tooth agenesis.
Analysis excluded six individuals with clefting, tooth agenesis, and additional major anomalies or facial dysmorphology.
Control data from Lidral et al. (1998).
Number of variants is number of chromosomes for rare allele/total number of alleles.
DISCUSSION
No specific variant of MSX1 has been directly implicated as a major causal allele for clefting thus far. In our study, we found association of MSX1 variants not only with clefting, but also with clefting and tooth agenesis (Table 3). The 101C>G variant is more frequent in individuals with both orofacial clefting and tooth agenesis than in controls. In contrast, the *6C>T variant is more common in individuals with isolated CL/P than in controls. Also, the frequency of the *6C>T variant is much lower in clefting with tooth agenesis cases. Similar results were found in the clefting and tooth agenesis study group, when individuals presenting additional major anomalies or facial dysmorphology were excluded from the analysis. Moreover, when all 52 individuals were compared with the controls, the p-value was not significant (p = 0.052) (data not shown). This provides evidence that future investigations should phenotypically distinguish clefting associated with tooth agenesis from clefting alone. It appears that the variant 101C>G is marking a specific genetic factor that contributes to the clefting and tooth agenesis phenotype, and the variant *6C>T is specific for isolated clefting.
Interestingly, the MSX1-CA 169 base-pair allele (allele 4)—the most common allele in all populations, and one that has been associated with isolated clefting in populations of European descent (Lidral et al., 1998; Beaty et al., 2001; Vieira et al., 2003)—is in linkage disequilibrium with the *6C>T variant (Jezewski et al., 2003). It is possible, therefore, that the *6C>T variant is the actual functional variant contributing to clefting in the study population. The cytosine of the *6C>T variant is conserved with bovines, but not with mice or rats. The variant 101C>G could also be functionally relevant to the development of clefting with tooth agenesis. This variant produces an amino acid change, Ala34Gly, which is conserved among mice, rats, bovines, and chickens (Jezewski et al., 2003). Functional analysis of the variants observed in this study would be of interest in further investigations of their consequences and role in clefting, with or without tooth agenesis.
In the present study, no mutations were found in MSX1 coding regions in individuals with isolated CL/P or both orofacial clefting and tooth agenesis. Similarly, De Muynck et al. (2004) found no mutations in 43 families with CL/P with or without tooth agenesis. To date, only one MSX1 mutation was reported in a family with both CL/P and hereditary tooth agenesis (van den Boogaard et al., 2000). One possible explanation could be that MSX1 mutations for this phenotype are in regulatory regions, which have not been well-characterized. Another explanation is that MSX1 microdeletions may be causal in our study participants, and such deletions would have been missed by our mutation screen. Previous reports have showed that oligodontia in individuals with Wolf-Hirschhorn syndrome is associated with deletion or inactivation of one copy of MSX1 (Hu et al., 1998; Nieminen et al., 2003), supporting the conclusion that hereditary tooth agenesis associated with mutations in MSX1 is caused by haploinsufficiency.
Gene-gene interactions could be the mechanism for developing CL/P with tooth agenesis. There is genetic evidence that MSX1 interacts with PAX9 in isolated tooth agenesis (Vieira et al., 2004), and apparently Pax9 regulates Msx1 expression in the mouse (Peters et al., 1998). PAX9 is also a transcription factor expressed in the face and tooth buds, and mice lacking Pax9 also present with both cleft palate and oligodontia (Peters et al., 1998). Future studies focusing on the MSX1 variants observed in our study and PAX9 variants are recommended to test the hypothesis that these two genes play a joint role in CL/P with tooth agenesis.
Finally, the phenotypes of the study participants may not be caused by MSX1 coding mutations. There is a typical pattern of tooth agenesis and a large number of missing teeth among the families reported to have MSX1 mutations. Specifically, the average number of missing teeth has been reported to be 11/person (Vastardis et al., 1996), 8/person (van den Boogaard et al., 2000), 16/person (Jumlongras et al., 2001), 12/person (Lidral and Reising, 2002), and 17/person (De Muynck et al, 2004). In our study, only two cases (10.5%) presented oligodontia (6 or more missing teeth) (Fig.). This suggests, again, that agenesis of only a few teeth is not associated with MSX1 mutations (Lidral and Reising, 2002).
ACKNOWLEDGMENTS
We thank all the individuals who participated in this study. We also thank Jeff Murray for the use of his laboratory facilities and equipment under the auspices of the Craniofacial Anomalies Research Center. This work has benefited greatly from discussions with Alex Vieira. We appreciate the insightful technical suggestions from Steven Bullard. This work was supported by NIH grant R01DE14677, March of Dimes grant #6-FY01-616, and start-up funds from the Ohio State University.
APPENDIX
MSX1 PRIMER PAIRS
| Name | Primer Sequence | Primer Locationa | Size (bp) |
|---|---|---|---|
| X1.1F | TGG CCA GTG CTG CGG CAG AA | 414-433 | 421 |
| X1.3R | TCT GGC AGC TTG AGG AGT CC | 815-834 | |
| X1.4F | CGC TCG GCC ATT TCT CGG TG | 792-811 | 152 |
| X1.4R | GCG CCT GGG TTC TGG CTA CT | 924-943 | |
| X2.1F | GGC TGA TCA TGC TCC AAT GCT | 3186-3205 | 493 |
| X2.3R | GTA CAT GCT GTA GCC CAC AT | 3658-3677 | |
| X2.3F | AGC TGG AGA AGC TGA AGA TG | 3478-3497 | 264 |
| X2.4R | GCA CCA GGG CTG GAG GAA TC | 3722-3741 |
Nucleotide numbers correspond to GenBank entry AF426432.
Footnotes
A supplemental appendix to this article is published electronically only at http://www.dentalresearch.org.
REFERENCES
- Beaty TH, Wang H, Hetmanski JB, Fan YT, Zeiger JS, Liang KY, et al. A case-control study of nonsyndromic oral clefts in Maryland. Ann Epidemiol. 2001;11:434–442. doi: 10.1016/s1047-2797(01)00222-8. [DOI] [PubMed] [Google Scholar]
- De Muynck S, Schollen E, Matthijs G, Verdonck A, Devriendt K, Carels C. A novel MSX1 mutation in hypodontia. Am J Med Genet A. 2004;128:401–403. doi: 10.1002/ajmg.a.30181. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Hetmanski JB, Park J, Scott AF, Ingersoll R, Fuernkranz HA, et al. Family-based analysis of MSX1 haplotypes for association with oral clefts. Genetic Epidemiol. 2003;25:168–175. doi: 10.1002/gepi.10255. [DOI] [PubMed] [Google Scholar]
- Graber LW. Congenital absence of teeth: a review with emphasis on inheritance patterns. J Am Dent Assoc. 1978;96:266–275. doi: 10.14219/jada.archive.1978.0054. [DOI] [PubMed] [Google Scholar]
- Hu G, Vastardis H, Bendall AJ, Wang Z, Logan M, Zhang H, et al. Haploinsufficiency of MSX1: a mechanism for selective tooth agenesis. Mol Cell Biol. 1998;18:6044–6051. doi: 10.1128/mcb.18.10.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet. 2003;40:399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Lie RT, Wilcox AJ, Murray JC, Taylor JA, Saugstad OD, et al. Variants of developmental genes (TGFA, TGFB3, and MSX1) and their associations with orofacial clefts: a case-parent triad analysis. Genet Epidemiol. 2003;24:230–239. doi: 10.1002/gepi.10223. [DOI] [PubMed] [Google Scholar]
- Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, et al. A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet. 2001;69:67–74. doi: 10.1086/321271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Reising BC. The role of MSX1 in human tooth agenesis. J Dent Res. 2002;81:274–278. doi: 10.1177/154405910208100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63:557–568. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno LM, Arcos-Burgos M, Marazita ML, Krahn K, Maher BS, Cooper ME, et al. Genetic analysis of candidate loci in non-syndromic cleft lip families from Antioquia-Colombia and Ohio. Am J Med Genet A. 2004;125:135–144. doi: 10.1002/ajmg.a.20425. [DOI] [PubMed] [Google Scholar]
- Mossey PA, Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski DF, editor. Cleft lip & palate: from origin to treatment. Oxford University Press; New York: 2002. pp. 127–158. [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen P, Kotilainen J, Aalto Y, Knuutila S, Pirinen S, Thesleff I. MSX1 gene is deleted in Wolf-Hirschhorn syndrome patients with oligodontia. J Dent Res. 2003;82:1013–1017. doi: 10.1177/154405910308201215. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta R. A review of tooth formation in children with cleft lip/palate. Am J Orthod Dentofacial Orthop. 1986;90:11–18. doi: 10.1016/0889-5406(86)90022-3. [DOI] [PubMed] [Google Scholar]
- Robert B, Sassoon D, Jacq B, Gehring W, Buckingham M. Hox-7, a mouse homeobox gene with a novel pattern of expression during embryogenesis. EMBO J. 1989;8:91–100. doi: 10.1002/j.1460-2075.1989.tb03352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romitti PA, Munger RG, Murray JC, Daack-Hirsch S, Hanson JW, Burns TL. The effect of follow-up on limiting non-participation bias in genetic epidemiologic investigations. Eur J Epidemiol. 1998;14:129–138. doi: 10.1023/a:1007406313703. [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Schliekelman P, Slatkin M. Multiplex relative risk and estimation of the number of loci underlying an inherited disease. Am J Hum Genet. 2002;71:1369–1385. doi: 10.1086/344779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RE, Cooper ME, Daack-Hirsch S, Shi M, Nepomucena B, Graf KA, et al. Targeted scan of fifteen regions for nonsyndromic cleft lip and palate in Filipino families. Am J Med Genet A. 2004;125:17–22. doi: 10.1002/ajmg.a.20424. [DOI] [PubMed] [Google Scholar]
- Shapira Y, Lubit E, Kuftinec MM. Congenitally missing second premolars in cleft lip and cleft palate children. Am J Orthod Dentofacial Orthop. 1999;115:396–400. doi: 10.1016/s0889-5406(99)70258-1. [DOI] [PubMed] [Google Scholar]
- Slayton RL, Williams L, Murray JC, Wheeler JJ, Lidral AC, Nishimura CJ. Genetic association studies of cleft lip and/or palate with hypodontia outside the cleft region. Cleft Palate Craniofac J. 2003;40:274–279. doi: 10.1597/1545-1569(2003)040<0274:GASOCL>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Jezewski PA, Machida J, Watanabe Y, Shi M, Cooper ME, et al. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet Med. 2004;6:117–125. doi: 10.1097/01.gim.0000127275.52925.05. [DOI] [PubMed] [Google Scholar]
- van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Orioli IM, Castilla EE, Cooper ME, Marazita ML, Murray JC. MSX1 and TGFB3 contribute to clefting in South America. J Dent Res. 2003;82:289–292. doi: 10.1177/154405910308200409. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Meira R, Modesto A, Murray JC. MSX1, PAX9, and TGFA contribute to tooth agenesis in humans. J Dent Res. 2004;83:723–727. doi: 10.1177/154405910408300913. [DOI] [PubMed] [Google Scholar]
- Wyszynski DF, Beaty TH, Maestri NE. Genetics of nonsyndromic oral clefts revisited. Cleft Palate Craniofac J. 1996;33:406–417. doi: 10.1597/1545-1569_1996_033_0406_gonocr_2.3.co_2. [DOI] [PubMed] [Google Scholar]


