Abstract
Guanidinium hydrochloride (GuHCl) at low concentrations significantly stabilizes the Fyn SH3 domain. In this work, we have demonstrated that this stabilizing effect is manifested through a dramatic (five- to sixfold) decrease in the unfolding rate of the domain with the folding rate being affected minimally. This behavior contrasts to the effect of NaCl, which stabilizes this domain by accelerating the folding rate. These data imply that the stabilizing effect of GuHCl is not predominantly ionic in nature. Through NMR studies, we have identified a specific binding site for guanidinium, and we have determined a dissociation constant of 90mMfor this interaction. The guanidinium-binding site overlaps with a functionally important arginine-binding pocket on the domain surface, and we have shown that GuHCl is a specific inhibitor of the peptide-binding activity of the domain. A different SH3 domain possessing a similar arginine-binding pocket is also thermodynamically stabilized by GuHCl. These data suggest that many proteins that normally interact with arginine-containing ligands may also be able to specifically interact with guanidinium. Thus, some caution should be used when using GuHCl as a denaturant in protein folding studies. Since arginine-mediated interactions are often important in the energetics of protein–protein interactions, our observations could be relevant for the design of small molecule inhibitors of protein–protein interactions.
Keywords: SH3 domain, protein folding kinetics, guanidinium-induced protein stabilization, peptide binding, arginine–protein interaction, specific guanidinium binding
A multitude of experimental studies have investigated the thermodynamic stability and folding kinetics of proteins. In the majority of these studies, chemical denaturants, generally urea or guanidine hydrochloride (GuHCl), have been utilized to induce protein unfolding. Despite the routine use of chemical denaturants in these studies, the mechanisms by which these agents affect protein stability are still not fully understood (Schellman 2002).
The effects of GuHCl on protein stability are particularly complicated due to its ionic character. Ions can either bind to the folded or unfolded states of proteins or influence stability through “screening” coulombic interactions, the Hofmeister effect, or changing the structure of the solvent (for a recent review, see Collins 2004). Thus, GuHCl can modulate protein stability through mechanisms related to both its denaturant and ionic properties. Although GuHCl is thought of primarily as a denaturant, there have been many cases reported where low concentrations of GuHCl actually stabilize proteins (Mayr and Schmid 1993; Monera et al. 1994; Makhatadze et al. 1998; Bhuyan 2002). In the majority of these cases, it has been shown that the ionic nature of GuHCl is the main factor in its stabilizing effect (Monera et al. 1994; Makhatadze et al. 1998). However, some studies have suggested that GuHCl may cause protein stabilization in a more specific manner (Mayr and Schmid 1993). Solution of a crystal structure of Ribonuclease A in GuHCl demonstrated direct binding of this compound to the folded state of the enzyme, resulting in a decrease in temperature factors (Dunbar et al. 1997). However, it was observed that guanidine bound to sites comprised of various amino acid residues with no obvious sequence or structural similarity among these sites detected. Guanidinium has been recently shown to be able to mimic the interactions mediated by the guanidino (H2N-CNH-NH2)+ moiety of an arginine side chain and restabilize a T4 Lysozyme variant with an arginine to alanine mutation (Yousef et al. 2004).
In a previous study performed in our laboratory examining the thermodynamic stabilities of several site-directed mutants of the SH3 domain of the Fyn tyrosine kinase, we found that the use of GuHCl as a denaturant in unfolding experiments resulted in measured unfolding free energy (ΔGu) values that were higher by ~1.1 kcal/mol compared with those obtained using urea-induced unfolding experiments or NMR spin relaxation dispersion spectroscopy in the absence of denaturants (Maxwell and Davidson 1998; Northey et al. 2002; Di Nardo et al. 2004). Our goal in the studies presented here is to elucidate the mechanism by which GuHCl stabilizes the Fyn SH3 domain. We have used folding kinetics studies, functional assays, mutagenesis, and NMR spectroscopy to systematically investigate the effects of GuHCl on the structure and stability of this domain, and we demonstrate the existence of a specific binding site for guanidinium on the surface of this protein.
Results and Discussion
GuHCl stabilizes the Fyn SH3 domain by slowing its unfolding rate
To obtain insights into the mechanism of stabilization by GuHCl of the Fyn SH3 domain, the folding (kf) and unfolding (ku) rates of a urea-denatured Fyn SH3 domain in the presence of low concentrations of GuHCl or NaCl were measured and are reported in Table 1. Since the complete unfolding of the wild-type Fyn SH3 domain requires nearly saturated solutions of urea, the unfolding arm of the chevron plot of this domain measured in urea is quite short, leading to larger than normal errors in determination of the unfolding rate (ku) of this protein and its dependence on urea concentration (mku) (Fig. 1; Table 1). To obtain more accurate measurements of ku values, we also used a destabilized mutant, R40N, with a ΔΔGu value of 2.61 kcal/mol with respect to wild type. The transition midpoint of urea denaturation of this mutant was shifted to a lower concentration of urea so that more data points could be collected along the unfolding arm of the chevron plot. For both the wild type and R40N mutant, it can be seen that GuHCl is indeed highly stabilizing at low concentrations with free energy of unfolding (ΔGu) of the wild-type domain increased by 1 kcal/mol in 0.4 M GuHCl and that of the R40N mutant increased by 0.6 kcal/mol at 0.2 M GuHCl (Table 1). It is clear that the stabilizing effect of GuHCl on this domain is manifested exclusively through slowing of the unfolding rate. For example, at 0.4MGuHCl, the unfolding rate of the wild-type domain is slowed by 6.5-fold, while the folding rate is affected very little. The R40N mutant is stabilized in the same manner. At concentrations greater than 0.4 M, GuHCl displays a net destabilizing effect, such that the change in the free energy of unfolding (ΔGu) of the R40N mutant assumes a strong linear relationship (r = 0.99) with GuHCl concentration (plot not shown). The slope of the best-fit line (1.71 kcal/mol.M) obtained using the R40N data is almost identical to the previously reported “meq value” (1.73 kcal/mol.M) for the Fyn SH3 domain (Maxwell and Davidson 1998).
Table 1.
The effect of GuHCl and NaCl on folding kinetics and thermodynamic stability of urea-denatured Fyn SH3 domain
| kf sec−1 | mkfa | kusec−1 | mkua | ΔGub kcal/mol | |
| Fyn WT [GuHCl] (M) | |||||
| 0 | 87.19 ± 4.61 | 0.82 ± 0.01 | 0.015 ± 0.009 | 0.40 ± 0.07 | 5.13 ± 0.30 |
| 0.2 | 81.40 ± 4.90 | 0.82 ± 0.02 | 0.004 ± 0.005 | 0.55 ± 0.14 | 5.83 ± 0.50 |
| 0.4 | 68.98 ± 5.40 | 0.81 ± 0.02 | 0.002 ± 0.003 | 0.63 ± 0.19 | 6.12 ± 0.70 |
| 0.6 | 55.90 ± 4.80 | 0.81 ± 0.03 | 0.009 ± 0.012 | 0.45 ± 0.20 | 5.19 ± 0.70 |
| Fyn R40N [GuHCl] (M) | |||||
| 0 | 5.46 ± 0.50 | 1.01 ± 0.06 | 0.076 ± 0.019 | 0.35 ± 0.04 | 2.52 ± 0.08 |
| 0.2 | 5.04 ± 0.50 | 0.95 ± 0.05 | 0.027 ± 0.009 | 0.50 ± 0.05 | 3.10 ± 0.12 |
| 0.4 | 3.76 ± 0.20 | 0.92 ± 0.04 | 0.031 ± 0.008 | 0.43 ± 0.04 | 2.84 ± 0.12 |
| 0.6 | 3.12 ± 0.16 | 0.98 ± 0.04 | 0.050 ± 0.010 | 0.32 ± 0.03 | 2.44 ± 0.08 |
| 0.8 | 2.07 ± 0.16 | 1.02 ± 0.07 | 0.063 ± 0.015 | 0.28 ± 0.04 | 2.06 ± 0.08 |
| 1.0 | 1.27 ± 0.11 | 0.99 ± 0.10 | 0.064 ± 0.018 | 0.30 ± 0.05 | 1.77 ± 0.10 |
| Fyn R40N [NaCl] (M) | |||||
| 0 | 5.46 ± 0.50 | 1.01 ± 0.06 | 0.076 ± 0.019 | 0.35 ± 0.04 | 2.52 ± 0.08 |
| 0.2 | 11.12 ± 1.27 | 1.09 ± 0.08 | 0.073 ± 0.020 | 0.26 ± 0.05 | 2.70 ± 0.12 |
| 0.4 | 15.28 ± 0.66 | 1.06 ± 0.03 | 0.064 ± 0.013 | 0.24 ± 0.03 | 3.24 ± 0.12 |
| 0.6 | 24.70 ± 1.60 | 1.14 ± 0.04 | 0.087 ± 0.047 | 0.17 ± 0.08 | 3.44 ± 0.08 |
Errors reported for kf, ku, mkf, and mku are fitting errors. Errors reported for ΔGu are by error propagation as described in Materials and Methods.
amkf and mku are the dependence of ln (kf) and ln (ku), respectively, on the concentration of urea.
b ΔGu = −RTln (ku/kf ).
Figure 1.
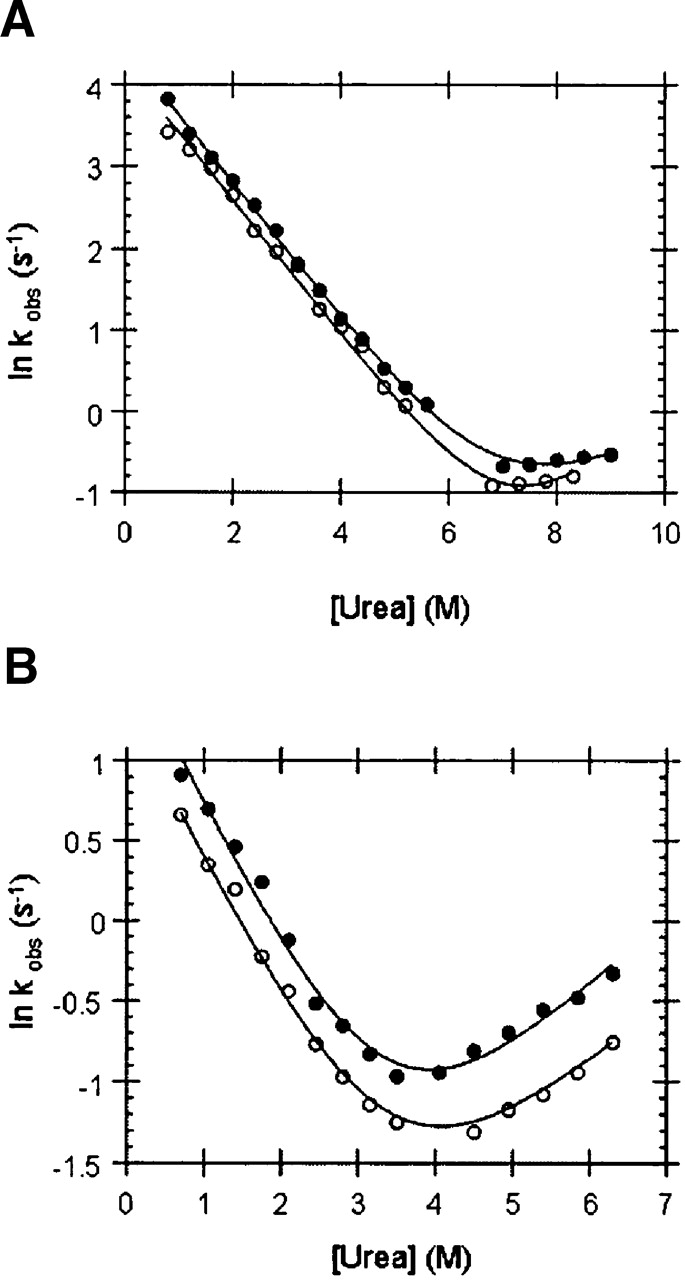
Chevron plots of (A) wild-type Fyn SH3 domain and (B) its R40N mutant in 0.4 M GuHCl. (•) Chevron plots in the absence of GuHCl.
To determine whether the stabilizing effect of GuHCl was due simply to its ionic character, we compared the stabilizing effect of this compound to that of NaCl. We have reported results for only the R40N mutant here mainly because of the difficulty associated with accurate measurements of the unfolding rate (ku) of the wild-type protein as described above. In addition, the wild-type protein folded at such a fast rate in high NaCl that it was difficult to measure accurately. The folding rate of the Fyn R40N mutant progressively increased in the presence of increasing concentrations of salt such that a plot of the natural logarithm of the folding rate (ln kf) assumes a strong linear dependence (r = 0.98) on the square root of the ionic strength (M1/2) of the NaCl concentration (plot not shown). Unfolding rates, on the other hand, essentially remained constant upon increasing salt concentration (see Table 1). In a recent study, a similar behavior for salt-mediated stabilization of the Fyn SH3 domain was observed (de Los Rios and Plaxco 2005). This behavior, where NaCl stabilizes predominantly by accelerating the folding rate, is a complete contrast to the response of the folding kinetics of the Fyn SH3 domain to increasing concentrations of GuHCl. These data imply that the mechanism of stabilization by GuHCl of this domain is not primarily ionic in nature.
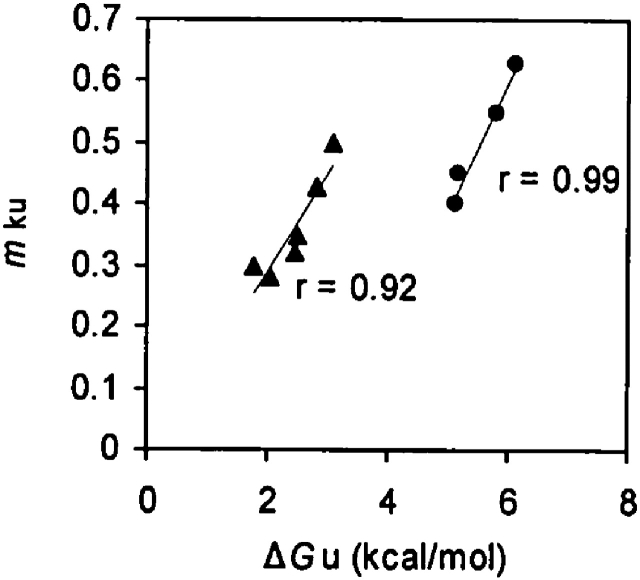
The deceleration of the unfolding rate in the presence of GuHCl was accompanied by no drastic change in the folding rate, which suggests that guanidinium exclusively interacts with the native state of this protein. Consistent with this notion, the mkf values remain unaltered upon increasing the concentrations of GuHCl (Table 1), whereas the mku values increase with increasing overall stability (ΔGu). As illustrated in Figure 2, the strong correlation (r = 0.99 for wild type and r = 0.92 for R40N) between mku and ΔGu argues for exclusive binding of guanidinium to the native state of the Fyn SH3 domain. Since the mku value is believed to be proportional to the difference in the exposed surface area between the native state and the folding transition state (Sanchez and Kiefhaber 2003), these results suggest that guanidinium binding buries part of the surface of the native state. Thus, a relatively larger surface area would become solvent exposed upon unfolding to the transition state of the guanidinium-bound Fyn SH3 domain. In addition, the lack of change of the mkf value suggests that GuHCl does not preferentially interact with either the unfolded or the folding transition state of this protein.
Figure 2.
The correlation between ΔGu and kinetic mku values for the wild-type Fyn SH3 domain (•) and the R40N mutant (▴) at different concentrations of GuHCl. The ΔGu values plotted were determined at the GuHCl concentrations shown in Table 1.
NMR peak shift analysis reveals the GuHCl binding site and allows for determination of its binding affinity
To explore the possibility of specific guanidinium binding to the folded state of the Fyn SH3 domain, we collected 15N-1H HSQC correlation spectra (Kay et al. 1992) of this protein in the presence of varying concentrations of GuHCl ranging from 0 to 0.85 M. As shown in Figure 3A, most amide peak positions are unaffected by the addition of GuHCl; however, the 1H and 15N chemical shifts of several residues exhibit dramatic changes. Notably, these peaks do not shift significantly in response to the addition of either 2Murea or 1MNaCl, as illustrated in Figure 3B. This suggests that the effect of GuHCl on the 15N-1H HSQC spectrum of the Fyn SH3 domain is caused by a specific interaction and is not simply due to the ionic or denaturant properties of this compound.
Figure 3.
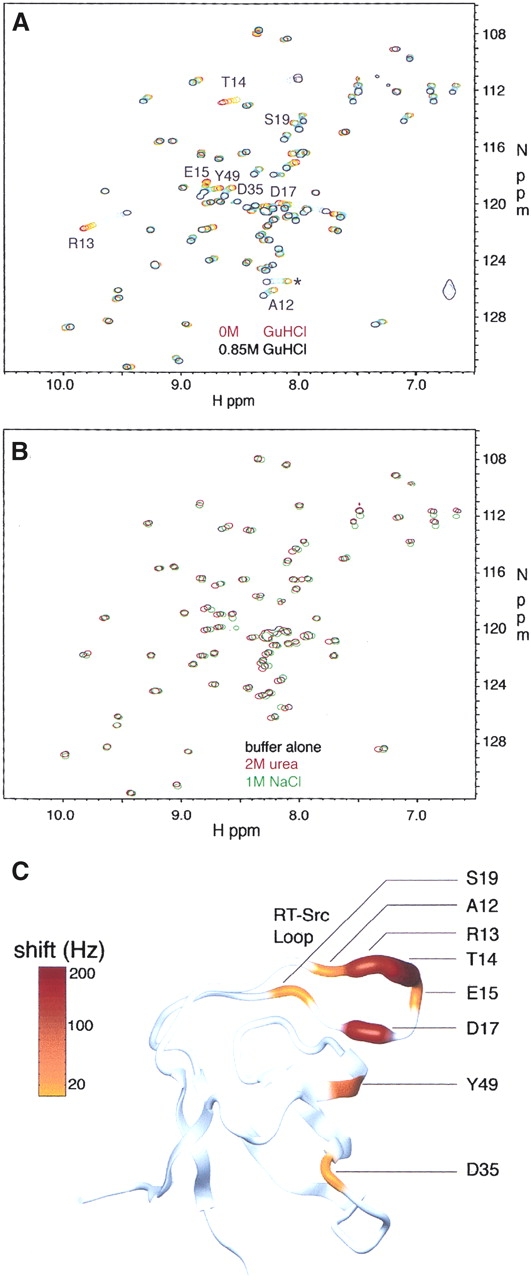
Overlays of 15N-1H correlation maps collected for wild-type Fyn SH3 domain samples containing (A) 0–0.85M GuHCl and (B) 2 M urea or 1MNaCl. The peak marked with an asterisk in A corresponds to a residue in the C-terminal 6-his tag region of the construct that shifted significantly upon GuHCl addition. Fitting the data from this peak yielded a Kd value of 560 ± 70 mM. This Kd value is only threefold different from that reported for the nonspecific protein :GuHCl interactions and is unlikely to report on a specific interaction between the 6-his tag and GuHCl. (C) Magnitude of total baseline-corrected peak shifts (ΔΩ = [ΔΩH2 + ΔΩN2]1/2), color coded on a ribbon-representation of the Fyn SH3 domain (generated using the coordinates of the X-ray crystal structure [Noble et al. 1993] and the program MOLMOL [Koradi et al. 1996]).
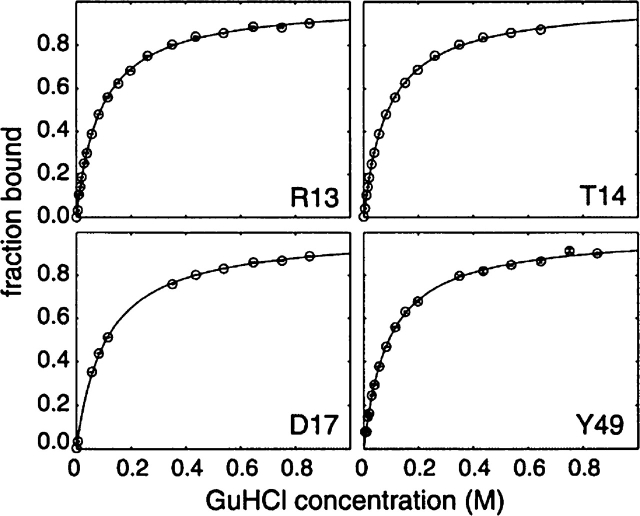
Little line-broadening is evident upon GuHCl addition, even for those peaks with the greatest chemical-shift changes, indicating that exchange between guanidinium-bound and unbound states is fast on the NMR chemical shift timescale. Under these conditions, the shift in peak position relative to the 0 M GuHCl spectrum is directly proportional to the fraction of the bound state, assuming a two-state binding model. We have used data for residues whose amide resonances change by more than one peak width to estimate the fraction of bound protein as a function of GuHCl concentration, as well as the dissociation constant (Kd) for guanidinium binding on a perresidue basis. The fraction of bound protein is plotted as a function of GuHCl concentration for several residues in Figure 4, together with best-fit hyperbolic curves. Values of Kd obtained for different peaks are in good agreement with an error-weighted mean of 90 ± 0.5 mM. Individual estimates for strongly shifting residues A12, R13, T14, E15, D17, and Y49 are within 40 mM of the mean value. This estimate of a Kd value of 90 mM for the Fyn SH3 domain :GuHCl interaction is 18-fold lower than the previously reported Kd value of 1.6 M for the nonspecific interaction between proteins or peptides and GuHCl (Makhatadze and Privalov 1992; Makhatadze 1999; Moglich et al. 2005).
Figure 4.
The guanidinium binding curves determined from NMR peak shift analysis. The fraction of protein bound is plotted as a function of GuHCl concentration with best-fit curves calculated according to Equation 2. Values were obtained from analyses of shifts in peak position in the 1H dimension. Results shown are for four different residues as indicated in the figure.
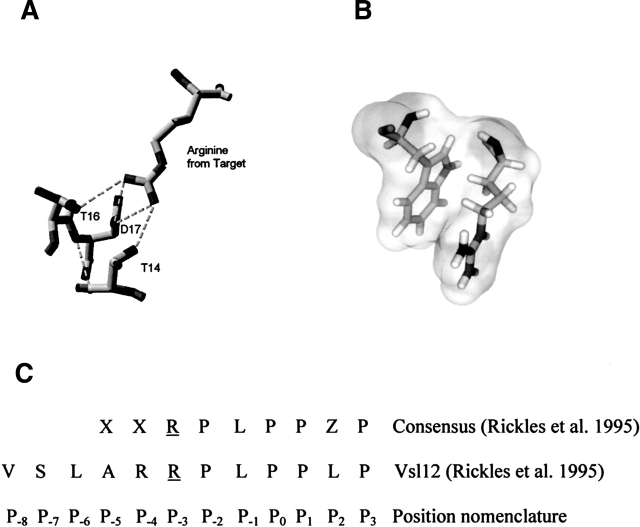
The magnitudes of peak shifts of residues that interact with guanidinium are color coded on the crystal structure of the Fyn SH3 domain in Figure 3C. It is clear that the largest changes in chemical shift are obtained for residues in the RT-Src loop and for residue Y49 in the fourth β strand. Interestingly, these residues are part of a conserved functional surface on SH3 domains that is often involved in binding to the side chain of a conserved arginine residue found in many SH3 target peptides. For example, the structure of a complex between the c-Src tyrosine kinase SH3 domain and the phage display-derived Vsl12 peptide ligand (Feng et al. 1995) reveals that the guanidino moiety of the target peptide arginine at the P−3 position (see Fig. 5C for position nomenclature) interacts with the RT-Src loop by forming hydrogen bonds with T14, T16, and D17 side chains (Fig. 5A). Since the Fyn SH3 domain also binds arginine-containing peptides, including Vsl12 (Rickles et al. 1995), and its sequence is 75% identical to that of the Src SH3 domain, it is likely that the same regions of this protein contact the conserved arginine side chain of target peptides. It is significant that both guanidinium and the arginine side chains contain a guanidino (H2N-CNH-NH2)+ moiety. Thus, related interactions to those between the SH3 domain and the target-peptide arginine side chain may also stabilize a complex with guanidinium.
Figure 5.
The interactions of the conserved arginine in SH3 domain ligands. (A) The guanidino moiety of the arginine side chain is involved in forming hydrogen bonds with the side chains of T14, T16, and D17 in the Src SH3 domain. (B) The aliphatic moiety of the arginine side chain donated by the target is also closely stacked against the side chain of W36. Figures were generated using the PDB file 1QWF (Feng et al. 1995) and the program PyMOL (DeLano Scientific). (C) The consensus sequence of phage display targets that have affinity for the Fyn SH3 domain is given along with the sequence of the Vsl12 ligand and the position nomenclature. The symbol X in the consensus sequence is any residue, and Z denotes hydrophobic residues. The conserved arginine residue at the P−3 position is underlined.
GuHCl inhibits the peptide-binding activity of the Fyn SH3 domain
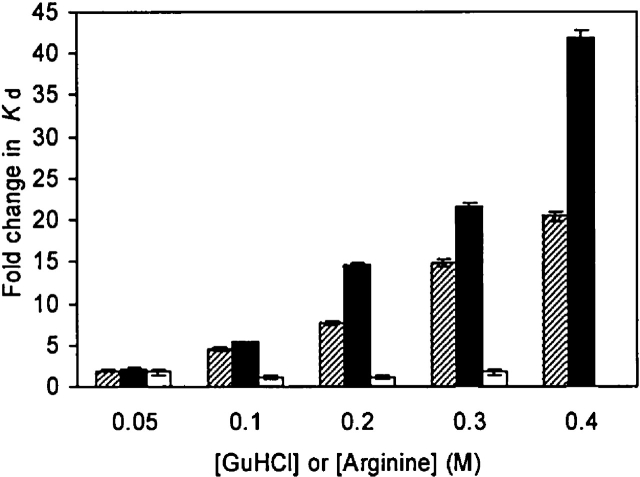
To directly demonstrate the binding of guanidinium to the site revealed by NMR peak shift analysis we took advantage of the overlap that existed between the proposed guanidinium-binding pocket and the binding site for the natural SH3 target peptide and measured the affinity of the Fyn SH3 domain for its target peptide in the presence of varying concentrations of GuHCl and arginine. As illustrated in Figure 6, increasing the concentration of GuHCl or arginine significantly decreased the affinity of the wild-type Fyn SH3 domain for its phage-display target peptide Vsl12. At 0.4 M GuHCl, the affinity of the wild-type Fyn SH3 domain for the Vsl12 peptide decreased 25-fold compared with the affinity in the absence of GuHCl. Urea at 0.8 M concentration and NaCl at 0.4 M, on the other hand, had no significant effects on the affinity, suggesting that the denaturant and the ionic properties of GuHCl play no role in its competition with target peptide for binding to the Fyn SH3 domain (data not shown). These observations are also consistent with NMR results suggesting no specific binding to the folded state of the Fyn SH3 domain of urea or NaCl.
Figure 6.
The effect of GuHCl on the affinity of the wild-type Fyn SH3 domain (hatched bars) and the T14R mutant (open bars) for the Vsl12 target peptide. Black bars indicate the effect of arginine on the affinity of the wild-type Fyn SH3 domain for this ligand. The reported “fold change” in Kd was calculated by normalizing the Kd value obtained at any concentration of GuHCl or arginine to the value obtained in the absence of GuHCl or arginine. Depicted error bars in each case indicate fitting errors. The Kd of T14R in 0.4 M GuHCl was not determined. The interaction between wild-type Fyn SH3 domain and target peptide in 0 M GuHCl has a Kd value of 0.22 ± 0.01 μM.
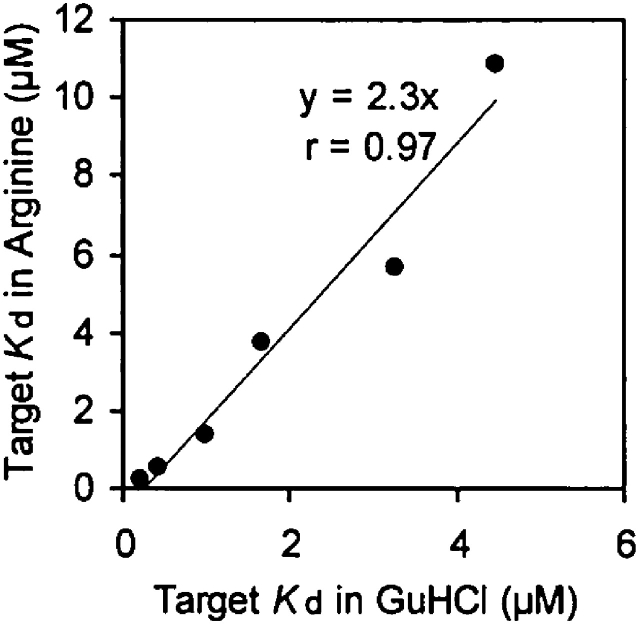
In Figure 7, we compare the efficacy on an equimolar basis of GuHCl and arginine in displacing target peptide. A plot of target Kd in corresponding concentrations of GuHCl and arginine is linear (r = 0.98). The excellent linear correlation of this fit supports the results of the NMR experiments, demonstrating that guanidinium utilizes the arginine-binding pocket on the surface of the Fyn SH3 domain for binding. Interestingly, the slope of the best-fit line (KdArg = 2.3·KdGuHCl) shows that arginine is a more potent competitor of the target peptide in binding to the Fyn SH3 domain. In the structure of the Src SH3 domain in complex with the Vsl12 peptide ligand (Feng et al. 1995), the aliphatic methylene groups of the arginine side chain in the target peptide are packed against side-chain atoms of W36 (Fig. 5B). The lack of aliphatic elements in guanidinium may explain its lower affinity for the arginine-binding pocket. Taking the above observations into consideration, we conclude that the displacement of target peptide by GuHCl is a direct result of the specific binding of guanidinium to the arginine-binding pocket of the Fyn SH3 domain.
Figure 7.
The influence of arginine and guanidinium binding on the affinity of the SH3 domain for its target peptide. Affinity of the wildtype Fyn SH3 domain for target peptide is plotted at corresponding concentrations of arginine and GuHCl.
Upon guanidinium binding, the T14 residue in the arginine-binding pocket exhibited the greatest amide proton resonance frequency shift ~200 Hz. To experimentally test whether T14 is critical for binding of guanidinium, we analyzed the binding of the Vsl12 target peptide to the T14R mutant in the presence of increasing concentrations of GuHCl ranging from 0 to 0.4 M. The T14R mutant lacking the critical T14 residue binds target peptide with a Kd value of 8.83 ± 0.48 μM, about 40-fold weaker than wild type. However, as illustrated in Figure 6, the addition of GuHCl has little effect on the affinity of T14R for its target peptide. Consistent with the results of the NMR experiments, these observations argue for a critical role of T14 in the binding of guanidinium, and confirm that the interaction between the Fyn SH3 domain and guanidinium is highly specific.
Does GuHCl stabilize other SH3 domains?
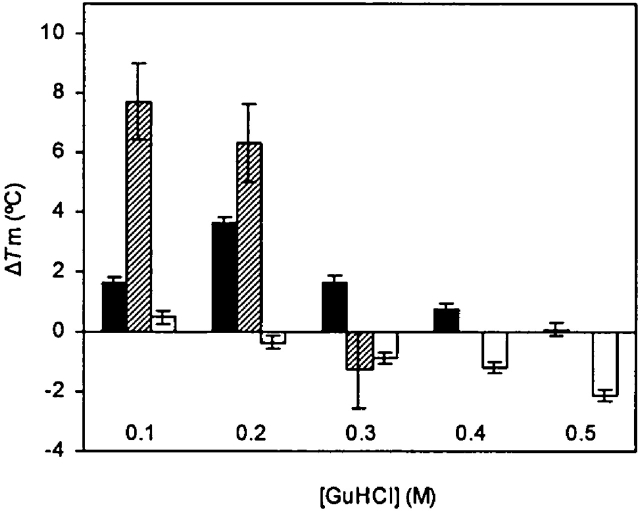
To address the question of whether stabilization by GuHCl might be a general feature of proteins that interact with arginine, we examined the ability of GuHCl to stabilize two other SH3 domains, one from the yeast Actin Binding Protein 1 (Abp1) protein and the other from the yeast Sho1 protein. As a simple means to test the stabilizing effect of GuHCl on these domains, we performed temperature-induced unfolding experiments in varying amounts of GuHCl. As expected, the wild-type Fyn SH3 domain exhibited an increase in the midpoint of its temperature-induced unfolding transition (Tm) in the presence of low concentrations of GuHCl, which was maximal at 0.2 M GuHCl, where the Tm was increased by 3.6°C (Fig. 8). The Sho1 SH3 domain exhibited an even greater degree of stabilization in the presence of GuHCl with a Tm increase of more than 7°C at 0.1 M GuHCl. In contrast, the Abp1 SH3 domain did not exhibit an increase in Tm at any concentration of GuHCl. Since the Sho1 SH3 domain has been shown to bind strongly to target peptides possessing arginine at the P−3 position (Marles et al. 2004), the stabilization of this domain by GuHCl is not surprising. On the other hand, the Abp1 SH3 domain binds tightly to a different class of peptide ligands containing lysine at the P−3 position (Rath and Davidson 2000; Zarrinpar et al. 2004), and this domain has never been directly shown to tightly bind peptides containing arginine at the P−3 position4. These data suggest that the ability of SH3 domains to be stabilized by GuHCl correlates with their ability to bind to peptides possessing arginine at the P−3 position.
Figure 8.
Influence of GuHCl on the Tm of the wild-type Fyn SH3 domain (black bars), the Sho1 SH3 domain (hatched bars), and the Abp1 SH3 domain (open bars). All of the ΔTm values are with respect to the Tm in 0 M GuHCl. A negative ΔTm value is indicative of destabilization. Error bars indicate uncertainties associated with fitting denaturation profiles to the appropriate equations as described in Materials and Methods. The Sho1 SH3 domain did not exhibit cooperative unfolding at GuHCl concentration of 0.4 M or higher. The wild-type Fyn, Abp1, and Sho1 SH3 domains in 0 M GuHCl have Tm values of 76.9 ± 0.11, 53.43 ± 0.20, and 47.75 ± 1.37 (°C), respectively.
Conclusions
We have demonstrated, for the first time, the presence of a specific binding pocket for guanidinium on the surface of a protein, which overlaps with a functionally important arginine-binding surface. Our data show that guanidinium binding to the Fyn SH3 domain results in increased thermodynamic stability and specific inhibition of function. Another SH3 domain, which binds arginine-containing peptides, is also stabilized by GuHCl. Our results raise the possibility that any protein with an arginine-binding site may be stabilized by GuHCl. Since arginine is one of the three most common amino acids that are both present and energetically important within protein–protein interaction interfaces (Jones and Thornton 1996; Bogan and Thorn 1998), many proteins must possess arginine-binding pockets on their surfaces. Thus, the phenomenon of specific protein stabilization by guanidinium may be considerably more widespread than has been previously recognized. This observation should provide a note of caution for investigators using GuHCl as a denaturant in quantitative studies of protein folding and stability. On the other hand, the knowledge that many protein–protein interaction interfaces will specifically bind to guanidinium could provide a new starting point for the rational design of small molecule inhibitors of these interactions.
Materials and methods
Chemicals
Optical grade solutions of 8 M GuHCl were purchased from Pierce Biotechnology. Bioshop Canada was the provider of biotechnology grade urea and NaCl used in this study.
Mutagenesis and protein purification
All of the proteins and protein variants used in this study were expressed as C-terminal hexahistidine fusions in Escherichia coli strain Bl21* (DE3). Recombinant proteins were purified using nickel affinity chromatography under denaturing conditions as described previously (Maxwell and Davidson 1998). Proteins were folded through dialysis and used as such without cleaving the hexahistidine tag. The dialysis buffer consisted of a 50 mM sodium phosphate solution (pH 7.0). Protein folding and functional assays were performed in this buffer.
Folding kinetics
Folding and unfolding rates were measured at 298 K on a Bio-Logic SFM-4 stopped-flow device equipped with a Photo Multiplier Tube (PMT) monitoring the recovery of intrinsic Trp fluorescence upon folding of urea denatured protein. Excitation was carried out at 295 nm and all of the fluorescence above 309 nm was collected. Traces were fit to appropriate single exponential functions using BioKine. At each concentration of urea, at least five separate shots of 3–5 μM protein were averaged. Assuming a linear dependence of ln kobs on urea concentration, kinetic chevron plots were fit to chevron equation:
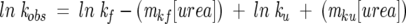 |
where kf and ku are the folding and unfolding rates at 0Murea and mkf and mku denote the dependence of kf and ku on the concentration of denaturant. Fitting was performed by a least squares method using Kaleidagraph (Synergy software).
NMR spectroscopy and quantitative peak shift studies
NMR experiments were performed on 0.25 mM uniformly 15N isotopically enriched protein samples containing 50 mM sodium phosphate (pH 7.0), 100 mM NaCl, 0.2 mM EDTA, and 4 mM DSS at 25°C using a Varian Inova spectrometer operating at 500 MHz 1H frequency. GuHCl was added to final concentrations of 0, 5, 10, 15, 20, 29, 38, 57, 83, 115, 153, 198, 260, 350, 436, 538, 646, 750, and 851 mM. Spectra were processed and peak positions quantitated using the NMRPipe/NMRDraw suite of programs (Delaglio et al. 1995), referencing 1H and 15N chemical shifts relative to the 1H methyl resonances of DSS (Wishart et al. 1995). Assignment of NMR spectra was accomplished with a 15N, 13C-labeled protein sample under identical buffer conditions using HNCACB (Wittekind and Mueller 1993) and TOCSY-HSQC (Montelione et al. 1992; Logan et al. 1993) pulse sequences.
Amide 15N and 1H chemical shifts showing changes greater than 0.4 or 0.08 ppm, respectively, were fit separately to the equation
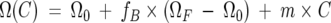 |
[1] |
where Ω (C) is the chemical shift at GuHCl concentration C, Ω0 = Ω (0), ΩF = Ω (∞), and m is a baseline correction factor discussed below. fB is the fraction of protein bound to GuHCl, which is given by
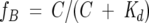 |
[2] |
The parameters Ω0, ΩF, m, and Kd were allowed to vary in least-squares fits of the data. The weighted mean of the dissociation constant was calculated according to
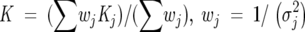 |
[3] |
where σj is the experimental uncertainty in the dissociation constant, Kj, calculated from a single set of 1H or 15N chemical shifts, and the sum extends over all individual Kd estimates. The uncertainty in the weighted mean was computed using a boostrap simulation (Efron and Tibshirani 1986).
Linear baseline corrections were found to be necessary for adequate fits of experimental data. Extracted values range from m = −0.30 ppm/M for A12 to 0.06 ppm/M for T14 1H chemical shifts; in the case of 15N chemical shifts, the range extends from −1.29 ppm/M for T14 to 0.39 ppm/M for D35. This linear dependence on GuHCl concentration is likely due to either slight noncooperative changes in protein conformation or very weak guanidinium binding in addition to the stronger (Kd ≈ 90 mM) interaction.
Binding studies
Dissociation constants were measured using the changes in the intrinsic fluorescence of the Trp residue located in the target binding pocket of the Fyn SH3 domain as a probe for binding. A λ CI fusion construct of a previously characterized Vsl12 target peptide (Maxwell and Davidson 1998) was used in these studies. Fluorescence signal was collected at equilibrium on an Aviv Spectrofluorometer Model ATF 105 (Aviv Associates). After subtraction of the fluorescence contribution from target alone from that of the complex, fluorescence data were fit as described previously (Maxwell and Davidson 1998).
Temperature melts
The changes in the ellipticity (mdeg) at 220 nm were recorded at equilibrium at different temperatures on an Aviv Circular Dichroism spectrometer Model 62A DS (Aviv Associates). Sigmoidal unfolding curves were fit by nonlinear least-squares methods using the program Igor Pro to the standard equations to obtain the Tm as described previously (Maxwell and Davidson 1998).
Error analysis
The errors reported for kobs, mkf, mku, Kd, and Tm are uncertainties associated with the least-squares fitting of the data to appropriate equations. Folding kinetics experiments, temperature melts, and binding assays were repeated at least twice. In all cases, the variations observed in kobs, mkf, mku, Kd, and Tm parameters were <5%. Uncertainties reported for ΔG, ΔTm are by error propagation according to the general equation:
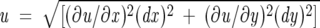 |
where u = f(x,y) and dx and dy are the residual errors of x and y, respectively.
Acknowledgments
A.Z-A. is supported by a doctorate Canada Graduate Scholarship (CGS) from the Natural Sciences and Engineering Research Council of Canada (NSERC). This work was supported by operating grants from the Canadian Institutes of Health Research to A.R.D. and L.E.K.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051829106.
Footnotes
A study by Fazi et al. (2002), however, has suggested that the Abp1 SH3 domain may still have an affinity for peptides containing arginine at the P−3 position, as assessed by phage display experiments, but the affinity of these interactions has never been measured. These interactions may not be as strong as those to peptides containing lysine at the P−3 position.
References
- Bhuyan, A.K. 2002. Protein stabilization by urea and guanidine hydrochloride. Biochemistry 41: 13386–13394. [DOI] [PubMed] [Google Scholar]
- Bogan, A.A. and Thorn, K.S. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280: 1–9. [DOI] [PubMed] [Google Scholar]
- Collins, K.D. 2004. Ions from the Hofmeister series and osmolytes: Effects on proteins in solution and in the crystallization process. Methods 34: 300–311. [DOI] [PubMed] [Google Scholar]
- de Los Rios, M.A. and Plaxco, K.W. 2005. Apparent Debye-Huckel electrostatic effects in the folding of a simple, single domain protein. Biochemistry 44: 1243–1250. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Di Nardo, A.A., Korzhnev, D.M., Stogios, P.J., Zarrine-Afsar, A., Kay, L.E., and Davidson, A.R. 2004. Dramatic acceleration of protein folding by stabilization of a nonnative backbone conformation. Proc. Natl. Acad. Sci. 101: 7954–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar, J., Yennawar, H.P., Banerjee, S., Luo, J., and Farber, G.K. 1997. The effect of denaturants on protein structure. Protein Sci. 6: 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron, B. and Tibshirani, R. 1986. Bootstrap methods for standard errors, confidence intervals and other measures of statistical accuracy. Stat. Sci. 1: 54–77. [Google Scholar]
- Fazi, B., Cope, M.J., Douangamath, A., Ferracuti, S., Schirwitz, K., Zucconi, A., Drubin, D.G., Wilmanns, M., Cesareni, G., and Castagnoli, L. 2002. Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: Structural and functional analysis. J. Biol. Chem. 277: 5290–5298. [DOI] [PubMed] [Google Scholar]
- Feng, S., Kasahara, C., Rickles, R.J., and Schreiber, S.L. 1995. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc. Natl. Acad. Sci. 92: 12408–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S. and Thornton, J.M. 1996. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 93: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, L.E., Keifer, P., and Saarinen, T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114: 10663–10665. [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Logan, T.M., Olejniczak, E.T., Xu, R.X., and Fesik, S.W. 1993. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3: 225–231. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I. 1999. Thermodynamics of protein interactions with Urea and Guanidinium Hydrochloride. J. Phys. Chem. B 103: 4781–4785. [Google Scholar]
- Makhatadze, G.I. and Privalov, P.L. 1992. Protein interactions with urea and guanidinium chloride. A calorimetric study. J. Mol. Biol. 226: 491–505. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., Lopez, M.M., Richardson 3rd, J.M., and Thomas, S.T. 1998. Anion binding to the ubiquitin molecule. Protein Sci. 7: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles, J.A., Dahesh, S., Haynes, J., Andrews, B.J., and Davidson, A.R. 2004. Protein–protein interaction affinity plays a crucial role in controlling the Sho1p-mediated signal transduction pathway in yeast. Mol. Cell 14: 813–823. [DOI] [PubMed] [Google Scholar]
- Maxwell, K.L. and Davidson, A.R. 1998. Mutagenesis of a buried polar interaction in an SH3 domain: Sequence conservation provides the best prediction of stability effects. Biochemistry 37: 16172–16182. [DOI] [PubMed] [Google Scholar]
- Mayr, L.M. and Schmid, F.X. 1993. Stabilization of a protein by guanidinium chloride. Biochemistry 32: 7994–7998. [DOI] [PubMed] [Google Scholar]
- Moglich, A., Krieger, F., and Kiefhaber, T. 2005. Molecular basis for the effect of urea and guanidinium chloride on the dynamics of unfolded polypeptide chains. J. Mol. Biol. 345: 153–162. [DOI] [PubMed] [Google Scholar]
- Monera, O.D., Kay, C.M., and Hodges, R.S. 1994. Protein denaturation with guanidine hydrochloride or urea provides a different estimate of stability depending on the contributions of electrostatic interactions. Protein Sci. 3: 1984–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione, G.T., Lyons, B.A., Emerson, S.D., and Tashiro, M. 1992. An efficient triple resonance experiment using carbon 13 isotropic mixing for determining sequence-specific resonance assignments of isotopically- enriched proteins. J. Am. Chem. Soc. 114: 10974–10975. [Google Scholar]
- Noble, M.E., Musacchio, A., Saraste, M., Courtneidge, S.A., and Wierenga, R.K. 1993. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 12: 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northey, J.G., Di Nardo, A.A., and Davidson, A.R. 2002. Hydrophobic core packing in the SH3 domain folding transition state. Nat. Struct. Biol. 9: 126–130. [DOI] [PubMed] [Google Scholar]
- Rath, A. and Davidson, A.R. 2000. The design of a hyperstable mutant of the Abp1p SH3 domain by sequence alignment analysis. Protein Sci. 9: 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickles, R.J., Botfield, M.C., Zhou, X.M., Henry, P.A., Brugge, J.S., and Zoller, M.J. 1995. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc. Natl. Acad. Sci. 92: 10909–10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, I.E. and Kiefhaber, T. 2003. Hammond behavior versus ground state effects in protein folding: Evidence for narrow free energy barriers and residual structure in unfolded states. J. Mol. Biol. 327: 867–884. [DOI] [PubMed] [Google Scholar]
- Schellman, J.A. 2002. Fifty years of solvent denaturation. Biophys. Chem. 96: 91–101. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Bigam, C.G., Yao, J., Abildgaard, F., Dyson, H.J., Oldfield, E., Markley, J.L., and Sykes, B.D. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6: 135–140. [DOI] [PubMed] [Google Scholar]
- Wittekind, M. and Mueller, L. 1993. HNCACB, a high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. Series B 101: 201–205. [Google Scholar]
- Yousef, M.S., Baase, W.A., and Matthews, B.W. 2004. Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme. Proc. Natl. Acad. Sci. 101: 11583–11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar, A., Bhattacharyya, R.P., Nittler, M.P., and Lim, W.A. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell 14: 825–832. [DOI] [PubMed] [Google Scholar]