Abstract
Symmetry, and in particular point group symmetry, is generally the rule for the global arrangement between subunits in homodimeric and other oligomeric proteins. The structures of fragments of tropomyosin and bovine fibrinogen are recently published examples, however, of asymmetric interactions between chemically identical chains. Their departures from strict twofold symmetry are based on simple and generalizable chemical designs, but were not anticipated prior to their structure determinations. The current review aims to improve our understanding of the structural principles and functional consequences of asymmetric interactions in proteins. Here, a survey of >100 diverse homodimers has focused on the structures immediately adjacent to the twofold axis. Five regular frameworks in α-helical coiled coils and antiparallel β-sheets accommodate many of the twofold symmetric axes. On the basis of these frameworks, certain sequence motifs can break symmetry in geometrically defined manners. In antiparallel β-sheets, these asymmetries include register slips between strands of repeating residues and the adoption of different side-chain rotamers to avoid steric clashes of bulky residues. In parallel coiled coils, an axial stagger between the α-helices is produced by clusters of core alanines. Such simple designs lead to a basic understanding of the functions of diverse proteins. These functions include regulation of muscle contraction by tropomyosin, blood clot formation by fibrin, half-of-site reactivity of caspase-9, and adaptive protein recognition in the matrix metalloproteinase MMP9. Moreover, asymmetry between chemically identical subunits, by producing multiple equally stable conformations, leads to unique dynamic and self-assembly properties.
Keywords: asymmetry, homodimer, half-of-site reactivity, junction bend, degeneracy, tropomyosin, fibrinogen, caspase, conformational changes, structure
Two important conceptual advances in the 1960s led to a new understanding of the significance of symmetry in biological structures. One was the recognition by Monod and colleagues (Monod et al. 1963, 1965) that certain oligomeric enzymes have the property of altering their structures in response to chemical stimuli and that this nonlinear, so-called allosteric behavior can be understood in terms of a switching between two or more symmetrical states of the molecule. Although other models for allostery exist, the symmetry model (Monod et al. 1965) accounts for a broad range of allosteric effects in many enzymes (Perutz 1989). Symmetry, and in particular point group symmetry, is indeed most commonly the rule for the global arrangement between subunits in homodimeric and other oligomeric proteins as it is thought to maximize stability and avoid unwanted aggregation (for a recent general overview, see Goodsell and Olson 2000). The other concept was that of quasiequivalence by Caspar and Klug (1962). In this and later studies, it was shown how identical protein subunits can form large globally symmetrical structures—such as virus particles—by dropping the requirement of strict (or sometimes any) (Liddington et al. 1991; Reinisch et al. 2000) equivalence between subunits. The conformational differences between the subunits in virus capsids, originally conceived as small variations in bonding patterns, often involve significant structural switches (Rossmann 1984; Johnson and Speir 1997). These departures from symmetry, which mediate the different quasi- or nonequivalent interactions, lead to a more stable structure than would strict equivalence.
It is well known that local asymmetry between chemically identical subunits is found in many proteins (Goodsell and Olson 2000). In various flexible proteins, these asymmetries may be induced by external factors, for example, an asymmetric crystal environment around the dimer or the binding of ligands such as DNA (King et al. 1999) or enzyme substrates (Koshland et al. 1966). Asymmetry in some other homodimers appears to be an intrinsic or “pre-existent” property of the protein (Seydoux et al. 1974). For a number of enzymes, biochemical experiments have helped distinguish between these sources of asymmetry (Wang and Pan 1996; Nagradova 2001). In this laboratory, however, we have used X-ray crystallography to reveal simple chemical designs at the dimer axis of tropomyosin (Brown et al. 2001) and fibrinogen (Madrazo et al. 2001) that provided visual evidence for the intrinsic natures of their asymmetric structures. These results, both the existence of the asymmetries and their underlying designs, were not anticipated until the high-resolution crystal structures were determined. We have therefore begun an analysis of homodimers in the Protein Data Bank in order to identify structural principles that can explain the making and breaking of symmetry at the twofold rotation axis in diverse proteins.
In the first part of the article we show that register slips between strands of repeating residues and the avoidance of steric clashes between preferred side-chain rotamers in antiparallel β-sheets, as well as axial shifts between parallel helices, break twofold symmetry between chemically identical subunits in a wide variety of proteins. These designs appear to derive from specific sequences of residues and have previously been described for similar situations within protein subunits (Gernert et al. 1995; Hutchinson et al. 1998; Eneqvist et al. 2000). Five common frameworks for accommodating symmetric twofold axes are also described. Functional implications of dimer asymmetry are reviewed in the second part of the article. These features result in the well-known half-of-sites reactivity of certain enzymes, and one type of design produces specific bent conformations in a number of fibrous proteins. Asymmetry between chemically identical subunits can also produce unique dynamic properties by generating multiple conformations of the dimer, variable self-assembly patterns, and adaptive protein recognition.
Asymmetric designs at the dimer axis
Antiparallel β-sheet
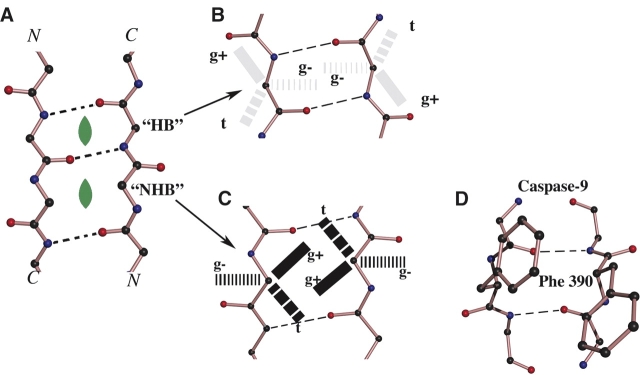
The antiparallel β-sheet shows in two distinct manners how the breaking of symmetry in a homodimeric molecule can be understood according to simple and regular design principles. β-Sheets differ from α-helices in that main-chain hydrogen bonds are formed between rather than within chains. Simply on the basis of the main-chain hydrogen-bonding pattern, parallel β-sheets can never incorporate a twofold rotation axis. An antiparallel β-sheet composed of two chemically identical halves, however, may be divided by a dyad, oriented perpendicularly to the sheet, as long as the axis is located directly between the α-carbons of two residues directly across from one another on adjacent strands (hollow ovals in Fig. 1A) (Fraser and McRae 1973; see also below). However, for symmetry to still apply, the side chains must cooperate: The register of the strands must be such that identical positions along the sequence (i and i′) from the two subunits are located across from each other at the dyad (red circle in Fig. 1A), and the conformations (rotameric forms) of their side chains must be the same. Either of these requirements for symmetry is sometimes broken (Figs. 1B, 2D), and examples of specific sequences that help produce such asymmetries are discussed below.
Figure 1.
Register asymmetry in a dimeric antiparallel β-sheet. (A) An antiparallel β-sheet (depicted schematically) composed of two chemically identical chains may be twofold symmetric when analogous positions along the amino acid sequence (e.g., i and i′) are located across from each other at the dimer axis; here, the midpoint (red circle) between analogous residues of the dimer coincides with a local twofold symmetry axis (hollow black oval) accorded by the geometry of the main chain (black). (Note that analogous residues in a symmetrical register may either be main-chain “hydrogen-bonded” to each other [as pictured here] or form a “non-hydrogen-bonded” pair as explained in Fig. 2; see also Table 1). (B) When the register of one of the chains is “shifted” by one position (or any odd number of positions) relative to that in A, only different positions along the amino acid sequence (e.g., i and i i′ – 1) are located directly across from each other; in this case the midpoint (red circle) between analogous residues (e.g., i and i′) of the dimer falls on a non-twofold symmetric location of the main-chain structure (i.e., the red circle does not coincide with a hollow black oval). Some of the chemical consequences of this register shift for any pair of analogous residues are that one of their main-chain carbonyls points into the dimer interface while the other points away, and that their side chains are found on opposite faces of the β-sheet (here, i [bold red] faces toward the reader and i′ [faded light red] faces away; see also E). (Note that in the language of Fig. 2, residue i in this asymmetric register is forming main-chain hydrogen bonds across the dimer interface [to i′ – 1], while the analogous residue i′ is not.) (C) In the N-terminal γ domain of the dimeric bovine fibrinogen molecule (Madrazo et al. 2001), the asymmetrically registered alignment is enforced in an antiparallel β-sheet-like structure by a rare arrangement of consecutive disulfides that covalently link residue γ8 (i) to residue γ′ 9 (i′ + 1), as well as γ′ 8 to γ9. The asymmetry of this arrangement is propagated throughout the N-terminal 14 residues of the two γ chains and may have important implications for the uniformity of the fibrin clot. (We also note that a pair of adjacent reciprocal disulfide bonds connects the subunits of seminal bovine ribonuclease [PDB 11BG], but in this case they are part of α-helices and the dimer is roughly symmetric.) (D,E) The dimeric interface of mannose-binding protein can adopt either the symmetric (D) or asymmetric (E) register, depending upon crystallization conditions (Weis et al. 1991, 1992). Note that the side-chain interactions between the strands nearest the central axis of the dimer are similar in the two cases, due to chemical similarity (common γ methyl group) of the consecutive valine and threonine side chains (see text). Other interactions (not shown) more distant from the central axis differ between the two crystal forms. (F) For antiparallel strands consisting only of alternating charged residues (e.g., the displayed polar zipper of Perutz and colleagues [De Baere et al. 1992; Perutz 1994]), a symmetrical register necessarily yields a residue pair of like charges at the twofold axis. (G) For such a peptide, a complementary network of oppositely charged residues can only be produced in an asymmetric register.
Figure 2.
Side-chain rotamer asymmetry in a dimeric antiparallel β-sheet. (A) An antiparallel β-pleated sheet may accommodate a perpendicularly oriented twofold axis (green filled oval) in one of two geometrically distinctive locations: at the center of a pair of residues that are hydrogen bonded to each other (HB) or at the center of a pair of residues that are not hydrogen bonded to each other (NHB). (B,C) On each of the two residue-pair geometries, these figures superimpose the three staggered rotameric conformations (about the Cα–Cβ bond) that are most commonly observed for side chains. In proteins in general, the relative populations of these rotamers to some extent depend both on the identity of the side chain and the φ–ψ angles of the main chain (Dunbrack and Karplus 1993), but usually the g+ rotamer is most favored. (B) For an HB residue pair, two side chains both having the g+ rotameric conformation face away from one another. (C) For the NHB pair, such side chains are in position to interact most closely with one another. (D) In caspase-9 (Renatus et al. 2001), the dimer axis is located at the center of an NHB pair of phenylalanines (390 and 390′). These bulky side chains would clash if they both adopted the g+ conformation, specifically because of the NHB geometry at the dimer axis. The asymmetry that in this case occurs (where one phenylalanine adopts the g+ conformation and the other is swung away) is propagated throughout the respective monomers (see Renatus et al. 2001) and apparently causes the dimer to display half-site reactivity (Renatus et al. 2001). Similar asymmetry occurs in insulin dimers at Phe25 (not shown).
Register asymmetry in antiparallel β-sheets
A register asymmetry between two chemically identical antiparallel β-strands—in which a residue (i) from one subunit is directly across from an adjacent residue along the sequence (e.g., i′ – 1) of the other subunit (Fig. 1B)—occurs in crystal structures of fibrinogen (Fig. 1C) and mannose binding protein (Fig. 1E). As discussed previously (Madrazo et al. 2001), the asymmetrically registered alignment in bovine fibrinogen is enforced by a rare arrangement of adjacent reciprocal disulfides (Fig. 1C) and could have important implications for the uniformity of the fibrin clot (Maddrazo et al. 2001; see below). In contrast, mannose binding protein can form either the symmetrically aligned (Fig. 1D) or register “shifted” (Fig. 1E) antiparallel β-strands, depending on crystallization conditions (Weis et al. 1991, 1992).
Both of these asymmetric structures retain a remnant of symmetry in that identical (cysteines in fibrinogen) or similar (threonine and valine in mannose binding protein) side chains are across from each other at the axis, even though they are in different positions of the sequence (Fig. 1C,E). The Matrix MetalloProtein MMP9 homodimer provides a related situation: Its intersubunit interface consists of two parallel chains (also see below) that pair up out of register (Cha et al. 2002) such that residues (in PDB numbering) 189 and 192—both valines—are across from each other; the offset between the subunits in this dimer is suggested to provide a novel mechanism for adaptive protein recognition (Cha et al. 2002; see expanded discussion below). The capacity for a β-strand to “slip” its register also has precedents within monomeric proteins. A two-residue difference in register occurs between the GDP-(Amor et al. 1994) and GTP-bound (Goldberg 1998) forms of ARF1; here, the pattern of side-chain hydrophobicity (Goldberg 1998), but not size, is roughly conserved at most positions. A three-residue difference in register of a β-strand in a mutant of transthyretin places a leucine at position 58 in a site normally occupied by native leucine 55; this difference results in new protein–protein interactions, a feature which is used to model transthyretin amyloid formation (Eneqvist et al. 2000).
As these examples suggest, having identical or similar residues near each other in sequence may be expected to promote register slips, and these could break symmetry upon oligomer formation. The 180° reorientation of a β-strand that shifts by an odd number of residues (Fig. 1, cf. down strands in A and B) would usually result in major differences in side-chain interactions in the protein; dynamic shifting of this sort between two or more registers would probably not readily occur unless the residues or important features of the residues repeat. β-Sheet-forming polypeptides such as polyglutamines are implicated in a number of neurodegenerative diseases (Perutz 1994; Ross et al. 2003), and one may speculate that variable strand registers may occur during their self-assembly.
Departures from a symmetric register may also be promoted in β-sheets consisting solely of alternating charged side chains (e.g., the pattern displayed in Fig. 1F,G). In such a case, the necessary identity of the two side chains nearest a twofold rotation axis in the symmetric register (i and i′ in Fig. 1A) imposes the positioning of two like charges across from each other (Fig. 1F). Certain rotameric conformations of these side chains can, of course, minimize the charged repulsion, but potentially destabilizing interactions may also be avoided by breaking the symmetry with a register shift (as in Fig. 1G). These symmetry considerations can provide additional detail to theoretical models of certain polar zippers, of the type + + − −, put forward by Perutz and colleagues (De Baere et al. 1992; Perutz 1994).
Rotamer asymmetry in antiparallel β-sheet
Even in antiparallel β-sheet dimers whose strands adopt the symmetrical register (Fig. 1A), conformational differences are occasionally observed between the two otherwise identical side chains nearest the dyad, and these differences sometimes derive from the residue pair’s underlying main-chain geometry. Because of the β-sheet’s pleated nature there are two alternating types of residue pairs (“HB” and “NHB” in Fig. 2A; see legend for explanations), each of which can accommodate a twofold axis at its center (filled green ovals in Fig. 2A). These two types of residue pairs have distinctive main-chain geometries, and a resulting major implication is that they often promote specific interactions for their constituent side chains (Wouters and Curmi 1995; Hutchinson et al. 1998; Fig. 2B,C). For example, in an NHB pair (Fig. 2C), two phenylalanine side chains would generally clash if they both adopted the same, otherwise favorable (Ponder and Richards 1987; Dunbrack and Karplus 1993), “g+” rotameric conformation (see Fig. 2); they instead, in this case, usually adopt different rotameric conformations (g+ for one phenylalanine; t or g− for the other) (Hutchinson et al. 1998). The juxtaposition of this bulky side chain with this specific main-chain geometry appears to explain the source of asymmetry in a number of homodimers, including insulin (see position 25 in PDB ID 4INS, 7INS, 9INS, and 1B9E) and the key cell-death protein caspase-9 (Fig. 2D). For this protein, the asymmetry is propagated from the dimer axis to distant parts of the enzyme, causing a nonfunctional active site in one monomer but not in the other (Renatus et al. 2001; see below).
α-Helical coiled coils
In addition to adjacent antiparallel strands of a β-sheet, two interacting α-helices also commonly accommodate a twofold rotation axis. In contrast to the β-sheet whose geometry is somewhat constrained by the main-chain hydrogen-bonding patterns, α-helices interact with one another predominantly through their side chains, and the distance, orientation, and relative position between the helices are found to vary greatly both within monomers and at the dimer interface. Here we focus our attention on the best-understood type of interaction between α-helices—that of the coiled coil—because its regular pattern permits a simple understanding of its symmetry properties. Two-stranded α-helical coiled coils occur in two orientations, parallel and antiparallel, which share certain features. Their amino acid sequences are characterized by a seven-residue repeat unit, a b c d e f g, where the a and d positions, which face most directly into the core, are generally apolar. These residues interlock in a “knobs-into-holes” fashion, which usually involves the interaction of a residue from one helix with four (Crick 1953) or sometimes three (Gernert et al. 1995) residues from the partner helix. In a two-stranded coiled coil, the orientation of the a and d knobs are distinctive (O’Shea et al. 1991) and in part on this basis relatively short coiled-coil regions in globular proteins can also be identified.
Parallel and antiparallel α-helical coiled coils are nevertheless distinguished in a number of basic ways (for review, see Lupas 1996), and these differences impact their capacities to accommodate a twofold rotation axis. Since the basic heptad repeat spans two turns of an α-helix, the canonical antiparallel coiled coil can accommodate a perpendicularly oriented twofold axis in one of two relatively discrete and geometrically distinctive axial positions: either nearest (in projection) the b or the f positions (see filled green ovals and legend in Fig. 3). Note that in antiparallel coiled coils, a small axial shift of one helix relative to the other does not necessarily break symmetry (the perpendicularly oriented symmetry axis could then also shift accordingly).
Figure 3.
Two types of locations accommodate a twofold rotation axis in antiparallel α-helical coiled coils. The number of residues in each chain that span consecutive core positions alternate between four (the a, b, c, and d positions) and five (the d, e, f, g, and a positions); the two-stranded coiled coil may thus be visualized as consisting of alternating “b-centered” ((abcd)2) and “f-centered” ((defga)2) rings of residues. Similar to the alternating hydrogen-bonded and non-hydrogen-bonded residue pairs in the antiparallel β-sheets, the b-centered and f-centered rings are geometrically distinctive, and in antiparallel coiled coils each of them can accommodate a perpendicularly oriented dyad at its center (green filled ovals).
Parallel α-helical coiled coils can also accommodate a twofold rotation axis, which in this case runs along the length of the coiled-coil axis (green double-headed arrow in Fig. 4A). For twofold symmetry to be maintained here, however, the two chains must be in register along the length of the coiled coil, i.e., identical residues—with identical side-chain conformations—must be found directly across from one another throughout the coiled coil. A symmetry-breaking (~1.2 Å) axial stagger of the helices (Fig. 4B) occurs in segments of tropomyosin and other homodimeric coiled coils containing a cluster of core (a- and d-position) alanines (Brown et al. 2001). The stagger occurs because, in contrastto coreleucines (Fig.4C), alanines preferentially fit into relatively small, axially shifted holes on an α-helix (Fig. 4D). This difference in binding sites for alanine and leucine side chains has also been described in antiparallel heteromeric coiled coils (Gernert et al. 1995) and has been evaluated statistically in an extensive survey of globular proteins (Walther et al. 1996). The asymmetry in the alanine-containing parallel coiled coils therefore appears to be an intrinsic consequence of their sequences. Helical staggering also occurs in a segment of the APC tumor suppressor that is unusually rich in core (polar) asparagines and in a-position leucines (in contrast to β-branched residues) (Dayand Alber 2000). Conformational (rotameric) differences also occur between the analogous side chains of this dimer, as well as between the asparagines of the leucine zipper that form buried hydrogen bonds (O’Shea et al. 1991). Rotameric differences also occur between analogous bulky core side chains in a number of coiled coils such as variable surface glycoprotein, cortexillin, and the C terminus of tropomyosin (described by Li et al. [2002]), perhaps because only one of the two paired residues can fit into the core.
Figure 4.
Axial-shift asymmetry in parallel α-helical coiled coils. (A) Parallel α-helical coiled coils may accommodate a twofold rotation axis running along the length of the coiled-coil axis (green double-headed arrow). (B) Symmetry is broken when the two chains are axially out of register. (C) In tropomyosin (and other proteins), a relatively symmetrical coiled-coil segment occurs with core leucines that fit into four-residue holes of the adjacent helix whereas (D) local axial staggering is promoted by core alanines (Brown et al. 2001) because of their preferential fit into axially shifted three-residue holes (Gernert et al. 1995; Walther et al. 1996).
In general, the twofold rotation axis imposes constraints on the precise geometry of nearby residues that otherwise would not interact symmetrically; indeed, the preservation of perfect (e.g., crystallographic) twofold symmetry appears to be much rarer for parallel coiled coils or other parallel arrangements of chains (which have all their interactions right at the axis and none far away) than for globular dimers containing antiparallel chains.
Consequences of asymmetry in homodimers
A common concern for crystallographers is to verify that the conformation observed in a crystal accurately represents the functionally relevant structure(s). By extension, one may ask whether an asymmetric conformation in a homodimer is the preferred (or major) intrinsic structure or it is merely induced, say, by different packing forces on the two monomers. Put another way, one may ask whether the observed asymmetric conformation is a relatively stable feature of the molecule or whether it is in rapid interchange with other conformations. (NMR studies in solution can sometimes distinguish between these possibilities, but this technique is less amenable to large [e.g., oligomeric] structures.) The intrinsic and structurally relevant nature of a crystallographically observed asymmetry may be supported when the asymmetry persists under different crystal conditions (e.g., space groups), is located at the dimer interface rather than exterior surface, involves a set of residues, and especially, as surveyed above, when a simple chemical basis for the asymmetry is identified. Further support may be provided by biochemical studies, especially as discussed now for certain enzymes.
Half-of-sites reactivity
The general capacity for two chemically identical monomers to adopt different conformations provides the structural basis for the functions of many dimeric enzymes that use “half-of-the sites” reactivity and that display (negatively) cooperative behavior between domains. A major objective in this field is to determine whether the asymmetry of the homodimeric enzyme being studied is ligand-induced (Koshland et al. 1966) or pre-existent (i.e., an intrinsic property of the unliganded protein) (Seydoux et al. 1974). (An intermediate option is where the unliganded enzyme is in equilibrium between asymmetric and symmetric conformations [Nagradova 2001].) Kinetic and chemical modification experiments have been used to distinguish between these models (Wang and Pan 1996; Nagradova 2001 and references therein).
Inspection of crystal structures using the guidelines enumerated above can also play a role in analyzing half-of-site reactive dimeric enzymes, and on this basis many such enzymes might appear to follow the ligand-induced sequential model more closely than the pre-existent asymmetry model. Crystal structures of phosphofructokinase (Shirakihara and Evans 1988), tryptophan-tRNA synthetase (Ilyin et al. 2000), and glyceraldehyde 3-phosphate dehydrogenase (Song et al. 1999), for example, show symmetric dimer interfaces (but see also Nagradova 2001). Murine glutathione S-transferase, which in one crystal structure does show a linkage between an arginine side-chain asymmetry at the dimer axis and different active site conformations and contents in the individual subunits (Xiao et al. 1999), nevertheless shows symmetric dimers in other crystals. Moreover a simple chemical design that would promote one arrangement of the arginine pair over the other is not apparent to us; a structural signal thus appears to be sent from one active site through the dimer axis using the arginine bipolar switch to the second active site (Xiao et al. 1999).
Pre-existent or intrinsic asymmetry generated from the dimer axis, however, may account for properties of aspartate transcarbamoylase and of caspase-9. In a relatively high resolution crystal structure of Escherichia coli aspartate transcarbamoylase, the N termini of adjacent regulatory chains adopt significantly different conformations and together form a highly distorted antiparallel-like β-sheet structure at the dimeric axis (Jin et al. 1999). This asymmetric structure is adjacent to the nucleotide (regulatory) binding sites of the enzyme and would thus appear to account for their separation into high- and low-affinity binding sites (Jin et al. 1999). Although this region of the molecule is not highly ordered, the multiple contacts between the two N-terminal chains and the lack of any available symmetric structures (to our knowledge) would suggest that the asymmetry here is a stabilized pre-existent feature of the molecule.
The connection between fundamental aspects of protein secondary and side-chain structure and half-site reactivity is perhaps best illustrated for caspase-9. This key protease in apoptosis has been studied extensively by Renatus and colleagues (Renatus et al. 2001). Their biochemical investigations have shown that the enzyme is self-activated by dimer formation, and that only half of the potentially available catalytic sites are labeled by a substrate analog. This observation is explained by their crystal structure of the dimer, which reveals that the active site is in a functional conformation in one but not the other subunit, a difference the investigators suggest is generated by different side-chain conformations of phenylalanine 390 and phenylalanine 390′ between two anti-parallel β-strands at the dimer interface. These differences in the phenylalanines cause the side chains of the two tyrosines 331 to adopt different conformations, which in turn appears to allow the catalytic conformation of cysteine 285 in only one of the subunits (see full description and figures in Renatus et al. 2001). As described above, caspase-9 exhibits asymmetry at the dimer axis because for a non-hydrogen-bonded (NHB) pair of residues of an antiparallel β-strand, as are residues 390 and 390′, two phenylalanine side chains cannot simultaneously adopt their most favored g+ rotamer without clashing (Fig. 2C,D). The simplicity of this design, also seen in other proteins, suggests that this “rotamer” asymmetry at the dimeric axis, as well as the resulting inactivation of one monomer’s binding site, are pre-existent and relatively stable features of this caspase.
In heterodimers there are, of course, always pre-existent asymmetry between the two subunits and extreme cases of “half-site reactivity” in which one chain is catalytically inactive is observed in a number of heterodimers of chemically homologous subunits (Todd et al. 2002). Many such heterodimers, as well as single-chain monomers with strong internal twofold symmetry (e.g., VAT-N) (Coles et al. 1999), are thought to have evolved from homodimeric precursors (Todd et al. 2002)—and one may ask whether they evolved preferentially from homodimers that themselves tended to display pre-existent or at least easily induced asymmetry. Homodimeric enzymes that already have one site inactivated may have “less to lose” than symmetric dimers from an event of gene duplication and subsequent mutational divergence into a more complex heterodimeric structure.
Junction bends
Dimers whose subunits adopt different conformations are necessarily asymmetric (at least locally). The converse does not have to be true: Two, say, relatively rigid subunits not related to each other by the same transformation (i.e., a twofold rotation axis) but asymmetrically by a single pair of different (reciprocally related) transformations may, in principle, retain identical folds, although the asymmetric environments for the analogous residues are likely, in practice, to induce some conformational change. Fairly significant local differences in the conformations of two flexible subunits, however, must occur when the subunits are related to each other by two or more pairs of transformations, with each pair applicable to a different segment of the chain. Furthermore, the local conformations at the junctions between such segments can, in some cases, predictably alter the global shape of the molecule.
Tropomyosin provides such a specific example. As described previously (Brown et al. 2001), the joining of an axially staggered asymmetric parallel coiled-coil segment (see above) and an in-register symmetric segment changes the global shape of the molecule by producing a bend of the coiled-coil axis away from the wedge created by the locally longer chain (Fig. 5, left); this bend is accommodated by different length main-chain H-bonds in the two α-helices. This design in tropomyosin promotes the supercoiled conformation necessary for its interaction with actin (see Brown et al. 2001). This type of so-called “junction bend” (term used by Goodsell et al. 1994) is found in various (otherwise unrelated) parallel homodimeric coiled coils: We have identified it at residue 332 of cortexillin (PDB ID 1d7m; Burkhard et al. 2000; Brown et al. 2001) and now at residue 83 of the DNA-binding protein PUT3 (PDB ID 1zme; Swaminathan et al. 1997). Moreover, a similar junction bend is also seen in the anti-parallel and heteromeric α-helical bundle of the ColE1 rop protein, at residues 20 and 40 (Banner et al. 1987; Brown et al. 2001). In these cases, the different axial relationships of the partner helices in each of the adjacent coiled-coil segments appears to be caused by the different residues (alanines vs. leucines) located at the helical interface (Fig. 4; see above), and the junction bend is thus an intrinsic preferred feature of the molecule.
Figure 5.
Junction bends in asymmetric homo-oligomers. The junction of symmetric (bottom half) and asymmetric (upper half) segments of a protein can predictably control the molecule’s overall shape. In a parallel dimeric molecule (left) such as tropomyosin, cortexillin, or the DNA-binding protein PUT3 (see text; Brown et al. 2001), a relatively small (~1.2 Å) axial staggering of relatively widely separated (8–10 Å) α-helices yields a small wedge (triangle) in one chain away from which the molecular axis (solid lines) thus bends by a small (~3–6) degree. This geometrical design applies to the trimeric collagen–foldon structure (Stetefeld et al. 2003) (right), where the relatively large (~5–6 Å) axial stagger between narrowly separated (~4–5 Å) collagen chains leads to a severe (~63°) bend (see text).
The type of junction bend observed in homodimeric α-helical coiled coils also occurs in homotrimeric proteins containing the axially staggered collagen triple helix. A recent crystal structure of a designed peptide consisting of collagen sequence stabilized by an adjacent threefold symmetric in-register foldon domain from T4 fibritin (Fig. 5, right) reveals a specific bend of the foldon domain away from the locally longer collagen chain(s) (Stetefeld et al. 2003)—precisely the same type of design first observed in two-stranded tropomyosin (Brown et al. 2001). In fact, compared to alanine-staggered dimeric α-helical coiled coil, the chains of the collagen triple helix are closer together (~4–5 Å using interchain main-chain H-bonds vs. ~8–9 Å using side-chain knobs into holes packing) and more axially staggered (up to ~5–6 Å vs. ~1 Å); these two geometrical features combine in a predictable fashion to specify a more severe bend in the collagen–foldon structure (~63°) than in tropomyosin (~6°). Collagen-like sequences are in fact usually stabilized by noncollagenous domains, and in some cases, such as the rat liver mannose-binding protein (Wallis and Drickamer 1997) and human class A scavenger receptors (Resnick et al. 1996) as well as the foldon, these domains include three-stranded α-helical coiled coils. The junction between the asymmetric collagenous and relatively symmetric coiled-coil regions of these other proteins are also likely to be stabilized in bent rather than straight conformations.
Conformational degeneracy
The half-site reactive enzymes and bent rods discussed above require producing and stabilizing different conformations for the two subunits of the dimer. One may ask then what advantage is afforded to proteins that can produce two different conformations of chemically identical subunits when simply using subunits with different amino acid sequences would directly yield the desired pre-existent asymmetry. One simple advantage may be frugality: Homodimers require less genetic material than do heterodimers (see Crick and Watson 1957). But of greater interest are potential functional niches of asymmetric homodimers derived from their distinctive dynamic, assembly, and recognition properties.
These properties derive from an unusual conformational landscape accessible for chemically identical subunits that upon interacting with one another adopt different conformations. Here, at any one node of asymmetry, the choice of one subunit, say the left one, adopting conformation “X” and the right subunit adopting conformation “X′” would be energetically indistinguishable from the alternative of the left subunit adopting conformation X′ and the right one adopting conformation X (Fig. 6A; see also Goodsell and Olson 2000). Such a dimer would thus have two lowest-energy conformations under identical environmental conditions. If two or more (separated) nodes of asymmetry are introduced (see below), then the dimer would have the intrinsic potential to adopt multiple distinguishable conformations of equally low energy (see, e.g., Fig. 6C,D). The energetic equivalence or “degeneracy” of these conformations is fundamentally different from the energy landscape of monomers or symmetric polymers, where there is generally only one most stable conformation, and is different from random coils (such as unfolded proteins or very long thin polymers) or even freely jointed chains where there is more of a continuum of accessible conformations.
Figure 6.
Conformational degeneracy in asymmetric homodimers. (A) When an object dimerizes asymmetrically, the choice of one subunit, say the left one, adopting conformation “X” and the right subunit adopting conformation “X′” would be energetically indistinguishable from the alternative of the left subunit adopting conformation X′ and the right one adopting conformation X. The rate of interchange between these degenerate states likely varies from case to case (see text for description of tropomyosin [“Tm”] and fibrinogen [“Fgen”] structures). (B) Piston-like motions between two equivalently stabilized pairs of axially staggered chains in a parallel homodimer. (C) Bistable joints in a homodimer containing symmetrically in-register (red) and asymmetrically staggered (blue) segments (such as tropomyosin). (D) Irregular self-assemblies arising from asymmetric contacts (such as in the protofilament backbone of fibrin displayed here schematically; see “Nonuniformities in self-assembly” section of text for details).
The actual realization of asymmetric homodimers, as well as flexible proteins in general, to sample each of its energetically equivalent conformations or configurations may of course be restricted by other factors, such as ligands that may preferentially select one (or some) of these states. Also, the kinetic ease at which the asymmetric homodimer can interchange or “reciprocate” (Goodsell and Olson 2000) between the degenerate states likely varies from case to case (Fig. 6A), and has been measured to occur on the order of seconds in the case of the Mnt repressor tetramerization domain (Noreen et al. 1999). The general capacity of an asymmetrical dimer to adopt and perhaps interchange between its degenerate conformations can find use in various physiological settings.
Consider, for example, two chemically identical parallel chains that are stabilized in a staggered out-of-register arrangement; an interchange between the two resulting degenerate structures, i.e., left chain up/right chain down and left chain down/right chain up, could resemble the motion of two pistons (Fig. 6B; Day and Alber 2000; Brown et al. 2001). Such an interchange may occur fairly readily between α-helices related by small staggers (Fig. 6A,B): 1.5 Å shifts between packed helices in different crystals have been observed in proteins (Lesk and Chothia 1984); also, core alanines, which generally promote ~1.2 Å –staggered α-helices in parallel homodimeric coiled coils such as tropomyosin (see “Designs,” above), are in certain crystal environments found in the symmetrically unstaggered arrangement (see supplemental material in Brown et al. 2001) that would be the likely intermediate between interchanging staggered helices.
In the aspartate receptor, two long antiparallel and nonidentical α-helices (α 1 and α 4) appear capable of interacting with two ~1 Å different axial registers, here under different conditions. One register is in the presence of an aspartate molecule and the other in its absence, and ligand-induced axial sliding of helices in this bacterial chemo-receptor appears to transmit a signal across the membrane (Ottemann et al. 1999). In this connection, studies have eliminated any axial sliding between the identical and the parallel helices (α 1 and α 1′) of this receptor as contributing to signal transmission (Falke and Hazelbauer 2001); note that an ~1 Å off-symmetry axial staggering between chemically identical helices would, according to the asymmetric degeneracy principle, yield two possible conformations regardless of the absence or the presence of ligand.
If in a homodimeric rod we combine the concepts of asymmetry-derived conformational degeneracy (Fig. 6A,B) and junction bends (Fig. 5), we may expect to see a “bistably jointed” segmentally flexible molecule (Fig. 6C). In contrast to rods that are stiff, purely flexible, or freely jointed (Howard 2001), such a bistably jointed rod may have distinctive dynamic and functional properties. For example, one contribution to the flexibility of tropomyosin is suggested by the crystal structure of its N terminus (Brown et al. 2001): At each of the alanine–leucine junctions in its sequence (see above) the molecule could flex between two stabilized oppositely bent conformations mediated by piston-like motions in the asymmetric axially staggered alanine sections. Since according to tropomyosin’s sequence there are a number of presumptively asymmetric regions separated from one another by relatively in-register symmetric regions, and assuming that the asymmetric regions are conformationally independent of one another, then this design alone could lead to multiple conformations for the molecule (Fig. 6C). The flexibility (or “semiflexibility”) of tropomyosin appears critical for its ability to move azimuthally on the actin surface and hence to regulate muscle contraction.
Nonuniformities in self-assembly
In addition to providing multiple equally accessible conformations within dimeric proteins, asymmetric nonequivalent contacts between chemically identical subunits also has implications for self-assembly. As discussed above (Fig. 6A), at any one such contact region there is, in principle, a choice of which subunit adopts which conformation and/or is in which nonequivalent position. The question that can then be asked is whether an assembly built up from a series of such asymmetric interfaces has a single overall structure or whether a variable and irregular pattern emerges. In other words, are the different asymmetric interfaces independent from one another?
Perhaps the most well-known self-assembly systems composed of chemically identical subunits in quasi- or nonequivalent positions are the icosahedrally shaped viral shells containing >60 identical units (Caspar and Klug 1962). Here the departures from equivalency are both geometrically mandated and mediated by the capacity of the individual subunits to adopt different conformations when interacting with one another. In the assembly of these protein subunits, it may be possible to go through a large number of configurations (see page 22 in Caspar and Klug 1962), but many of these would be considered mistakes (see also Berger et al. 1994) that would have to be rectified or avoided by interactions with other agents (e.g., nucleic acid, cations) (Johnson and Speir 1997) before a complete closed shell of desired sized can result (Caspar and Klug 1962). In other words, the various nonequivalent contacts must ultimately be organized in some manner to close the shell, as long as these contacts are necessary for keeping the shell intact (see Harrison et al. 1978). Organized patterns of quasi-equivalent interactions between chemically identical subunits adopting two different conformations have also been used to construct models of helically symmetrical bacterial flagellar filament (Calladine 1975; see also Macnab and Aizawa 1984). The double-helically symmetric F-actin filament can display multiple conformational states, but under a specified condition generally “anneals” into one dominant and uniform polymer (Orlova et al. 2004), and “adjacent subunits do not act independently” (Egelman 2003). Constraints of closing a shell or of forming a defined helically symmetrical array, however, would not occur in a more open and linear self-assembly.
The protofilament of the fibrin blood clot provides an exceptional example of a self-assembly system that may be intrinsically and discretely nonuniform. The symmetry properties and architecture of fibrin’s subunit and its protofilament are quite unlike those of the viral shells or helical filaments described above. Fibrin (~340 kDa) is a long (~450 Å) chemical homodimer of three nonidentical polypeptide chains. The regions near the N termini of these three chains from the two halves of the molecule come together in the center to form a small globular “disulfide knot” domain that is for the most part twofold symmetric. However, two reciprocal disulfides near the dimer axis (Fig. 1C; see above) cause the N-terminal 14 residues of the two γ chains to adopt significantly different conformations in bovine fibrinogen (asymmetric triangle near “Y” and “Y′” in Fig. 6D) (see also Madrazo et al. 2001). In addition, the C termini of each of the three chains end in globular domains, and the C-terminal γ domains of adjacent fibrin (ogen) molecules form end-to-end contacts containing a screw-like offset from strict twofold rotational symmetry (where “X” and “X′” meet in Fig. 6D) (Spraggon et al. 1997; Brown et al. 2000; see also Yang et al. 2002). Since this offset between the C termini is well defined and completely conserved in numerous crystal structures and the deviation from twofold symmetry at the N termini is apparently enforced by covalent bonds, it would appear that for each case there might be a relatively high kinetic barrier to interchange between the degenerate asymmetric states (Fig. 6A).
A key architectural feature of the fibrin protofilament is that these two sources of asymmetry, which alternate in the end-to-end bonded filament of fibrin (Fig. 6D), are separated by a large (~225 Å) distance. This feature suggests that, in contrast to the viral shells or helical polymers described above, the choice of which fibrin subunit adopts which conformation (or is in which non-equivalent screw environment) at any one asymmetric interface may be independent of the choice at the next interface. Under these circumstances alone, the extended filament would adopt any one of many degenerate conformations with respect to these asymmetries (Fig. 6D). Whether such a variable self-assembly pattern is actually realized in the mature fibrin clot is not known; it is possible that constraints may be imposed by the lateral association of these filaments into two-stranded half-staggered protofibrils or higher order fibers, and direct experiments on polymerized fibrin would be necessary. Any configurational variability in the self-assembly of fibrin could contribute in some measure to the adaptability of the fibrin clot.
Adaptive protein recognition
A related consequence of asymmetry in homodimers, in particular where the individual subunits form identifiable domains that interact with a global offset, may be a predisposition for the monomer to be able to bind various proteins (Cha et al. 2002). The haemopexin-like (PEX) domain of the matrix metalloproteinase MMP9 dimerizes asymmetrically where one subunit is offset along the dimer axis relative to the other PEX subunit. In contrast to twofold symmetric homodimers, in which each subunit binds identical surfaces of the partner subunit, in the offset homodimer each subunit recognizes and binds to a different (and in PEX9 overlapping) surface. The capacity to recognize different protein targets is thus built-in, and PEX9 binds with high affinity haemopexin-like domains from different matrix metalloproteinases as well as endogenous inhibitors of MMPs (Cha et al. 2002). The possible chemical cause of the asymmetric dimerization and adaptive protein recognition properties of PEX9 was not elaborated upon by these investigators. Inspection of the PEX9 dimer structure shows the offset interface to include extended strands in which valine residues 189 (numbering in PDB file) from one subunit and 192 from the other subunit are closest to each other at the dimer axis; in fact, this strand consists of a conserved repeating pattern of polar and valine side chains in MMP9s; as suggested above, repeating residues in a sequence may facilitate slippage from a symmetrical register.
Perspective
Relating primary sequence to three-dimensional structure is one of the great challenges of molecular biology, and advances have been made in understanding the folding of some single polypeptide chains and simple protein assemblies. Previous analyses of the structures of protein dimers (e.g., Miller 1989; Jones and Thornton 1996) have described the common folds that contact each other throughout the subunit interface. These studies, however, could not have benefited from insights obtained from recent tropomyosin and fibrinogen crystal structures (Brown et al. 2001; Madrazo et al. 2001) and thus did not focus on the structures occurring immediately adjacent to the dimer axis nor on their functional implications. Motivated by the unanticipated but simply designed departures from symmetry in these crystal structures, we have begun a visual survey of homodimers in the Protein Data Bank, examining the structures immediately adjacent to the axis. These preliminary results show at least five commonly used regular frameworks that (among others) accommodate a twofold symmetric axis. Based on our analyses, we conclude that these frameworks can provide a rational basis for relating primary sequence to the maintenance or breaking of symmetry in quaternary structure. These molecular asymmetries, in turn, have important functional consequences in diverse physiological systems.
There are also more general consequences. Intrinsic uncertainty and indeterminism need not be limited to the subatomic quantum realm; as described above, by producing distinct multiple conformations of identical energy, asymmetric contacts between two (or more) copies of an object may provide a basic structural method for injecting such unpredictability and irregularity into the macromolecular world. The oft-found capacity of some proteins to take on two (or more) different shapes (of similar enough energy to be both observed) has already broadened the classical sequence determined structure doctrine (Anfinsen 1973), and asymmetric contacts between identical or similar amino acid sequences may provide a conceptual basis for such occurrences. Moreover, asymmetry-induced degeneracy in self-associations is, in principle, not limited to proteins or even other molecules, and may well extend to structures of entirely different dimensions.
Acknowledgments
I thank C. Cohen, T. Yeates, and D. Goodsell and for reading the manuscript and making comments, A. Yarovoy for assistance with compiling the data for Table 1, and K. Allen for alerting me to the relevance of half-site reactivity. C. Cohen also wrote part of the introduction and provided generous encouragement for this work, which has been supported by her NIH grant (AR017346).
Table 1.
Five geometrically distinct regular frameworks for accommodating a twofold symmetry axis are found in diverse proteins

We have visually inspected over 100 protein homodimers, representing an initial survey to investigate the designs used to accommodate a twofold axis. Ninety of these proteins are derived from two lists of non- (or low) homologous homodimers solved by X-ray crystallography: 30 entries from a survey of Jones and Thornton (1996) who derived some general principles of protein–protein interactions, and a list of 60 2.0 Å resolution structures deposited between 1997 and 2002 culled from a list of (<50%) nonhomologous homodimers created using the PDB Web server. (Since the coordinates deposited in the data bank are normally only for an asymmetric unit, the coordinates for homodimers consisting of crystallographically related monomers were generated using the Protein Quaternary Structure query form [http://pqs.ebi.ac.uk].) Structures were visually inspected using O (Jones et al. 1991), with the focus being on the conformations of the proteins at the dimer axis. In the 90 protein structures examined, 213 interfaces (roughly defined as a contact region spanned by sequentially contiguous residues) were identified about the dimer axis. Fifty-seven of these interfaces (# row in table header) may be described as single β-sheets that extend across the axis or α-helical coiled coils (in which a heptad assignment can be made for the residues) (see table headers), and are the main focus of this article. Future, more extensive studies will classify the other less regular and less well-understood interfaces.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051658406.
References
- Amor, J.C., Harrison, D.H., Kahn, R.A., and Ringe, D. 1994. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 372: 704–708. [DOI] [PubMed] [Google Scholar]
- Anfinsen, C.B. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. [DOI] [PubMed] [Google Scholar]
- Berger, B., Shor, P.W., Tucker-Kellogg, L., and King, J. 1994. Local rule-based theory of virus shell assembly. Proc. Natl. Acad. Sci. 91: 7732–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.H., Volkmann, N., Jun, G., Henschen-Edman, A.H., and Cohen, C. 2000. The crystal structure of modified bovine fibrinogen. Proc. Natl. Acad. Sci. 97: 85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.H., Kim, K.-H., Jun, G., Greenfield, N.J., Dominguez, R., Volkmann, N., Hitchcock-DeGregori, S.E., and Cohen, C. 2001. Deciphering the design of the tropomyosin molecule. Proc. Natl. Acad. Sci. 98: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard, P., Kammerer, R.A., Steinmetz, M.O., Bourenkov, G.P., and Aebi, U. 2000. The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure 8: 223–230. [DOI] [PubMed] [Google Scholar]
- Calladine, C.R. 1975. Construction of bacterial flagella. Nature 255: 121–124. [DOI] [PubMed] [Google Scholar]
- Caspar, D.L.D. and Klug, A. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27: 1–24. [DOI] [PubMed] [Google Scholar]
- Cha, H., Kopetzki, E., Huber, R., Lanzendörfer, M., and Brandstetter, H. 2002. Structural basis of the adaptive molecular recognition by MMP9. J. Mol. Biol. 320: 1065–1079. [DOI] [PubMed] [Google Scholar]
- Coles, M., Diercks, T., Liermann, J., Groger, A., Rockel, B., Baumeister, W., Koretke, K.K., Lupas, A., Peters, J., and Kessler, H. 1999. The solution structure of VAT-N reveals a “missing link” in the evolution of complex enzymes from a simple βαββ element. Curr. Biol. 9: 1158–1168. [DOI] [PubMed] [Google Scholar]
- Crick, F.H.C. 1953. The packing of α-helices: Simple coiled coils. Acta Crystallogr. 6: 689–697. [Google Scholar]
- Crick, F.H.C. and Watson, J.D. 1957. Virus structure: General principles. In CIBA Foundation symposium on the nature of viruses (eds. G.E.W. Wolstenholme and E.C.P. Millar), pp. 5–13. Little, Brown, Boston.
- Day, C.L. and Alber, T. 2000. Crystal structure of the amino-terminal coiled-coil domain of the APC tumor suppressor. J. Mol. Biol. 301: 147–156. [DOI] [PubMed] [Google Scholar]
- De Baere, I., Liu, L., Moens, L., Van Beeumen, J., Gielens, C., Richelle, J., Trotman, C., Finch, J., Gerstein, M., and Perutz, M. 1992. Polar zipper sequence in the high-affinity hemoglobin of Ascaris suum: Amino acid sequence and structural interpretation. Proc. Natl. Acad. Sci. 89: 4638–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbrack Jr., R.L. and Karplus, M. 1993. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 230: 543–574. [DOI] [PubMed] [Google Scholar]
- Egelman, E.H. 2003. A tale of two polymers: New insights into helical filaments. Nat. Rev. Mol. Cell Biol. 4: 621–630. [DOI] [PubMed] [Google Scholar]
- Eneqvist, T., Andersson, K., Olofsson, A., Lundgren, E., and Sauer-Eriksson, A.E. 2000. The β-slip: A novel concept in transthyretin amyloidosis. Mol. Cell 6: 1207–1218. [DOI] [PubMed] [Google Scholar]
- Falke, J.J. and Hazelbauer, G.L. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, R.D.B. and McRae, T.P. 1973. Conformations in fibrous proteins. Academic Press, New York.
- Gernert, K.M., Surles, M.C., Labean, T.H., Richardson, J.S., and Richardson, D.C. 1995. The alacoil: A very tight, antiparallel coiled-coil of helices. Protein Sci. 4: 2252–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, J. 1998. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell 95: 237–248. [DOI] [PubMed] [Google Scholar]
- Goodsell, D.S. and Olson, A.J. 2000. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29: 105–153. [DOI] [PubMed] [Google Scholar]
- Goodsell, D.S., Kaczor-Grzeskowiak, M., and Dickerson, R.E. 1994. The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T-A steps. J. Mol. Biol. 239: 79–96. [DOI] [PubMed] [Google Scholar]
- Harrison, S.C., Olson, A.J., Schutt, C.E., Winkler, F.K., and Bricogne, G. 1978. Tomato bushy stunt virus at 2.9 Å resolution. Nature 276: 368–373. [DOI] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of motor proteins and the cytoskeleton. Sinauer Associates, Sunderland, MA.
- Hutchinson, E.G., Sessions, R.B., Thornton, J.M., and Woolfson, D.N. 1998. Determinants of strand register in antiparallel β-sheets of proteins. Protein Sci. 7: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin, V.A., Temple, B., Hu, M., Li, G., Yin, Y., Vachette, P., and Carter Jr., C.W. 2000. 2.9 Å crystal structure of ligand-free tryptophanyl-tRNA synthetase: Domain movements fragment the adenine nucleotide binding site. Protein Sci. 9: 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., Stec, B., Lipscomb, W.N., and Kantrowitz, E.R. 1999. Insights into the mechanisms of catalysis and heterotropic regulation of Escherichia coli aspartate transcarbamoylase based upon a structure of the enzyme complexed with the bisubstrate analogue N-phosphonacetyl-L-aspartate at 2.1 Å. Proteins 37: 729–742. [PubMed] [Google Scholar]
- Johnson, J.E. and Speir, J.A. 1997. Quasi-equivalent viruses: A paradigm for protein assemblies. J. Mol. Biol. 269: 665–675. [DOI] [PubMed] [Google Scholar]
- Jones, S. and Thornton, J.M. 1996. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 93: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- King, D.A., Zhang, L., Guarente, L., and Marmorstein, R. 1999. Structure of a HAP1–DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat. Struct. Biol. 6: 64–71. [DOI] [PubMed] [Google Scholar]
- Koshland Jr., D.E., Nemethy, G., and Filmer, D. 1966. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry 5: 365–385. [DOI] [PubMed] [Google Scholar]
- Lesk, A.M. and Chothia, C. 1984. Mechanisms of domain closure in proteins. J. Mol. Biol. 174: 175–191. [DOI] [PubMed] [Google Scholar]
- Li, Y., Mui, S., Brown, J.H., Strand, J., Reshetnikova, L., Tobacman, L.S., and Cohen, C. 2002. The crystal structure of the C-terminal fragment of striated-muscle α-tropomyosin reveals a key troponin T recognition site. Proc. Natl. Acad. Sci. 99: 7378–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddington, R.C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T.L., and Harrison, S.C. 1991. Structure of simian virus 40 at 3.8 Å resolution. Nature 354: 278–284. [DOI] [PubMed] [Google Scholar]
- Lupas, A. 1996. Coiled coils: New structures and new functions. Trends Biochem. Sci. 21: 375–382. [PubMed] [Google Scholar]
- Macnab, R.M. and Aizawa, S. 1984. Bacterial motility and the bacterial flagellar motor. Annu. Rev. Biophys. Bioeng. 13: 51–83. [DOI] [PubMed] [Google Scholar]
- Madrazo, J., Brown, J.H., Litvinovich, S., Dominguez, R., Yakovlev, S., Medved, L., and Cohen, C. 2001. Crystal structure of the central region of bovine fibrinogen (E5 fragment) at 1.4- Å resolution. Proc. Natl. Acad. Sci. 98: 11967–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. 1989. The structure of interfaces between subunits of dimeric and tetrameric proteins. Protein Eng. 3: 77–83. [DOI] [PubMed] [Google Scholar]
- Monod, J., Changeux, J.P., and Jacob, F. 1963. Allosteric proteins and cellular control systems. J. Mol. Biol. 6: 306–329. [DOI] [PubMed] [Google Scholar]
- Monod, J., Wyman, J., and Changeux, J.P. 1965. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 12: 88–118. [DOI] [PubMed] [Google Scholar]
- Nagradova, N.K. 2001. Interdomain interactions in oligomeric enzymes: Creation of asymmetry in homo-oligomers and role in metabolite channeling between active centers of hetero-oligomers. FEBS Lett. 487: 327–332. [DOI] [PubMed] [Google Scholar]
- Noreen, I.M.A., Kaptein, R., Sauer, R.T., and Boelens, R. 1999. The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils. Nat. Struct. Biol. 6: 755–759. [DOI] [PubMed] [Google Scholar]
- Orlova, A., Shvetsov, A., Galkin, V.E., Kudryashov, D.S., Rubenstein, P.A., Egelman, E.H., and Reisler, E. 2004. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl. Acad. Sci. 101: 17664–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea, E.K., Klemm, J.D., Kim, P.S., and Alber, T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254: 539–544. [DOI] [PubMed] [Google Scholar]
- Ottemann, K.M., Xiao, W., Shin, Y.-K., and Koshland D.E. Jr., 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285: 1751–1754. [DOI] [PubMed] [Google Scholar]
- Perutz, M. 1989. Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 22: 139–237. [DOI] [PubMed] [Google Scholar]
- ———. 1994. Polar zippers: Their role in human disease. Protein Sci. 3: 1629–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder, J.W. and Richards, F.M. 1987. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 193: 775–791. [DOI] [PubMed] [Google Scholar]
- Reinisch, K.M., Nibert, M.L., and Harrison, S.C. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404: 960–967. [DOI] [PubMed] [Google Scholar]
- Renatus, M., Stennicke, H.R., Scott, F.L., Liddington, R.C., and Salvesen, G.S. 2001. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. 98: 14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick, D., Chatterton, J.E., Schwartz, K., Slayter, H., and Krieger, M. 1996. Structures of class A macrophage scavenger receptors. Electron microscopic study of flexible, multidomain, fibrous proteins and determination of the disulfide bond pattern of the scavenger receptor cysteine-rich domain. J. Biol. Chem. 271: 26924–26930. [DOI] [PubMed] [Google Scholar]
- Ross, C.A., Poirier, M.A., Wanker, E.E., and Amzel, M. 2003. Polyglutamine fibrillogenesis: The pathway unfolds. Proc. Natl. Acad. Sci. 100: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann, M.G. 1984. Constraints on the assembly of spherical virus particles. Virology 134: 1–11. [DOI] [PubMed] [Google Scholar]
- Seydoux, F., Malhotra, O.P., and Bernhard, S.A. 1974. Half-site reactivity. CRC Crit. Rev. Biochem. 2: 227–257. [DOI] [PubMed] [Google Scholar]
- Shirakihara, Y. and Evans, P.R. 1988. Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J. Mol. Biol. 204: 973–994. [DOI] [PubMed] [Google Scholar]
- Song, S.Y., Xu, Y.B., Lin, Z.J., and Tsou, C.L. 1999. Structure of active site carboxymethylated D-glyceraldehyde-3-phosphate dehydrogenase from Palinurus versicolor. J. Mol. Biol. 287: 719–725. [DOI] [PubMed] [Google Scholar]
- Spraggon, G., Everse, S.J., and Doolittle, R.F. 1997. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature 389: 455–462. [DOI] [PubMed] [Google Scholar]
- Stetefeld, J., Frank, S., Jenny, M., Schulthess, T., Kammerer, R.A., Boudko, S., Landwehr, R., Okuyama, K., and Engel, J. 2003. Collagen stabilization at atomic level: Crystal structure of designed (GlyPro-Pro) 10 foldon. Structure 11: 339–346. [DOI] [PubMed] [Google Scholar]
- Swaminathan, K., Flynn, P., Reece, R.J., and Marmorstein, R. 1997. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat. Struct. Biol. 4: 751–759. [DOI] [PubMed] [Google Scholar]
- Todd, A.E., Orengo, C.A., and Thornton, J.M. 2002. Sequence and structural differences between enzyme and nonenzyme homologs. Structure 10: 1435–1451. [DOI] [PubMed] [Google Scholar]
- Wallis, R. and Drickamer, K. 1997. Asymmetry adjacent to the collagen-like domain in rat liver mannose-binding protein. Biochem. J. 325: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, D., Eisenhaber, F., and Argos, P. 1996. Principles of helix–helix packing in proteins: The helical lattice superposition model. J. Mol. Biol. 255: 536–553. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-X. and Pan, X.-M. 1996. Kinetic differentiation between ligand-induced and pre-existent asymmetric models. FEBS Lett. 388: 73–75. [DOI] [PubMed] [Google Scholar]
- Weis, W.I., Kahn, R., Fourme, R., Drickamer, K., and Hendrickson, W.A. 1991. Structure of the calcium-dependent lectin domain from a rat mannose-binding protein determined by MAD phasing. Science 254: 1608–1615. [DOI] [PubMed] [Google Scholar]
- Weis, W.I., Drickamer, K., and Hendrickson, W.A. 1992. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature 360: 127–134. [DOI] [PubMed] [Google Scholar]
- Wouters, M.A. and Curmi, P.M. 1995. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins 22: 119–131. [DOI] [PubMed] [Google Scholar]
- Xiao, B., Singh, S.P., Nanduri, B., Awasthi, Y.C., Zimniak, P., and Ji, X. 1999. Crystal structure of a murine glutathione S-transferase in complex with a glutathione conjugate of 4-hydroxynon-2-enal in one subunit and glutathione in the other: Evidence of signaling across the dimer interface. Biochemistry 38: 11887–11894. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Pandi, L., and Doolittle, R.F. 2002. The crystal structure of fragment double-D from cross-linked lamprey fibrin reveals isopeptide linkages across an unexpected D-D interface. Biochemistry 41: 15610–15617. [DOI] [PubMed] [Google Scholar]