Abstract
A docking-and-alignment protocol was devised in order to build amyloid protofilaments of Transthyretin (TTR), starting from partially disrupted TTR monomeric subunits and based on experimentally available information. The docking approach is driven by a combination of shape complementarity and energetic criteria, and uses constraints derived from experimental data obtained for the fibrillar state. The dimeric structures obtained were then subjected to an alignment scheme followed by clustering analysis, producing a collection of protofilaments with distinct geometric properties. The selected protofilament model presented here does agree with known experimental data and general amyloid properties; it is formed by two extended continuous β-sheets with the β-strands perpendicular to the main axis of the protofilament and a helical twist with a period of ~48 β-strands. This TTR proto-filament model may be an important step in the understanding of the molecular mechanisms of TTR aggregation, as well as, a valuable instrument in drug design strategies against amyloid diseases.
Keywords: Transthyretin, amyloid, protofilament, protein docking, supramolecular assembly
Transthyretin (TTR) is one of several proteins known to be involved in human amyloid diseases. TTR is a homotetrameric protein mostly found in the plasma and the cerebral spinal fluid, and has been identified as the causative agent of such diseases as Familial Amyloidotic Polyneuropathy, Familial Amyloidotic Cardiomyopathy, and Senile Systemic Amyloidosis. It is believed that, in the process of amyloid formation, TTR dissociates to non-native monomeric units, which may act as the building blocks of the amyloid fibrils (Lai et al. 1996; Quintas et al. 1999, 2001; for review, see Brito et al. 2003). The structural characterization of these fibrils and the identification of the entities involved in fibril assembly are crucial for understanding of the mechanisms of pathogenesis in amyloid diseases, and for the development of appropriate therapeutic strategies. Experimental techniques have not yet been able to produce a high-resolution structure of an amyloid fibril of TTR. The work presented here proposes a high-resolution working model of the elementary units that constitute the fibrils, the protofilaments.
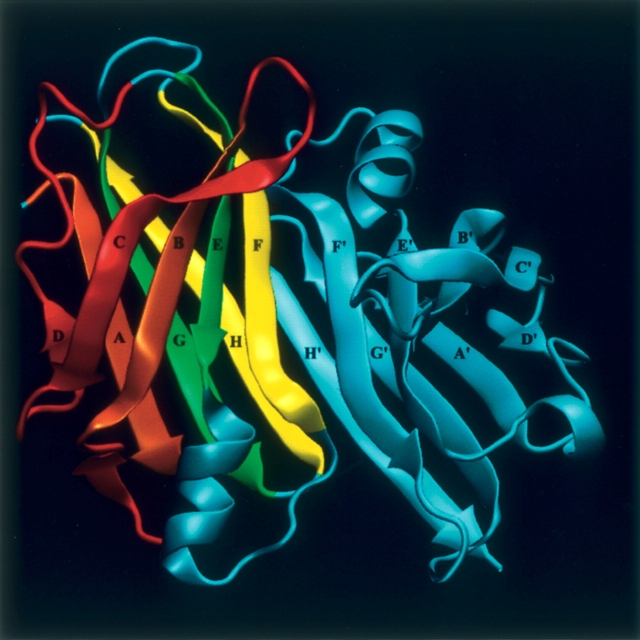
Each TTR subunit has a β-sandwich fold composed of two four-stranded β-sheets labeled DAGH and CBEF, as shown in Figure 1. In the native protein, the β-sheets from two monomers associate edge-to-edge through β-strands H/H′ and F/F′ to produce a dimer composed of two extended β-sheets formed by strands DAGHH′G′A′D′ and CBEFF′E′B′C′. Association of two of these dimers mainly through hydrophobic interactions mediated by the AB and GH loops forms the functional homotetramer.
Figure 1.
Schematic view of the crystallographic dimer of TTR (PDB entry 1F41; Hornberg et al. 2000), with the identification of the β-strands. Amyloid formation by TTR implies tetramer dissociation, structural alteration of the monomer, and subunit association into aggregates. The monomer–monomer interaction is believed to be mediated by two types of interfaces: a near-native interface (NearNI) comprising strands F/F′ and H/H′ (in yellow) of adjacent subunits, and a non-native interface (NonNI) constituted by strands A/A′ and B/B′ (in brown) of adjacent subunits. In order to expose strands A and B, a partially disrupted monomer was built by removing strands C and D and the loops AB and CD (in red). The figure was produced with the program VMD (Humphrey et al. 1996).
Recently, efforts have been made to determine the three-dimensional arrangement of the TTR subunits in the amyloid fibrils, based on site-directed spin labeling EPR studies (Serag et al. 2001, 2003) and H/D exchange NMR studies (Olofsson et al. 2004). Yeates and collaborators (Serag et al. 2003) proposed an anti-parallel head-to-head/tail-to-tail arrangement of the TTR subunits in the fibril, with the native intersubunit contact between β-strands F and F′ maintained, and a new intersubunit interface formed between β-strands B and B′, in order to build the continuous extended cross-β structure characteristic of amyloid. The formation of this new interface implies the displacement of strands C and D from the β-sandwich. Additionally, Olofsson et al. (2004) identified a core region of the TTR subunit with large solvent protection factors. The data indicate that strands B, E, and F and strands G and H of the TTR subunit are, if not totally, partially maintained in the fibrils. The presence of solvent-exposed residues in strands C and D, as well as in the connecting and the following loops, clearly shows that this region is not part of the fibrillar core. Taken together, these results support a protofilament structure formed by a core of two three-stranded β-sheets (BEF and AGH) (Fig. 1), where strands F/F′ and H/H′ are in a native-like arrangement, forming a near-native interface, and strands A and B participate in a new non-native interface.
To build a molecular model in accordance with the structural characteristics mentioned above, two sets of docking computations were performed using the program HADDOCK (Dominguez et al. 2003): one set to recreate the near-native interface (NearNI) and another set to build the new non-native interface (NonNI) (Table 1). The docking procedure is driven by a combination of shape complementarity and energetic criteria, and uses ambiguous interaction restraints, defined between any atom of the active residues of the ligand protein and all atoms of all active and passive residues of the receptor protein. Active residues are those residues experimentally identified to be involved in the interaction and solvent-accessible in the monomeric form of the protein. Passive residues are all solvent-accessible neighbors of active residues. Active and passive residues were defined based on NMR H/D exchange data (Olofsson et al. 2004) and solvent-accessibility calculations (Table 1). Additional restraints for the NearNI and NonNI interfaces were defined according to distances experimentally determined by EPR (Serag et al. 2001, 2003; Table 1). To generate the NearNI, two native monomers were docked against each other at their native interface (F/F′ and H/H′), generating near-native dimeric structures. To generate the NonNI, two partially disrupted monomers (Fig. 1) were created by deleting β-strands C and D and loops AB and CD. These partially disrupted monomers were then docked at the A/A′–B/B′ interface, generating non-native dimeric structures. Two thousand docked structures were calculated for each docking run, and the best dimeric structures, with the lowest intermolecular energies and obeying all experimental constraints, were kept.
Table 1.
Restraints used in the docking procedure
| Restraints | ||||||
| Ambiguous | Unambiguous | |||||
| Interface | Residues exposed to the solvent in the monomeric forma | NMR protected residues in the fibrillar stateb | HADDOCK active residuesc | HADDOCK passive residuesd | Interresidue distances (Å)e | |
| NonNI | 12–20, 28–35 | 18, 19, 28–34 | 18, 19, 28–34 | 12–17, 20, 35 | 33(B)–33(B′) | 14 |
| 29(B)–29(B′) | 12 | |||||
| 31(B)–31(B′) | 8 | |||||
| NearNI | 86, 87, 89, 92, 94, 96, 99, 114–125 | 89–98, 118–124 | 89, 92, 94, 96, 118–124 | 86, 87, 99, 114–117, 125 | 89(1EF)–96(F′) | 12 |
| 94(F)–94(F′) | 15 | |||||
| 96(F)–89(lE′F′) | 12 | |||||
| 96(F)–96(F′) | 25 | |||||
a Solvent accessibilities were calculated with NACCESS (S.J. Hubbard and J.M. Thornton, University College London). Residues with accessibilities >45% are considered exposed to the solvent.
b Residues belonging to the interfaces between subunits with detectable H/D exchange protection factors in the fibrillar state (Olofsson et al. 2004).
c Residues accessible to the solvent in the monomeric form and solvent-protected in the fibrillar state.
d Residues accessible to the solvent and/or neighbors of active residues.
e Distances experimentally determined by EPR (Serag et al. 2001, 2003). B, B′, F, F′, lEF, and lE′F′ identify the β-strands and loop-EF.
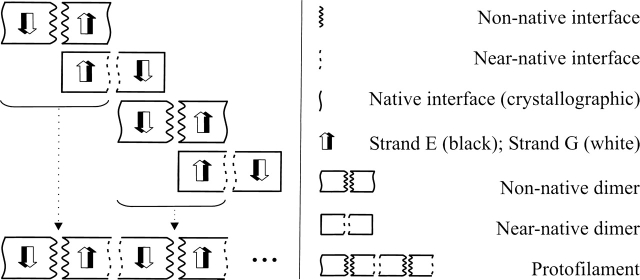
In order to build protofilaments, an alignment scheme was devised using the non-native dimers as building blocks in the protofilament assembly, and the near-native dimers as a template to build the interface between adjacent non-native dimers (Fig. 2). Combining all the best non-native dimeric structures with all the best near-native dimeric structures in the alignment procedure, we obtained thousands of protofilament structures. These were then clustered from the pairwise RMSD-matrix using the gromos algorithm implemented in the GROMACS package (Berendsen et al. 1995; Daura et al. 1999; Lindahl et al. 2001), producing characteristic structural families with different geometric properties. Most of the protofilaments obtained with this procedure are elongated structures with helical topology and formed by two extended β-sheets. These structures have helical periods in the range of 7–10 dimers, showing that subtle structural differences in the interfaces are of paramount importance for overall supramolecular geometry. A detailed analysis of the differences among protofilament structural families will be presented elsewhere.
Figure 2.
Scheme for the alignment of the docked dimers used to build the protofilament structures. The black and white arrows represent the two β-strands used to align the structures, since their native structure was maintained throughout the docking procedure.
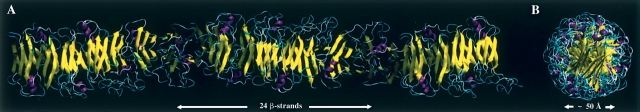
One of the protofilament structural families obtained from the docking-and-alignment procedure is characterized by linear fibrillar structures with β-strands perpendicular to the main protofilament axis, which is generally accepted as a characteristic of amyloid, the so-called cross-β structure (Eanes and Glenner 1968). In order to rebuild the full polypeptide chain in each TTR subunit, the peptide fragments initially removed to create the partially disrupted monomers were added. These peptide sequences, comprising β-strands C and D and loops AB and CD, were energy-minimized and subjected to a short molecular dynamics run, without interfering with the subunit interfaces or the subunit core. The final protofilament structure obtained is formed by two extended continuous β-sheets, (BEFF′E′B′)n and (AGHH′G′A′)n, with the β-strands nearly perpendicular to the main axis of the protofilament. The protofilament, with a diameter of ~50 Å, presents a helical twist with a period of ~48 β-strands, that is, 16 monomeric units with two three-stranded β-sheets each (BEF and AGH) (Fig. 3).
Figure 3.
(A) Schematic representation of the TTR protofilament model obtained, showing the size of half of the repeating unit. (B) Protofilament cross-section dimension including only the core β-strands.
Finally, to relax the protofilament structure, we carried out a short molecular dynamics simulation using the program NAMD (Kale et al. 1999) running on a cluster of 100 commodity computers (Centopeia at the Universidade de Coimbra). After energy minimization of the protofilament in vacuum, water molecules and ions were added to a final ionic strength of 150 mM, resulting in a system of >240,000 atoms. After equilibration of the molecular system at 310 K with the ensemble NVT, a production run of 100 psec was carried out, using periodic boundary conditions. Long-range electrostatics were computed at every step using the Particle Mesh Ewald method. The aggregation state and global fold of the protofilament were not affected during the simulation. The overall stereochemical quality of the final structure was assessed with PROCHECK (Laskowski et al. 1993), revealing that >75% of the residues are in the most favorable regions of the Ramachandran plot and the very few residues (<1%) in disallowed regions belong to exposed loops.
It is interesting to note that, based on X-ray fiber diffraction studies, Blake and Serpell (1996) proposed a model for TTR amyloid consisting of extended β-sheets with a helical twist and a fiber repeating unit of ~115 Å, corresponding to 24 β-strands. It must be more than coincidental that our model, based on energetic and shape complementarity criteria, has a helical period double than Blake’s model. In fact, it is possible that the X-ray diffraction pattern observed for the fiber could be the result of lateral association of protofilaments shifted by half-period. If this is not the case, to build a protofilament with a period of 24 β-strands a much more twisted helical structure is required, and a large conformational rearrangement of the β-sheets in the TTR subunit is necessary. Interestingly, our protofilament model shows surface segregation of charged residues in a helical arrangement, which could be responsible for the half-period pairing of protofilaments, in order to avoid electrostatic repulsions.
In summary, our results show that docking two TTR subunits to recreate a non-native interface involving β-strands A and B requires full solvent exposure of these strands. The docking and alignment procedure generated a range of protofilament structures, in agreement with the structural polymorphism observed for amyloid fibrils and recently reported (Cardoso et al. 2002; Jansen et al. 2005). The protofilament structure proposed here has geometric properties in close agreement with the known characteristics of TTR amyloid. Several experimental studies have shown that TTR amyloid fibrils are formed by continuous β-sheet helices (Blake and Serpell 1996) and protofilaments with diameters in the order of 40–60 Å, as revealed by electron microscopy and X-ray fiber diffraction (Serpell et al. 1995; Sunde et al. 1997). Additionally, all the EPR distance restraints and NMR protection factors initially imposed are observed in the final structures, and the generated structures have good stereochemical properties. This model may be refined in the future by the introduction of other experimentally derived constraints, but at this stage may become a valuable instrument in the rational design of compounds with therapeutic potential to inhibit amyloid fibril formation by TTR.
Acknowledgments
We acknowledge the computer resources provided by the Centre for Computational Physics, Universidade de Coimbra, and the support of “Fundação para a Ciência e Tecnologia” and the program FEDER, Portugal, through grants POCTI/BME/49583/2002 (to R.M.M.B.) and SFRH/BD/1354/2000 (to N.L.-F.).
Abbreviations
EPR, electron paramagnetic resonance
NearNI, near-native interface
NonNI, non-native interface
TTR, Transthyretin
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051787106.
References
- Berendsen, H.J.C., van der Spoel, D., and van Drunen, R. 1995. GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91: 43–56. [Google Scholar]
- Blake, C. and Serpell, L. 1996. Synchroton X-ray studies suggest that the core of the transthyretin amyloid fibril is a continuous β-sheet helix. Structure 4: 989–998. [DOI] [PubMed] [Google Scholar]
- Brito, R.M.M., Damas, A.M., and Saraiva, M.J. 2003. Amyloid formation by Transthyretin: From protein stability to protein aggregation. Curr. Med. Chem. Immun. Endoc. Metab. Agents 3: 349–360. [Google Scholar]
- Cardoso, I., Goldsbury, C.S., Müller, S.A., Olivieri, V., Wirtz, S., Damas, A.M., Aebi, U., and Saraiva, M.J. 2002. Transthyretin fibrillogenesis entails the assembly of monomers: A molecular model for in vitro assembled Transthyretin amyloid-like fibrils. J. Mol. Biol. 317: 683–695. [DOI] [PubMed] [Google Scholar]
- Daura, X., Gademann, K., Jaun, B., Seebach, D., van Gunsteren, W.F., and Mark, A.E. 1999. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 38: 236–240. [Google Scholar]
- Dominguez, C., Boelens, R., and Bonvin, A.M.J.J. 2003. HADDOCK: A protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125: 1731–1737. [DOI] [PubMed] [Google Scholar]
- Eanes, E.D. and Glenner, G.G. 1968. X-ray diffraction studies on amyloid filaments. J. Histochem. Cytochem. 16: 673–677. [DOI] [PubMed] [Google Scholar]
- Hornberg, A., Eneqvist, T., Olofsson, A, Lundgren, E., and Sauer-Eriksson, A.E. 2000. A comparative analysis of 23 structures of the amyloidogenic protein Transthyretin. J. Mol. Biol. 302: 649–669. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. 1996. VMD—Visual Molecular Dynamics. J. Mol. Graph. 14: 33–38. [DOI] [PubMed] [Google Scholar]
- Jansen, R., Dzwolak, W., and Winter, R. 2005. Amyloidogenic self-assembly of insulin aggregates probed by high resolution atomic force microscopy. Biophys. J. 88: 1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K., and Shulten, K. 1999. NAMD2: Greater scalability for parallel molecular dynamics. J. Comp. Phys. 151: 283–312. [Google Scholar]
- Lai, Z., Colon, W., and Kelly, J.W. 1996. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry 35: 6470–6482. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Lindahl, E., Hess, B., and van der Spoel, D. 2001. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Mod. 7: 306–317. [Google Scholar]
- Olofsson, A., Ippel, J.H., Wijmenga, S.S., Lundgren, E., and Öhman, A. 2004. Probing solvent accessibility of transthyretin amyloid by solution NMR spectroscopy. J. Biol. Chem. 279: 5699–5707. [DOI] [PubMed] [Google Scholar]
- Quintas, A., Saraiva, M.J., and Brito, R.M.M. 1999. The tetrameric protein transthyretin dissociates to a non-native monomer in solution. A novel model for amyloidogenesis. J. Biol. Chem. 274: 32943–32949. [DOI] [PubMed] [Google Scholar]
- Quintas, A., Vaz, D.C., Cardoso, I., Saraiva, M.J., and Brito, R.M.M. 2001. Tetramer dissociation and monomer partial unfolding precedes protofibril formation in amyloidogenic transthyretin variants. J. Biol. Chem. 276: 27207–27213. [DOI] [PubMed] [Google Scholar]
- Serag, A.A., Altenbach, C., Gingery, M., Hubbell, W.L., and Yeates, T.O. 2001. Identification of a subunit interface in transthyretin amyloid fibrils: Evidence for self-assembly from oligomeric building blocks. Biochemistry 40: 9089–9096. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Arrangement of subunits and ordering of β-strands in an amyloid sheet. Nat. Struct. Biol. 9: 734–739. [DOI] [PubMed] [Google Scholar]
- Serpell, L.C., Sunde, M., Fraser, P.E., Luther, P.K., Morris, E.P., Sangren, O., Lundgren, E., and Blake, C.C.F. 1995. Examination of the structure of the Transthyretin amyloid fibril by image reconstruction from electron micrographs. J. Mol. Biol. 254: 113–118. [DOI] [PubMed] [Google Scholar]
- Sunde, M., Serpell, L.C., Bartlam, M., Fraser, P.E., Pepys, M.B., and Blake, C.C.F. 1997. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273: 729–739. [DOI] [PubMed] [Google Scholar]