Abstract
TATA-binding protein (TBP)-interacting protein from the hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1 (Tk-TIP26) is a possible transcription regulatory protein in Thermococcales. Here, we report the crystal structure of Tk-TIP26 determined at 2.3 Å resolution with multiple-wavelength anomalous dispersion (MAD) method. The overall structure of Tk-TIP26 consists of two domains. The N-terminal domain forms an α/β structure, in which three α-helices enclose the central β-sheet. The topology of this domain is similar to that of holliday junction resolvase Hjc from Pyrococcus furiosus. The C-terminal domain comprises three α-helices, six β-strands, and two 310-helices. In the dimer structure of Tk-TIP26, two molecules are related with the crystallographic twofold axis, and these molecules rigidly interact with each other via hydrogen bonds. The complex of Tk-TIP26/Tk-TBP is isolated and analyzed by SDS-PAGE and gel filtration column chromatography, resulting in a stoichiometric ratio of the interaction between Tk-TIP26 and Tk-TBP of 4:2, i.e., two dimer molecules of Tk-TIP26 formed a complex with one dimeric TBP. The electrostatic surfaces of Tk-TIP26 and TBP from Pyrocuccus woesei (PwTBP) allowed us to build a model of the Tk-TIP26/TBP complex, and to propose the inhibition mechanism where two dimer molecules of Tk-TIP26 bind to a dimeric TBP, preventing its binding to TATA-DNA.
Keywords: transcription, TATA-binding protein (TBP), TBP-interacting protein, hyperthermophilic archaeon, X-ray crystallography, Cys2–His2 type zinc finger motif, gel filtration chromatography, protein structure/folding, structure/function studies, protein crystallization, docking proteins
TATA-box binding protein (TBP) plays a crucial role in basal transcription in eukaryotic and archaeal cells (Roeder 1996; Qureshi et al. 1997). In eukaryotic cells, RNA polymerases require multiple, accessory factors to interact with their respective promoters. In the case of the RNA polymerase II transcription system, at least six general transcription factors (GTFs)—TFIIA, TFIIB, TFIID (which is a large multisubunit complex consisting of TBP and TBP-associated factors [TAFs]), TFIIE, TFIIF, and TFIIH—are required to form a preinitiation complex (PIC) (Roeder 1996). In archaeal cells, archaeal RNA polymerase requires at least two additional GTFs: TBP and transcription factor B (TFB), which are homologs of eukaryotic TBP, and transcription factor IIB (TFIIB), respectively (Bell and Jackson 2001; Bell et al. 2001b). Moreover, recent studies revealed that archaea have an eukaryal TFIIEα homolog (TFE), and TFE could play a stimulatory role in transcription initiation (Bell and Jackson 2001; Bell et al. 2001a,b; Hanzelka et al. 2001). These results suggested that archaeal transcription shares high degrees of similarity with that of eukarya, and that it is simpler than eukaryal transcription. This simplicity of archaeal transcription could be a prototype in order to increase our understanding of eukaryotic transcription mechanisms.
To further our understanding of transcription mechanisms, it is very important to understand transcription factors other than GTFs. Recent studies suggested that several new transcription factors could interact with TBP. Some of them are referred to as “TBP-interacting proteins” (TIPs) because they were isolated using the affinity chromatography with TBP (Makino et al. 1996). TIPs have been identified in both eukaryal and archaeal cells. In eukarya, the most studied TIPs are TIP120 (Yogosawa et al. 1996) and TIP49 (Kanemaki et al. 1997). TIP120 was isolated from rat liver, and sequence analyses showed that the N-terminal region of TIP120 exhibited 44% of sequence similarity to that of Drosophila TAF80 (Yogosawa et al. 1996). Kinetic studies and electrophoretic mobility shift assays showed that TIP120 activated the basal levels of transcription from various kinds of promoters and also stimulated the DNA-binding activity of RNA polymerase II (Makino et al. 1999). TIP49 was also isolated from rat liver nuclear extracts using the affinity chromatography with TBP. The amino acid sequence of TIP49 contains an ATP-/GTP-binding P-loop, and exhibits sequence similarity of bacterial DNA helicase RuvB (Yamada et al. 2001). In archaea, eukaryal TIP49 homologs were identified in the genomes of Archeaoglobus fulgidus and Pyrococcus horikoshii (Soppa 1999). TIP26 was also isolated and characterized in several species of Thermococcales (Matsuda et al. 1999).
In 1999, Matsuda et al. first identified TIP26 in the Thermococcus kodakaraensis strain KOD1 (Tk-TIP26). KOD1 was isolated from a solfatara at a wharf on Kodakara Island, Kagoshima, Japan. The growth temperature of this strain ranges from 65° to 100°C, with an optimal growth temperature of 95°C (Morikawa et al. 1994; Fujiwara et al. 1998). Tk-TIP26 was isolated from cell lysates of this strain by affinity chromatography with TBP-agarose (Matsuda et al. 1999). Cloning of the gene encoding this protein showed that Tk-TIP26 is composed of 224 amino acid residues (molecular weight 25,558), and exists in a dimeric form. Database analyses indicate that the orthologous genes are only present in the genomes of two genera, Pyrococcus and Thermococcus. The recombinant Tk-TIP26 and Tk-TBP interact with each other with an equilibrium dissociation constant, KD, of 1.24–1.46 μM (Matsuda et al. 1999). Further studies on this protein revealed that in the absence of Tk-TFB, Tk-TIP26 prevents Tk-TBP from binding to TATA-DNA (Matsuda et al. 1999). On the other hand, in the presence of Tk-TFB, Tk-TIP26 does not inhibit the formation of the TFB/TBP/TATA-DNA ternary complex but interacts with this complex to form the TIP26/TFB/TBP/TATA-DNA quaternary complex (Matsuda et al. 2001). These results suggest that TIP26 plays a role as a new possible transcription regulation factor in Thermococcales.
Biological or biochemical studies have identified and characterized TIPs, but no three-dimentional structures of TIPs have been determined, while this information is required for further understanding of the interaction between TIPs and TBP.
We report here the first crystal structure of TBP-interacting protein (Tk-TIP26) from the hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1. In addition, we report the isolation of the Tk-TIP26/Tk-TBP complex. Characterization of this complex, and structural information about Tk-TIP26 and archaeal TBP gave us the insights on molecular mechanism of the interaction between TIP26 and TBP.
Results and Discussion
Overall structure
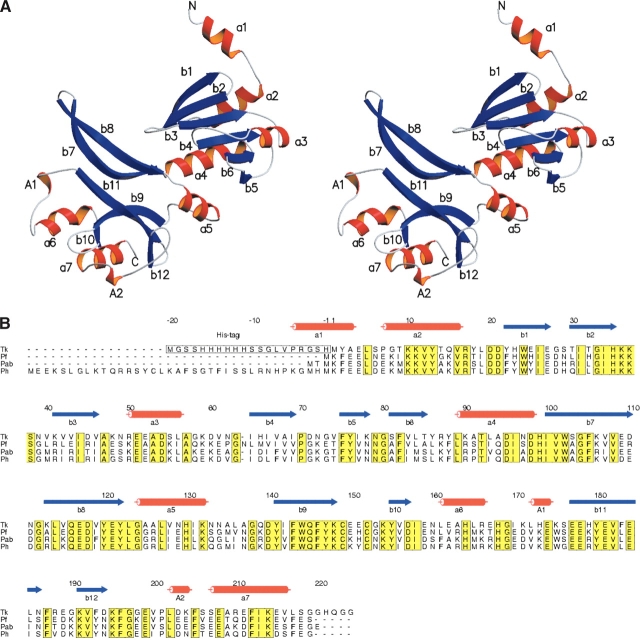
We obtained P43212 crystals of Tk-TIP26, and deter-mined its structure at 2.3 Å resolution. The asymmetric unit of the crystal lattice contains one molecule of Tk-TIP26. The overall structure of Tk-TIP26 is comprised of 7 α-helices (a1–a7), 12 β-strands (b1–b12), and 2 310-helices (A1 and A2), with overall dimensions of 64 × 49 × 28 Å (Fig. 1A). Additional electron densities derived from a part of a histidine tag (his-tag) are located before Met1. The last five residues of the his-tag (Pro5, Arg4, Gly3, Ser2, and His1) and the first three residues of Tk-TIP26 (Met1, Tyr2, and Ala3) form α-helix “a1” (Fig. 1A,B). Side residues of Arg4 and Ser2 form hydrogen bonds with those of Glu109 and Arg188 in the counterparts of the symmetrically equivalent molecules, respectively. Previously, we obtained the crystals of Tk-TIP26 without the his-tag. However, they diffracted to only 3.2 Å or worse, and they were not isomorphous (data not shown). The intermolecular interactions derived from the his-tag could contribute to the packing stabilization of molecules in the crystal and the improvement of resolution limit.
Figure 1.
(A) Stereodiagram of Tk-TIP26 molecule in an asymmetric unit. The character “a” (a1–a7) stands for α-helix; “A” (A1, A2), for 310-helix; “b” (b1–b12), for β-strand. (B) Sequence alignments of TBP-interacting proteins among Thermococcales. Tk, Thermococcus kodakaraensis; Pfu, Pyrococcus furiosus; Pab, Pyrococcus abyssi; Ph, Pyrococcus horikosii. The residues in the white box (−20–−1) represent the additional histidine-tag derived from pET28a. The residues in the yellow box represent the identical residues among these four species. The last five residues of the histidine-tag and the first three residues of Tk-TIP26 (1~3) form α-helix a1.
Tk-TIP26 is composed of two domains: N-terminal domain (−8~98) and C-terminal domain (99~224). Proteins whose folding is similar to that of Tk-TIP26 were searched with DALI (Holm and Sander 1995). As a result, the N-terminal domain of Tk-TIP26 shared a folding similarity to proteins with the topology of type II restriction endonucleases (see below). On the other hand, the C-terminal domain does not share folding similarity with any proteins whose structures have been determined to date.
N-terminal domain of Tk-TIP26
The N-terminal domain of Tk-TIP26 comprises four α-helices (a1–a4) and six β-strands (b1–b6), forming an α/β structure, in which the central β-sheet, formed by six β-strands, is surrounded by three α-helices: a2, a3, and a4 (Fig. 2A). As mentioned above, the folding of the N-terminal domain of Tk-TIP26 is similar to that of proteins with the topology of type II restriction endonucleases. Among these, archaeal holliday junction resolvase Hjc from Pyrococcus furiosus (Pfu-Hjc; PDB ID 1GEF; Nishino et al. 2001a, b) is the most topologically similar (Fig. 2A,C). Interestingly, the superimposition between the N-terminal domain of Tk-TIP26 and monomeric Pfu-Hjc shows an RMSD value for Cα carbon atoms of 3.5 Å , even though their sequence similarity is <10%. However, the electrostatic surface of the N-terminal domain of Tk-TIP26 is remarkably different from that of Pfu-Hjc. The N-terminal domain of Tk-TIP26 is mainly negatively charged, while Pfu-Hjc is positively charged (Fig. 2B). In addition, the arrangements of Tk-TIP26 in the dimer are notably different from that of Pfu-Hjc (Nishino et al. 2001a,b). Tk-TIP26 forms a dimer via the interaction between the N-terminal domain and the C-terminal domain in the counterpart of the dimer (Fig. 4A, below), while Pfu-Hjc is dimerized with a single domain. The contact of the dimeric -TIP26 involves hydrogen bonds (see below), while that of Pfu-Hjc involves hydrophobic interaction (Nishino et al. 2001a,b). These results suggest that Tk-TIP26 does not have the same function as in Pfu-Hjc, even though these two proteins share similar topology.
Figure 2.
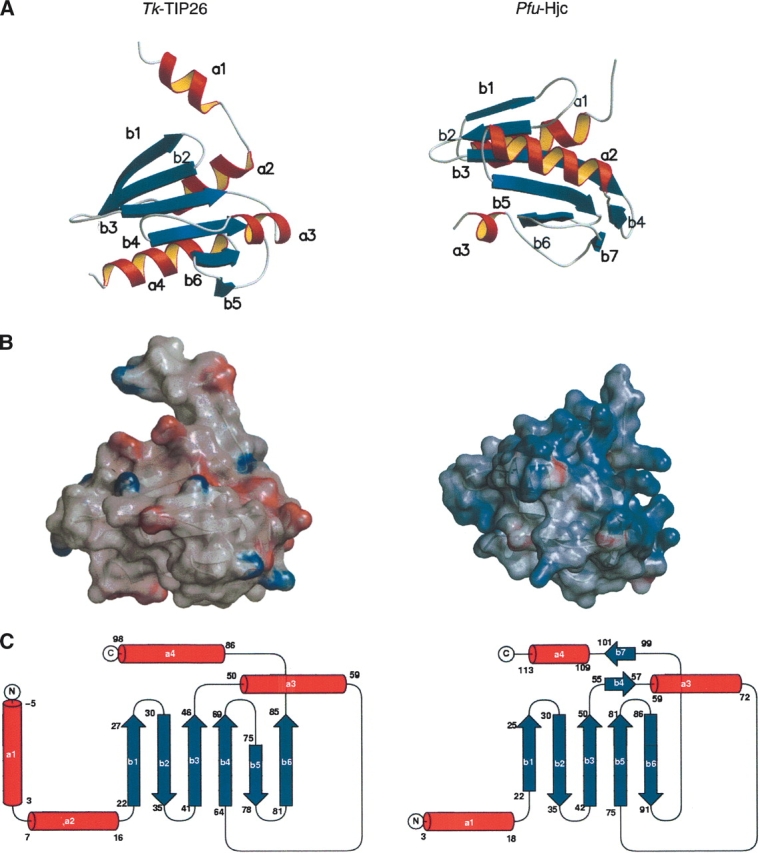
Structural comparison between the N-terminal domain of Tk-TIP26 (left image), and Pfu-Hjc (right image). (A) Ribbon diagrams. (B) Electrostatic surfaces generated by GRASP. Coloring is according to the local electrostatic potential range from −10 kT−1 (red) to +10 kT−1 (blue). (C) Topology diagrams.
Figure 4.
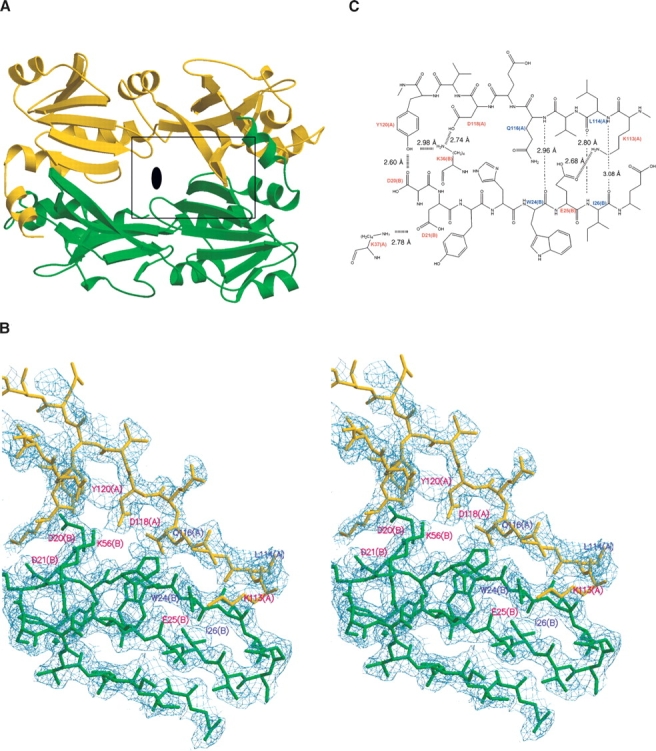
(A) Dimer structure of Tk-TIP26. The black ellipse in the middle of the dimer represents a crystallographic twofold axis. The area in the black box is magnified in B. (B) Close-up view of the dimer interface in stereo projection. The 2|Fo|−|Fc| electron density map was contoured at 1.0 σ. (C) The details of the dimer interactions. The residues involving the interaction between side residues are colored in red, and residues involving the interaction between main chains are colored in blue.
C-terminal domain
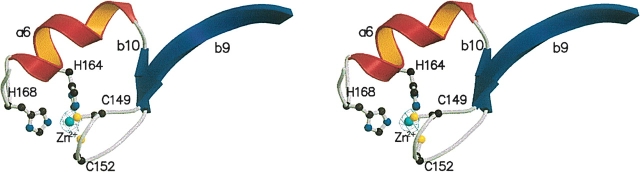
The C-terminal domain is comprised of three α-helices (a5–a7), six β-strands (b7–b12), and two 310-helices (A1 and A2). A zinc ion is found in the vicinity of the loop region between b9 and b10 (Fig. 3). The ion is coordinated by two cysteine (Cys149 and Cys152) and two histidine residues (His164 and His168), forming a tetrahedral coordination architecture. The secondary structures around these four residues are composed of β-strands b9 (from Tyr140 to Lys147), b10 (from Tyr154 to Asp156), and α-helix a6 (from Leu160 to Arg165), forming the classical Cys2His2-type zinc finger motif.
Figure 3.
Ribbon diagram of Zn-finger motif. Electron density map covered zinc ion is Simulated Annealed Omit Map contoured at 5.0 σ.
Most of the proteins containing the classical Cys2His2-type zinc finger motif are transcription factors that function by recognizing specific DNA sequences (Laity et al. 2001). The number of residues between the second cysteine and the first histidine is invariantly 12 (Lee et al. 1989; Pavletich and Pabo 1991). In the case of Tk-TIP26, the number of spacing residues between Cys-X2-Cys and His-X3-His is 11 rather than 12. Tk-TIP26 binds to DNA in the presence of the zinc ion, but the interaction between Tk-TIP26 and DNA is weak and seems to be non-specific (Matsuda et al. 1999). However, the topology of this domain is composed of “exactly” a β-hairpin and an α-helix.
The zinc finger motif in Tk-TIP26 is an unusual Cys2His2-type zinc finger and is not functional as a DNA-binding module, probably because Tk-TIP26 possesses only a single zinc finger motif. Zinc finger proteins usually contain multiple zinc finger motifs, which are required for specific interaction with DNA (Pavletich and Pabo 1991). In addition, this motif should not have the critical role for the stability of overall structure, because its mutant protein C149A/C152A could be over-expressed and purified as in wild-type protein (Matsuda et al. 1999).
This zinc finger motif is the first structure of Cys2His2-type zinc finger motif among archaeal proteins. However, the role of the motif in Tk-TIP26 is not unknown, and should be elucidiated.
Dimer structure of Tk-TIP26
Previously, it was reported that Tk-TIP26 exists in a dimeric form (Matsuda et al. 1999). The structure of dimeric Tk-TIP26 is shown in Figure 4A. Two molecules of Tk-TIP26 are related by the crystallographic twofold axis and the contact surface area is calculated to be 2260 Å2. The dimer interface is mainly composed of the β-strands b1 and b8 in the counterpart of the dimer (Fig. 4B,C). The contact involves hydrogen bonds between side residues of Lys37, Tyr120, Asp118, and Lys113 in one molecule, and Asp21, Asp20, Lys36, and Glu25 in the other, respectively. Hydrogen bonds are formed between main chains of Gln116 and Leu114 in one molecule, and Trp24 and Ile26 in the other, respectively (Fig. 4B,C).
Formation of a Tk-TIP26/Tk-TBP complex
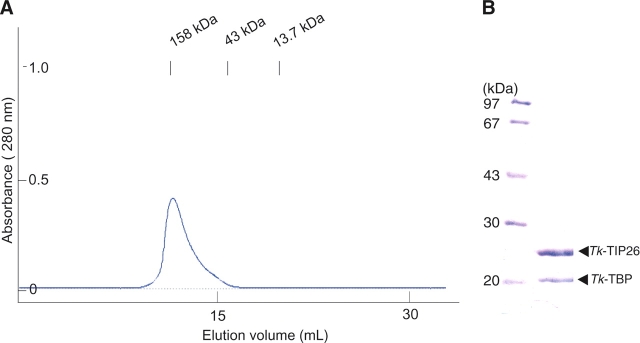
Purified Tk-TIP26 and Tk-TBP were mixed and incubated at 303 K for 30 min. After incubation, this mixture was applied to gel filtration column chromatography to purify. The chromatograph shows a single peak with molecular weight of ~158 kDa, showing that a Tk-TIP26/Tk-TBP complex is obtained (Fig. 5A). This value is in agreement with an estimate of molecular weight of this complex using dynamic light scattering (DLS) (data not shown). The peak fraction was applied to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resultant gel shows just two bands (Fig. 5B): one derived from Tk-TIP26 (molecular mass of ~27 kDa), and the other, from Tk-TBP (molecular mass of ~23 kDa). For the estimation of the stoichiometry of the interaction between Tk-TIP26 and Tk-TBP, several concentrations of Tk-TIP26, Tk-TBP, and the Tk-TIP26/Tk-TBP complex were loaded to SDS-PAGE on the same gel. The resultant gel was analyzed for band intensities in order to produce calibration curves of Tk-TIP26 and Tk-TBP (data not shown). The calibration curves indicate that in the lane of the Tk-TIP26/Tk-TBP complex, 0.126 pmol of Tk-TIP26 and 0.0675 pmol of Tk-TBP were present, i.e., the molar ratio of the interaction between Tk-TIP26 and Tk-TBP is ~2:1. Together with the estimation of molecular weight of the Tk-TIP26/Tk-TBP complex, Tk-TIP26 and Tk-TBP interact with stoichiometry of 4:2.
Figure 5.
Interaction between Tk-TIP26 and Tk-TBP. (A) The elution pattern of Tk-TIP26/Tk-TBP complex in gel filtration column chromatography. (B) SDS-PAGE of peak fractions of the chromatograph in A.
As mentioned above, Tk-TIP26 usually exists in dimeric form (Matsuda et al. 1999). On the other hand, biochemical and crystallographic studies reported that TBP usually exists in a dimeric form in solution and crystal structures (Nikolov and Burley 1994; Coleman et al. 1995; DeDecker et al. 1996). These results suggest that two dimer molecules of Tk-TIP26 interact with one dimer molecule of Tk-TBP.
Inhibition of the interaction between TBP and TATA-DNA by Tk-TIP26
In the absence of Tk-TFB, Tk-TIP26 prevents Tk-TBP from binding to TATA-DNA (Matsuda et al. 1999, 2001). We compared the structures and the electrostatic surfaces of Tk-TIP26 and TBP from Pyrococcus woesei (DeDecker et al. 1996) (PwTBP; PDB ID 1PCZ), which shares 83% of amino acid similarity with Tk-TBP. In the structure of dimeric Tk-TIP26, a negatively charged cleft is formed after formation of the dimer (Fig. 6A, left). On the other hand, dimeric TBP possesses specifically positively charged surfaces composed of Arg78, Lys82, Lys86, Lys88, Lys93, and Lys95 on both ends of the surface of the dimeric PwTBP (Fig. 6A, right). Subsequently, a model of the Tk-TIP26/TBP complex was predicted (Fig. 6B). In this model, dimeric TBP is sandwiched by two molecules of dimeric Tk-TIP26 from both ends of the surface, forming a 4:2 complex. Recent studies suggested that the equilibrium between TIP26/TBP complex and TBP/TATA-DNA complex inclines toward the fraction of the former complex in the absence of TFB(Matsuda et al. 2001). In addition, the 4:2 complex cannot access TATA-DNA in the absence of TFB (Matsuda et al. 1999), while dimeric TBP can interact with TATA-DNA by the dissociation to monomers (Coleman et al. 1995). These results suggest that the formation of the 4:2 complex would stabilize TBP in dimeric form, although it has not been elucidated how the interaction between the monomers of dimeric TBP would be affected by the formation of the 4:2 complex yet. This is the first structural basis of the inhibition mechanism where Tk-TIP26 prevents TBP from binding to TATA-DNA in the absence of TFB (Fig. 6C).
Figure 6.
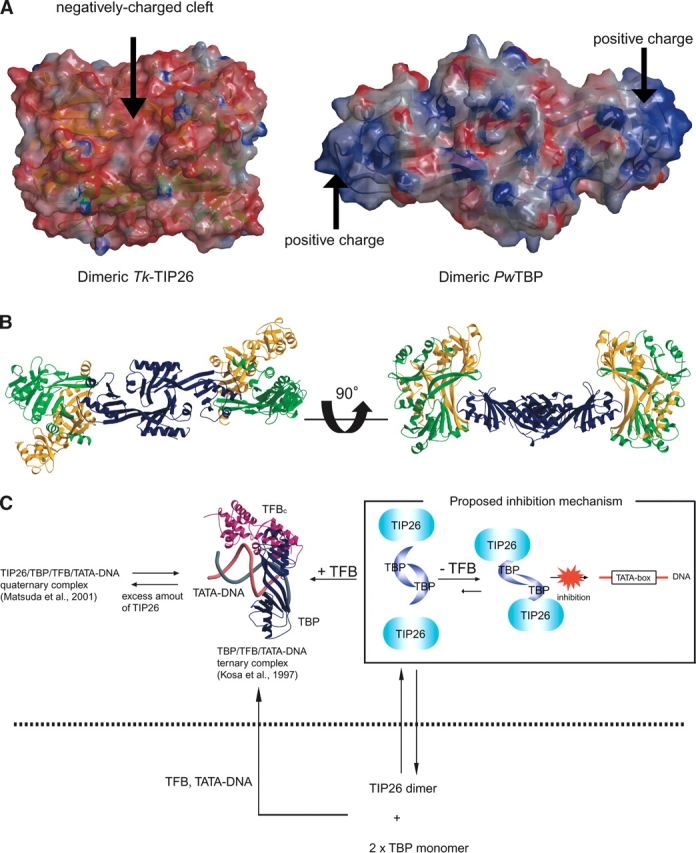
Proposed inhibition mechanism of the interaction between TBP and TATA-DNA by Tk-TIP26. (A) Electrostatic surfaces of dimeric Tk-TIP26 and dimeric PwTBP generated by GRASP (Nicholls et al. 1993). Coloring is according to the local electrostatic potential range from ~10 kT−1 (red) to +10 kT−1 (blue). (B) Model of Tk-TIP26/TBP complex. Two dimer molecules of Tk-TIP26 were colored as in Figure 4A. Dimeric PwTBP was colored with navy. The view of PwTBP in the left side was same as the one in Figure 6A. (C) Schematic draw of the inhibition mechanism in which Tk-TIP26 prevents TBP from binding to TATA-DNA. The ribbon model in the center of the figure is the structure of TBP/TFBc/TATA-DNA complex from P. woesei (Kosa et al. 1997).
On the other hand, it is also suggested that TFB shifts the equilibrium between TBP/TATA-DNA and TIP26/ TBP complexes so as to dramatically increase the fraction of TBP/TATA-DNA complex (Matsuda et al. 2001). In the presence of TFB and TATA-DNA, the fraction of the TBP/TATA-DNA complex would increase, via the dissociation of dimeric TBP to its monomers, forming a TFP/TBP/TATA-DNA ternary complex (Fig. 6C). In the presence of excess amounts of Tk-TIP26, the quaternary complex of TIP26/TFB/TBP/ TATA-DNA is formed, and finally, equilibrium between the ternary and the quaternary complex would be achieved (Fig. 6C).
Conclusions
The structural studies on Tk-TIP26 allowed us to build a putative model of the Tk-TIP26/TBP complex, providing insights in the molecular mechanisms of the interaction between TIP26 and TBP. These insights will give us a clue for the understanding of the interaction between TBP and other TBP-interacting proteins. Crystallographic studies on the Tk-TIP26/Tk-TBP complex and the Tk-TIP26/Tk-TBP/Tk-TFB/TATA-DNA quaternary complex are in progress to increase our understanding of the transcription regulation mechanism by TIP26.
Materials and methods
Data collection and structure determination
Overexpression, purification, crystallization, and X-ray data collection of native and selenomethionine (SeMet)-labeled Tk-TIP26 were described previously (Yamamoto et al. 2003).
The initial phases were estimated by the MAD method using the SeMet data. Two of the three selenium sites in an asymmetric unit were found using Bijvoet difference Patterson methods. These heavy atom parameters were refined, and phase calculation and density modification were processed using the program CNS (Brünger et al. 1998). The quality of the corresponding electron density map was excellent. An initial model was built using the program O (Jones et al. 1994) and TURBO-FRODO (Jones 1985). Diffraction data of SeMet remote wavelength between 40 and 2.8 Å resolution were included in simulated annealing refinement with bulk solvent correction using CNS (Brünger et al. 1998). Ten percent of the reflections were kept separate to monitor Rfree, and were not used in the refinement. When an R-factor reached ~28%, the protein model was used to refine the native data up to 2.3 Å . After several rounds of manual rebuilding and refinement, the individual B factors were refined, along with the addition of solvent molecules. A zinc ion was placed on the site with ~5 σ peak in an omit map. Zinc is the most probable metal because it shows a tetrahedral coordination geometry, and the temperature factor of the zinc ion modeled at the site had been refined to a value similar to those of the ligand atoms. The final refinement statistics are shown in Table 1. The current model contains regions of residues (−8–218) for a molecule in the asymmetric unit. No electron densities for the first 14 residues of the histidine-tag or C-terminal six residues were defined. These residues were eliminated from the refinement. Secondary structures were assigned with DSSP (Kabsch and Sander 1983).
Table 1.
Summary of crystallographic data of Tk-TIP26
| Se-Edge | Se-Peak | Se-Remote | Native | |
| Data collection | ||||
| X-ray source | SPring-8 | SPring-8 | SPring-8 | SPring-8 |
| BL40B2 | BL40B2 | BL40B2 | BL41XU | |
| Wavelength (Å) | 0.9795 | 0.9793 | 0.9873 | 0.9686 |
| Resolution range (Å)a | 40–2.8 (2.95–2.80) | 40–3.0 (3.16–3.00) | 40–2.8 (2.95–2.80) | 40–2.30 (2.41–2.30) |
| Unique reflections | 6407 | 5243 | 6246 | 11,625 |
| Multiplicity | 20.2 | 31.6 | 20.1 | 4.6 |
| Completeness (%)a | 100 (100) | 100 (100) | 100 (100) | 93.0 (94.1) |
| Rsyma,b | 7.1 (19.6) | 7.6 (16.7) | 7.3 (20.2) | 5.6 (20.9) |
| I/σa | 9.0 (3.5) | 8.4 (3.0) | 9.0 (3.5) | 10.2 (2.4) |
| Se atoms found (total) | 2 (3) | 2 (3) | 2 (3) | |
| Figure of meritc | 0.6151 | |||
| Refinement statistics | ||||
| Resolution range (Å) | 40–2.30 | |||
| Rcryst (%)d /Rfree (%)e | 19.1/24.1 | |||
| Water molecules | 110 | |||
| Metal ions | 1 | |||
| Glycerol molecules | 2 | |||
| RMSD Bond length (Å) | 0.005 | |||
| RMSD Bond angles (°) | 1.2 | |||
| Ramachandran (%) | ||||
| Favored | 90.1 | |||
| Allowed | 9.9 | |||
a Numbers in parentheses refer to the highest resolution shell.
bRsym = ∑|I − 〈I〉|/∑ 〈I〉, where I is the intensity of observation I and 〈I〉 is the mean intensity of the reflection.
c Figure of Merit =|F(hkl)best|/F(hkl).
dRcryst = ∑||Fo|−|Fc||/∑|Fc|, where Fo and Fc are the observed and the calculated structure factor amplitudes, respectively.
eRfree was calculated using a randomly selected 10% of the data set that was omitted through all stages of refinement.
Overexpression and purification of histidine-tagged Tk-TBP
The gene encoding Tk-TBP was cloned into pET28a (Novagen). The resultant plasmid was used to transform Escherichia coli BL21(DE3)pLysS (Novagen). Cells were grown in NZCYM medium at 303 K comprising 100 μg/mL kanamycin. When an OD660 reached 0.6, 1 mM IPTG was added to induce gene expression and cultivation was continued for an additional 12 h at 293 K. Cells were harvested by centrifugation at 15,000g for 20 min. Cells were suspended in 50 mM NaH2PO4 (pH 8.0) containing 300 mM NaCl, 10 mM imidazole, and 1 mM 2-mercaptoethanol (2-Me); disrupted by sonication; and centrifuged at 15,000g for 30 min. The supernatant was applied to a HiTrap Chelating HP column (GE Healthcare), equilibrated with the same buffer as that for sonication lysis. The sample was eluted along a 50–300 mM imidazole gradient. The active fractions were combined and concentrated with Centriprep YM-10 (Millipore) and applied onto a HiLoad 26/60 Super-dex 75 prep-grade column (GE Healthcare), equilibrated with 50 mM Tris-HCl (pH 6.8), 150 mM NaCl, and 1 mM DTT. The fractions containing Tk-TBP were pooled and concentrated with Centriprep YM-10 (Millipore) to 8.0 mg/mL. The purified Tk-TBP was frozen in liquid nitrogen and stored at 193 K.
Preparation of Tk-TIP26/Tk-TBP complex and SDS-PAGE analysis
In order to prepare the Tk-TIP26/Tk-TBP complex, equal volumes of purified Tk-TIP26 (0.6 mM) and Tk-TBP (0.3 mM) were mixed and incubated at 303 K for 30 min. The mixed solution was applied to the gel filtration column Super-dex 200 10/30 HR column (GE Healthcare), equilibrated with 50 mM Tris-HCl (pH 6.8), 150 mM NaCl, and 1 mM DTT. The active fractions (shown in Fig. 4A) were pooled, and used in the SDS-PAGE analysis. The stoichiometry between Tk-TIP26 and Tk-TBP was estimated by SDS-PAGE analysis under the following conditions: Several concentrations of purified Tk-TIP26 (8.81, 3.26, 2.29, and 2.21 μM), Tk-TBP (7.70, 3.01, 2.57, and 1.92 μM), and purified Tk-TIP26/Tk-TBP complex were applied to 12.5% polyacrylamide gel, PAGEL (ATTO), and proceeded to SDS-PAGE analysis at 20 mA constant current. The resultant gel was stained with Coomassie brilliant blue (CBB) and intensities of the bands in the gel were estimated by the program Lane & Spot Analyzer (ATTO). Using these intensities, the calibration curves of Tk-TIP26 and Tk-TBP were prepared, and these curves were used to estimate the concentrations of Tk-TIP26 and Tk-TBP in the complex.
Preparation of figures
Figures 1A, 2A and B, 3, 4A and B, and 6A and B were prepared with the programs MOLSCRIPT (Kraulis 1991), RASTER3D (Merritt and Murphy 1994), and GRASP (Nicholls et al. 1993). Figure 1B was prepared with ALSCRIPT (Barton 1993); Figure 2C, with TopDraw (Collaborative Computing Project, Number 4 1994; Bond 2003).
Protein Data Bank accession code
Refined coordinates and structure factor have been deposited in the RCSB Protein Data Bank under the accession code 2CZR.
Acknowledgments
We are grateful to K. Miura, M. Kawamoto, K. Hasegawa, N. Shimizu, and H. Sakai for their kind help in the collection of the X-ray diffraction data at BL38B1, BL40B2, and BL41XU in SPring-8; and to Prof. A. Nakagawa and Dr. E. Yamashita, Institute for Protein Research, Osaka University, for their help in fundamental data collection at BL44XU in SPring-8. The synchrotron radiation experiments were performed at the SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (2001A0336-CL-np and 2004B0538-NL1-np) and of the Institute for Protein Research, Osaka University (C00A44XU-73B-N). We are grateful to S. Hashima, ATTO Co. Ltd., for his kind help in the data analyses of SDS-PAGE. This study was partially supported by Grant-in-Aid from the Ministry of Education, Science, Sports and Culture, and also supported by the National Project on Protein Structural and Functional Analyses. T.Y. expresses his special thanks to the center of excellence (21COE) program “Creation of Integrated EcoChemistry of Osaka University.”
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051788906.
References
- Barton, G.J. 1993. Alscript: A tool to format multiple sequence alignments. Protein Eng. 6: 37–40. [DOI] [PubMed] [Google Scholar]
- Bell, S.D. and Jackson, S.P. 2001. Mechanism and regulation of transcription in archaea. Curr. Opin. Microbiol. 4: 208–213. [DOI] [PubMed] [Google Scholar]
- Bell, S.D., Brinkman, A.B., van der Oost, J., and Jackson, S.P. 2001a. The archaeal TFIIEα homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2: 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.D., Magill, C.P., and Jackson, S.P. 2001b. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 29: 392–395. [DOI] [PubMed] [Google Scholar]
- Bond, C.S. 2003. TopDraw: A sketchpad for protein structure topology cartoons. Bioinformatics 19: 311–312. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., et al. 1998. Crytallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 904–925. [DOI] [PubMed] [Google Scholar]
- Coleman, R.A., Taggart, A.K., Benjamin, L.R., and Pugh, B.F. 1995. Dimerization of the TATA binding protein. J. Biol. Chem. 270: 13842–13849. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project, Number 4 (CCP4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- DeDecker, B.S., O’Brien, R., Fleming, P.J., Geiger, J.H., Jackson, S.P., and Sigler, P.B. 1996. The crystal structure of a hyperthermophilic archaeal TATA-box binding protein. J. Mol. Biol. 264: 1072–1084. [DOI] [PubMed] [Google Scholar]
- Fujiwara, S., Takagi, M., and Imanaka, T. 1998. Archaeon Pyrococcus kodakaraensis KOD1: Application and evolution. Biotechnol. Annu. Rev. 4: 259–284. [DOI] [PubMed] [Google Scholar]
- Hanzelka, B.L., Darcy, T.J., and Reeve, J.N. 2001. TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEα. J. Bacteriol. 183: 1813–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20: 478–480. [DOI] [PubMed] [Google Scholar]
- Jones, T.A. 1985. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 115: 157–171. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1994. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., Makino, Y., Yoshida, T., Kishimoto, T., Koga, A., Yamamoto, K., Yamamoto, M., Moncollin, V., Egly, J.M., Muramatsu, M., et al. 1997. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235: 64–68. [DOI] [PubMed] [Google Scholar]
- Kosa, P.F., Ghosh, G., DeDecker, B.S., and Sigler, P.B. 1997. The 2.1-Å crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc. Natl. Acad. Sci. 94: 6042–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. Molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- Laity, J.H., Lee, B.M., and Wright, P.E. 2001. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11: 39–46. [DOI] [PubMed] [Google Scholar]
- Lee, M.S., Gippert, G.P., Soman, K.V., Case, D.A., and Wright, P.E. 1989. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245: 635–637. [DOI] [PubMed] [Google Scholar]
- Makino, Y., Yoshida, T., Yogosawa, S., and Tamura, T. 1996. Detection of TBP-interacting proteins (TIPs) and demonstration of a novel complex containing TBP and ATPases. Tanpakushitsu Kakusan Koso 41: 1170–1177. [PubMed] [Google Scholar]
- Makino, Y., Yogosawa, S., Kayukawa, K., Coin, F., Egly, J.M., Wang, Z., Roeder, R.G., Yamamoto, K., Muramatsu, M., and Tamura, T. 1999. TATA-binding protein-interacting protein 120, TIP120, stimulates three classes of eukaryotic transcription via a unique mechanism. Mol. Cell. Biol. 19: 7951–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, T., Morikawa, M., Haruki, M., Higashibata, H., Imanaka, T., and Kanaya, S. 1999. Isolation of TBP-interacting protein (TIP) from a hyperthermophilic archaeon that inhibits the binding of TBP to TATA-DNA. FEBS Lett. 457: 38–42. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., Fujikawa, M., Haruki, M., Tang, X.F., Ezaki, S., Imanaka, T., Morikawa, M., and Kanaya, S. 2001. Interaction of TIP26 from a hyperthermophilic archaeon with TFB/TBP/DNA ternary complex. Extremophiles 5: 177–182. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E. 1994. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 50: 869–873. [DOI] [PubMed] [Google Scholar]
- Morikawa, M., Izawa, Y., Rashid, N., Hoaki, T., and Imanaka, T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60: 4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, A., Bharadwaj, R., and Honig, B. 1993. GRASP: Graphical representation and analysis of surface properties. Biophys. J. 64: A166. [Google Scholar]
- Nikolov, D.B. and Burley, S.K. 1994. 2.1 Å resolution refined structure of a TATA box-binding protein (TBP). Nat. Struct. Biol. 1: 621–637. [DOI] [PubMed] [Google Scholar]
- Nishino, T., Komori, K., Ishino, Y., and Morikawa, K. 2001a. Dissection of the regional roles of the archaeal Holliday junction resolvase Hjc by structural and mutational analyses. J. Biol. Chem. 276: 35735–35740. [DOI] [PubMed] [Google Scholar]
- Nishino, T., Komori, K., Tsuchiya, D., Ishino, Y., and Morikawa, K. 2001b. Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition. Structure (Camb.) 9: 197–204. [DOI] [PubMed] [Google Scholar]
- Pavletich, N.P. and Pabo, C.O. 1991. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 Å . Science 252: 809–817. [DOI] [PubMed] [Google Scholar]
- Qureshi, S.A., Bell, S.D., and Jackson, S.P. 1997. Factor requirements for transcription in the Archaeon Sulfolobus shibatae. EMBO J. 16: 2927–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, R.G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21: 327–335. [PubMed] [Google Scholar]
- Soppa, J. 1999. Transcription initiation in Archaea: Facts, factors and future aspects. Mol. Microbiol. 31: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Kunishima, N., Mayanagi, K., Ohnishi, T., Nishino, T., Iwasaki, H., Shinagawa, H., and Morikawa, K. 2001. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. 98: 1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Matsuda, T., Sakamoto, N., Matsumura, H., Inoue, T., Morikawa, M., Kanaya, S., and Kai, Y. 2003. Crystallization and preliminary X-ray analysis of TBP-interacting protein from the hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1. Acta Crystallogr. D Biol. Crystallogr. 59: 372–374. [DOI] [PubMed] [Google Scholar]
- Yogosawa, S., Makino, Y., Yoshida, T., Kishimoto, T., Muramatsu, M., and Tamura, T. 1996. Molecular cloning of a novel 120-kDa TBP-interacting protein. Biochem. Biophys. Res. Commun. 229: 612–617. [DOI] [PubMed] [Google Scholar]