Abstract
A good approach to test our current knowledge on formation of protein β-sheets is de novo protein design. To obtain a three-stranded β-sheet mini-protein, we have built a series of chimeric peptides by taking as a template a previously designed β-sheet peptide, Betanova-LLM, and incorporating N- and/or C-terminal extensions taken from WW domains, the smallest natural β-sheet domain that is stable in absence of disulfide bridges. Some Betanova-LLM strand residues were also substituted by those of a prototype WW domain. The designed peptides were cloned and expressed in Escherichia coli. The ability of the purified peptides to adopt β-sheet structures was examined by circular dichroism (CD). Then, the peptide showing the highest β-sheet population according to the CD spectra, named 3SBWW-2, was further investigated by 1H and 13C NMR. Based on NOE and chemical shift data, peptide 3SBWW-2 adopts a well defined three-stranded antiparallel β-sheet structure with a disordered C-terminal tail. To discern between the contributions to β-sheet stability of strand residues and the C-terminal extension, the structural behavior of a control peptide with the same strand residues as 3SBWW-2 but lacking the C-terminal extension, named Betanova-LYYL, was also investigated. β-Sheet stability in these two peptides, in the parent Betanova-LLM and in WW-P, a prototype WW domain, decreased in the order WW-P > 3SBWW-2 > Betanova-LYYL > Betanova-LLM. Conclusions about the contributions to β-sheet stability were drawn by comparing structural properties of these four peptides.
Keywords: antiparallel β-sheet, NMR, peptide design, peptide structure, WW domain
Now that the genomes of many organisms, from very small bacteria to mammals, including the human, have been sequenced, a new goal is to understand the biological function of the coded proteins as well as the interactions among them. A first step toward this goal is to determine the structures of all coded proteins. However, crystallographic and NMR experimental determination of protein structures are slow processes while prediction of protein structure from the amino acid sequence fails in the absence of similar proteins of known three-dimensional structures. Understanding the principles underlying stability and folding of protein structures will greatly contribute to enhance reliability of protein structure prediction. Among protein secondary structures, β-sheets are more difficult to predict than α-helices. A good approach to test our current knowledge on formation of protein β-sheets is de novo protein design. Hence, the design of small antiparallel β-sheet motifs is a subject of great current interest.
The WW domain constitutes one of the smaller globular domains that are folded in the absence of metal ions or disulfide bridges (Sudol 1996). This small domain, about 40 amino acids long, consists of a three-stranded antiparallel β-sheet in which the side chains in one of its two faces interact with residues located at the N- and C-terminal tails to form a small hydrophobic core (Fig. 1A). Some WW domains have been taken as models to investigate kinetics and thermodynamics of β-sheet folding (Koepf et al. 1999; Crane et al. 2000; Jager et al. 2001; Jiang et al. 2001; Deechongkit and Kelly 2002; Deechongkit et al. 2004, 2006). Sequence alignment within this domain family has shown the existence of several conserved residues, which seem to be essential for structure stability. Based on this information, Oschkinat and colleagues designed a consensus WW domain sequence, denominated WW prototype, which turned out to have similar structure and stability to natural WW domains (Macías et al. 2000).
Figure 1.
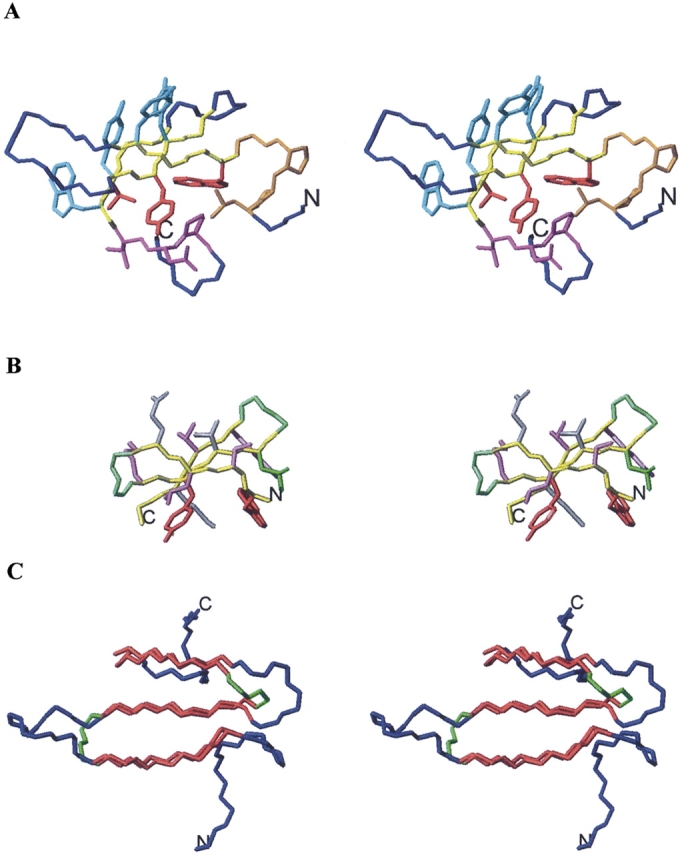
(A) Stereoscopic view of WW-P structure. Backbone atoms for β-strand residues aligned to Betanova, Betanova-LLM, and taken for the design of the chimeric peptides (Table 1) are shown in yellow. Backbone and side chain atoms for N-terminal and C-terminal residues incorporated in some of the chimeric peptides are shown in orange and magenta, respectively. Side chain atoms for β-strand residues common to WW-P and Betanova are indicated in red. Side chain atoms for β-strand residues incorporated in some of the designed peptides are shown in cyan. The remaining WW-P backbone atoms are colored in blue. (B) Stereoscopic view of Betanova-LLM structure. Side chain atoms for β-strand residues incorporated in all designed peptides are shown in purple. Side chain atoms for β-strand residues common to WW-P and Betanova are indicated in red. All displayed backbone atoms are included in the chimeric peptides. β-strand backbone atoms are shown in yellow and β-turn backbone atoms in green. (C) Stereoscopic view of WW-P (in blue) and Betanova-LLM (in green) backbone atoms superposed over β-strand residues (in red). N and C termini are labeled.
Several research groups have designed three-stranded antiparallel β-sheet miniproteins that are shorter than the WW domain and lack the relatively long N- and C-terminal tails present in WW domains (Das et al. 1998; Kortemme et al. 1998; Schenck and Gellman 1998; Sharman and Searle 1998; de Alba et al. 1999; Griffiths-Jones and Searle 2000; Santiveri et al. 2003). All of these designed peptides while adopting the target antiparallel β-sheet lacked the stability of natural WW domains. Incorporation of a non-natural dPro amino acid at the turn regions was shown to increase β-sheet stability in some of these designed peptides (Schenck and Gellman 1998; Santiveri et al. 2004). β-sheet was stabilized in the designed peptide named Betanova by substituting up to three residues at the strands (Table 1; López de la Paz et al. 2001). Residues were selected by using a protein design algorithm, PERLA (Fisinger et al. 2001). Betanova-LLM (Table 1) is one of those redesigned peptides exhibiting high β-sheet population (López de la Paz et al. 2001).
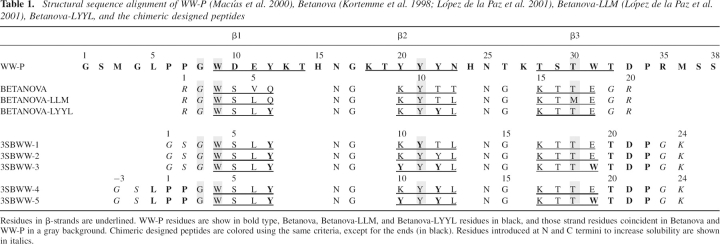
Table 1.
Structural sequence alignment of WW-P (Macías et al. 2000), Betanova (Kortemme et al. 1998; López de la Paz et al. 2001), Betanova-LLM (López de la Paz et al. 2001), Betanova-LYYL, and the chimeric designed peptides
Here, we explore the possibility of obtaining stable three-stranded antiparallel β-sheets by building hybrid peptides, that is, peptides whose sequences combine characteristics of WW domains and of a de novo designed peptide. In particular, we intended to enhance β-sheet stability in Betanova-LLM (López de la Paz et al. 2001). To that end, we built a series of Betanova-LLM/WW prototype chimeric peptides that contain the Betanova β-sheet backbone and incorporate extensions at either the C terminus or both the N and C termini taken from the WW prototype domain. Some strand residues are also taken from the WW prototype domains. Turns in the chimeric peptides come from Betanova-LLM. Strands are shorter in the chimeric peptides than in WW prototype, but formation of a small hydrophobic core, as the one present in WW domains, should be possible. Five chimeric peptides (Table 1) were cloned, expressed, and purified. β-Sheet formation in all chimeric peptides was examined by circular dichroism. Structure of the chimeric peptide exhibiting the circular dichroism (CD) spectra indicative of the highest β-sheet population, 3SBWW-2, was further investigated by NMR. To distinguish between the contributions to stability of the strand residues and the C-terminal extension, we designed a new Betanova-derived peptide with strand residues identical to 3SBWW-2. This control peptide, Betanova-LYYL (Table 1) was also studied by CD and NMR. Structural features of the β-sheet adopted by peptide 3SBWW-2 as well as its stability will be analyzed in comparison with those of Betanova-LLM and WW prototype (WW-P), as well as with that of the new Betanova-LYYL.
Results
Peptide design
We followed a double strategy: First we increased progressively the sequence identity in the three-stranded β-sheet motif between Betanova-LLM and WW-P, while trying to keep changes to a minimum. Second, we added C- and/or N-terminal WW-P sequences, which have been shown to be important for structure and/or stability, to Betanova. In this way, a series of chimeric peptides, named 3SBWW-X, were generated (Table 1).
First we consider the changes in the β-strands. The W and Y strand residues that are identical in WW-P, in Betanova, and in Betanova-LLM, as well as the T strand residue that is identical in WW-P and in Betanova, are maintained in all the designed peptides (Table 1; Fig. 1). Residues Q6, T11, K9, and E18 of Betanova-LLM were successively changed into those at their equivalent positions in WW-P, i.e., Y12, Y22, Y20, and W31, respectively, to yield peptides 3SBWW-1, 3SBWW-2, and 3SBWW-3 (Table 1). Those four residues are part of a solvent-exposed hydrophobic cluster in WW-P (Fig. 1). Residues Y22 and W31 are critical for the stability of WW domains (Macías et al. 2000; Jager et al. 2001). In homology with the C-terminal tail present in WW-P, a T21–D22–P23 C-terminal extension (Fig. 1A, residues in magenta) was also incorporated into these three designed peptides 3SBWW-1, 3SBWW-2, and 3SBWW-3. P23 corresponds to P34 in the WW domain. As described, this residue is part of a delocalized hydrophobic core, together with W9 and Y21, both of which have an equivalent residue in Betanova-LLM (see Fig. 1A,B). The P33–W9 contact joins the first and third strands of the β-sheet, and efforts to truncate the C terminus of WW domains beyond the invariant P34 residue have proved unsuccessful, resulting in an ensemble of non-native conformers with a high propensity to aggregate (Macías et al. 2000; Nguyen et al. 2003). In addition, point mutations of P34 have yielded unfolded conformations (Jager et al. 2001). G23 and K24 were maintained at the C terminus to reduce possible intermolecular aggregation of mutants.
The N terminus of WW domains contains two prolines (P6-P7) that are thought to restrict the conformational freedom of the system (Macías et al. 2000). In addition, P7 has been shown to be involved in a H-bonded network in some WW domains (Jager et al. 2001). A Leu residue precedes the prolines (L5 in Fig. 1A) and usually forms part of the hydrophobic cluster composed of the side chains of W9, Y21, and P34 (Fig. 1A). To evaluate if this N-terminal sequence can also contribute to the stability/structure of the designed peptides, we added it to 3SBWW-2 and 3SBWW-3 to obtain 3SBWW-4 and 3SBWW-5, respectively (Table 1). G1 and S2 were maintained at the N terminus to reduce possible intermolecular aggregation.
CD study identifies peptide 3SBWW-2 as the most successful design
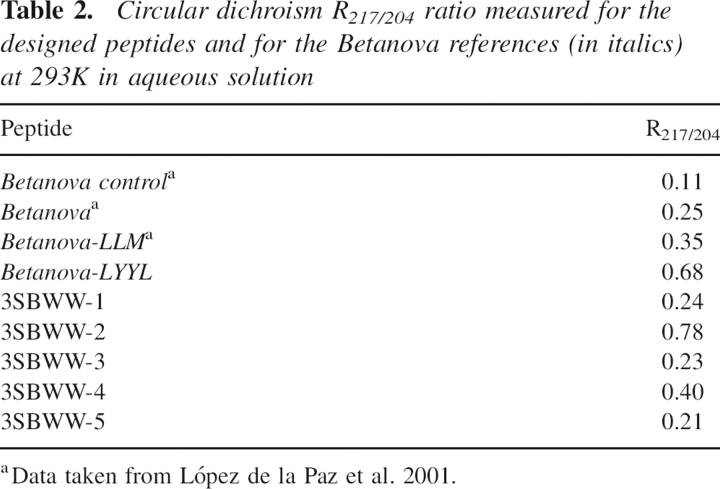
3SBWW peptides were expressed in Escherichia coli as GST fusions and purified as described in Materials and Methods. All mutations produced monomeric soluble protein as analyzed by analytical ultracentrifugation (data not shown) and in the case of 3SBWW-2 by NMR (see Materials and Methods). The secondary structure of mutants in aqueous solution was characterized by far-UV CD spectroscopy (Fig. 2). Three bands were considered: 204 nm (random coil contribution to the CD spectra), 217 nm (β-sheet contribution), and 230 nm (aromatic contribution). To compare the β-sheet content of the peptides, we calculated the ratio between the ellipticity at 217 nm and 204 nm (see ratio R217/204 in Table 2). This ratio is concentration-independent and in principle is related to the β-sheet content in solution (López de la Paz et al. 2001). As references, we used an unstructured variant of Betanova (Betanova Control in Table 2), Betanova and Betanova-LLM (one of the most stable Betanova sequences; López de la Paz et al. 2001). The CD data of the three peptides in aqueous solution show that peptide 3SBWW-2 is the most structured, exhibiting a clear local minimum 217 nm. In fact the R217/204 ratio in 3SBWW-2 was significantly higher than that in the two structured Betanova references (Table 2). This could reflect a higher β-sheet content of 3SBWW-2 relative to the reference molecules. Regarding 3SBWW-1 and 3SBWW-3, the other two variants with a C-terminal extension, comparison with the Betanova references indicates that both peptides show a β-sheet population very similar to Betanova but lower than that of the most stable Betanova-LLM (Table 1).
Figure 2.
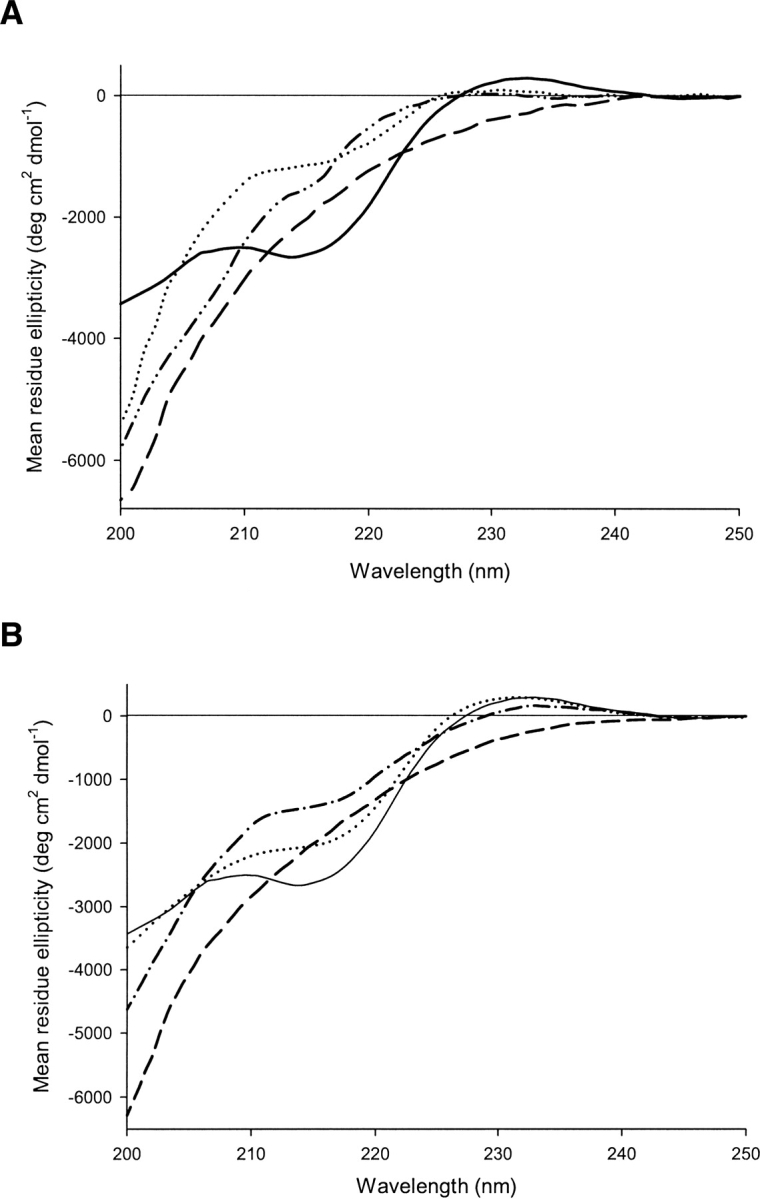
Far-UV circular dichroism spectra of the designed peptides as well as those of Betanova-LLM and Betanova-LYYL in 50 mM sodium phosphate at pH 7.0 and 293 K. Mean residue ellipticity is plotted against wavelength. (A) CD spectra of 3SBWW-1 (dashed-dotted line), 3SBWW-2 (solid line), 3SBWW-3 (dashed line), and Betanova-LLM as reference (dotted line) are shown. (B) CD spectra of 3SBWW-2 (solid line), 3SBWW-4 (dashed-dotted line), 3SBWW-5 (dashed line), and Betanova-LYYL (dotted line) are shown.
Table 2.
Circular dichroism R217/204 ratio measured for the designed peptides and for the Betanova references (in italics) at 293K in aqueous solution
Analysis of the CD spectra of 3SBWW-4 and 3SBWW-5 (Fig. 2B) indicates that the addition of the N-terminal extension to either 3SBWW-2 or 3SBWW-3 does not result in an increase in their respective β-sheet content. The spectrum of 3SBWW-5 corresponds to that of a mostly unfolded peptide. In the spectra of 3SBWW-4 the characteristic minimum at 217 nm is detected and, whereas it appears to be more folded than the Betanova-LLM reference variants, it is less structured than 3SBWW-2 (Fig. 2B; Table 2).
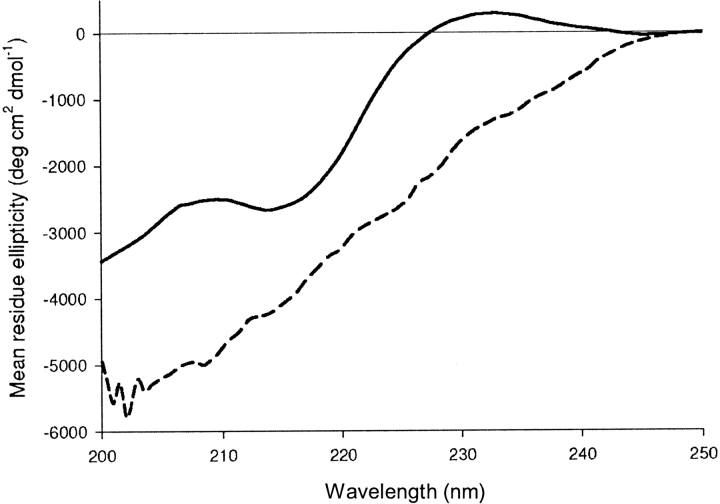
The aromatic signature at 230 nm present in 3SBWW-2 mutant is also a characteristic of WW domains and has been attributed to an ordered arrangement of the aromatic residues in the structure, since it is lost upon protein destabilization or unfolding (Macías et al. 2000). This band also disappears from 3SBWW-2 CD spectrum upon heating (see Fig. 3). Based on these results, we decided to further characterize this variant by NMR.
Figure 3.
Far-UV circular dichroism spectra of 3SBWW-2 in 50 mM sodium phosphate (pH 7.0) at 293 K (solid line) and 368 K (dashed line). Mean residue ellipticity is plotted against wavelength.
Peptide 3SBWW-2 adopts a three-stranded antiparallel β-sheet
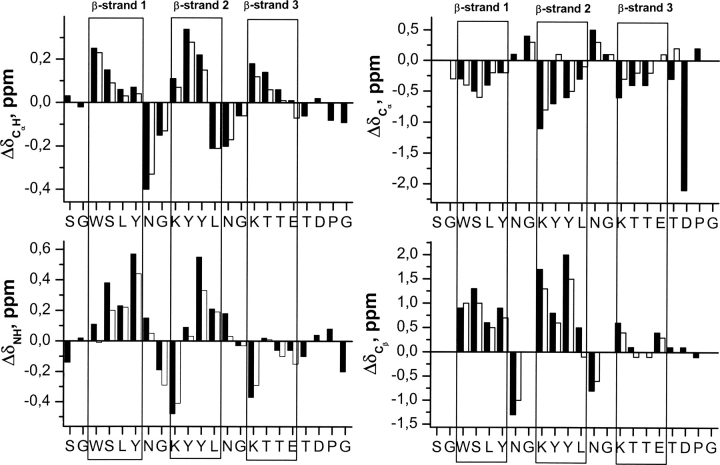
The profiles of ΔδCαH, ΔδNH, ΔδCα, and ΔδCβ conformational shift values (Δδ = δobserved − δrandom coil, ppm; Fig. 4) exhibited by peptide 3SBWW-2 in aqueous solution are consistent with those characteristic of the target three β-stranded antiparallel β-sheet (Spera and Bax 1991; Wishart et al. 1991; Santiveri et al. 2001; Fesinmeyer et al. 2005). The three stretches of residues with negative ΔδCα values and positive ΔδCαH, ΔδNH, and ΔδCβ values (residues 4–7, 10–13, and 16–19) correspond to the three β-strands. The negative ΔδCαH shift observed for residue L13, instead of the positive value expected for a strand residue, is probably due to the anisotropic effect of the aromatic ring currents of residues Y12, adjacent to L13, and W4, that faces L13 in the β-sheet (Fig. 5). Apart from this, the only deviations from the characteristic β-sheet pattern are observed for residues at the end of the third strand (T18 and E19; Fig. 4). Two regions (residues 8, 9 and 14, 15) that displayed the pattern of ΔδCα and ΔδCβ values characteristic of type I′ β-turns separate the β-strands (Santiveri et al. 2001). Residues K9 and K15 show the negative and large in absolute value ΔδNH shifts characteristic of the first strand residue after the β-turn (Fesinmeyer et al. 2005). The ΔδCα value observed for residue D21 that is negative and large in absolute value is that characteristic of Pro-preceding residues (Wishart et al. 1995; Schwarzinger et al. 2001).
Figure 4.
Histograms of ΔδCαH (ΔδCαH = δCαH observed − δCαH random coil, ppm), ΔδNH (ΔδNH = δNH observed − δNH random coil, ppm), ΔδCα (ΔδCα = δCα observed − δCα random coil, ppm), and ΔδCβ (ΔδCβ = δCβ observed − δCβ random coil, ppm) values as a function of sequence for peptides 3SBWW-2 (filled bars) and Betanova-LYYL (open bars) at pH 5.5 and 10°C. Random coil values for the 1H chemical shifts of CαH protons and for the 13C chemical shifts of Cα and Cβ carbons were taken from Wishart et al. (1995). ΔδNH values were obtained by using the CSDb program (Fesinmeyer et al. 2005). N and C termini residues that may be affected by charge-end effects are not plotted. β-Strand regions are boxed.
Figure 5.
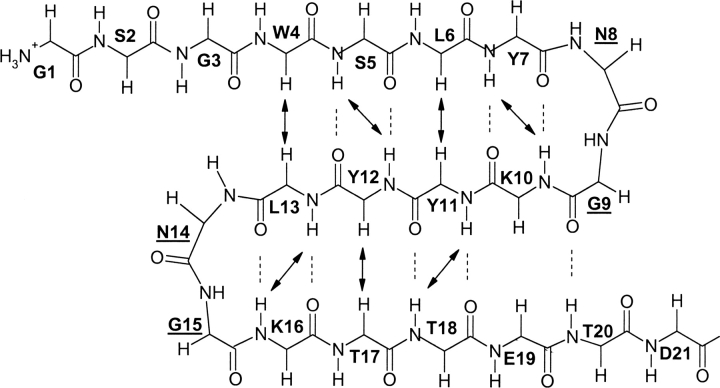
Schematic representation of the peptide backbone conformation of the target three-stranded antiparallel β-sheet. Dotted lines indicate the β-sheet hydrogen bonds; arrows indicate the observed long-range NOEs involving backbone protons.
The most definitive evidence for peptide 3SBWW-2 forming the target β-sheet comes from the set of observed NOEs. Most of the NOEs involving backbone protons expected for the target β-sheet, as well as a large number of NOEs between side chain protons of residues at adjacent strands on the same β-sheet face, were observed in the NOESY spectra of peptide 3SBWW-2 (Figs. 5, 6). Moreover, the presence of two-long-range NOEs between side chain protons of W4 and T18 residues located in β-sheet 1 and 3, respectively, demonstrates the formation of the desired three-stranded β-sheet (Figs. 5, 6).
Figure 6.
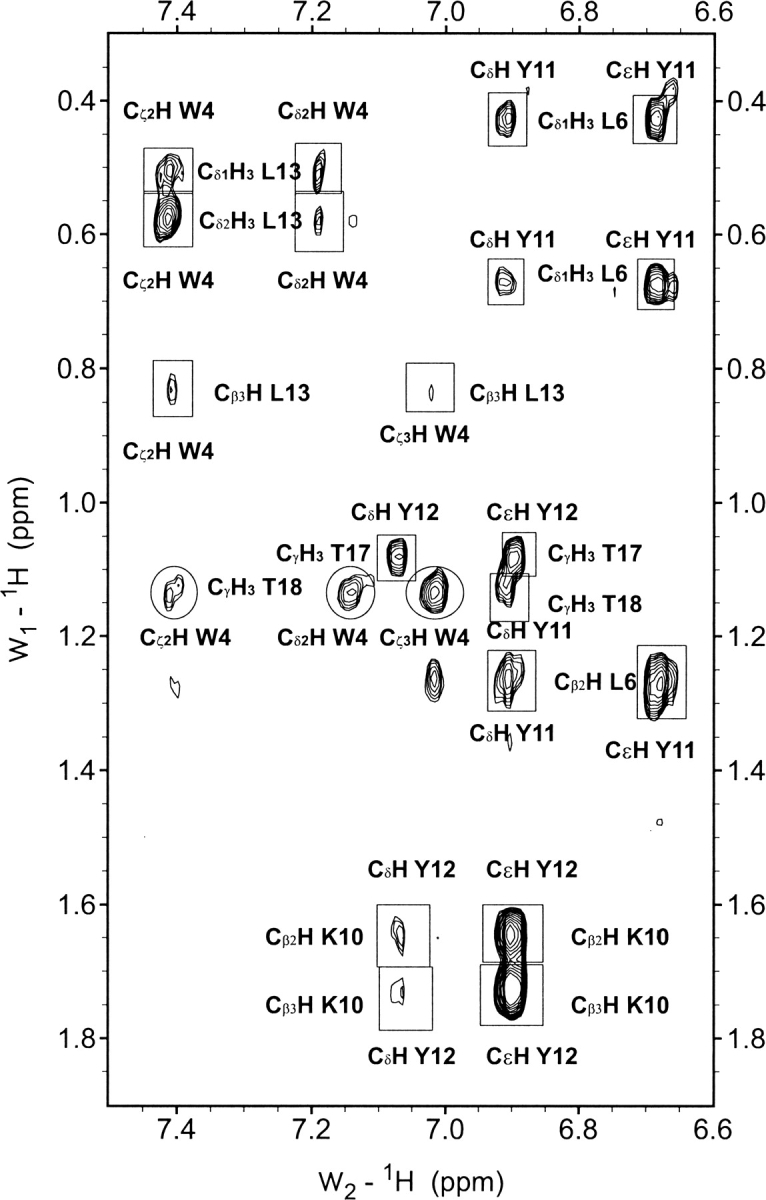
Selected region of the NOESY spectra of peptide 3SBWW-2 in H2O/D2O 9:1 (v/v) at pH 5.5 and 10°C. Non-sequential NOE cross-peaks are boxed and labeled, except for those between side chain protons of a residue located in the first and a residue in the third strand, W4 and T18, which are circled.
To further characterize the β-sheet adopted by peptide 3SBWW-2, we calculated its structure by using distance restraints (100 in total; 17 of them are sequential, 39 of medium range, and 44 of long range) derived from the intensities of the observed NOEs and dihedral ϕ and ψ angle constraints derived from CαH, 13Cα, and 13Cβ chemical shifts (28 in total). Intraresidual and sequential NOEs, apart from the CαHi–NHi+1 NOEs involving strand residues and NHi–NHi+1 NOEs involving turn residues, were excluded due to the contribution of random coil conformations to their intensities. This contribution is negligible in the case of the sequential strand NOEs considered because of the short CαHi–NHi+1 distance in the β-sheet conformation, 2.2 Å, and the short NHi–NHi+1 distances in turns, 2.4–2.6 Å. The calculated structures have a large well defined middle region and disordered N- and C-terminal residues (Fig. 7; pairwise RMSD values for residues 3–18 are 0.6 ± 0.2 Å for backbone atoms and 1.4 ± 0.3 Å for all heavy atoms). The side chains of most residues in the 3–18 region can be considered as well defined since their χ1 angles are in a range <30° (W4, S5, L6, Y7, K10, Y11, Y12, N14).
Figure 7.
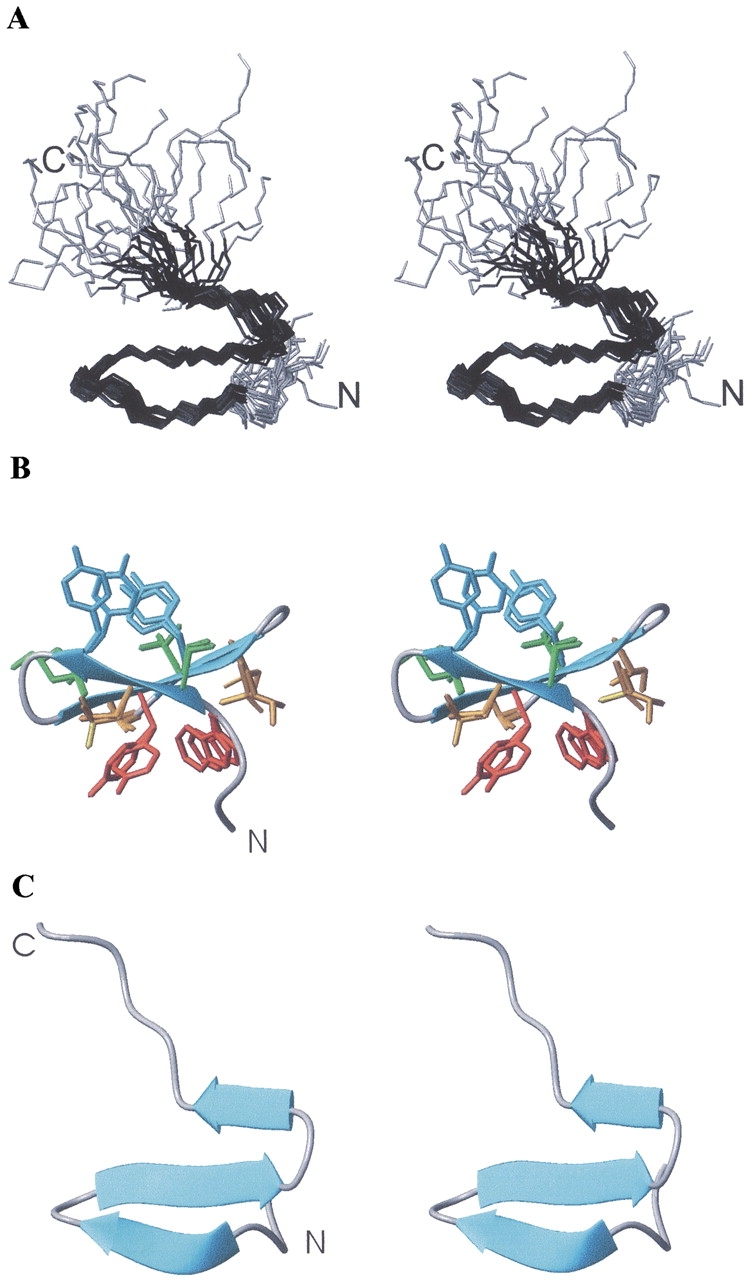
Stereoscopic views of the β-sheet structure calculated for peptide 3SBWW-2 in aqueous solution. (A) Superposition of backbone atoms of the best 30 calculated structures in an orientation similar to that of WW domain of Figure 1C. Residues 1–3 and 20–24 are in gray. N and C termini are labeled. (B) Backbone atoms are shown as a ribbon and in an orientation similar to that of Figure 1A,B. Superposition of the heavy side chain atoms of the best five calculated structures are also shown. Those of Y7 and Y12 are colored in cyan and all other side chains in the upper face are in green, those of W4 and Y11 are in red and all other in the bottom face are in orange. (C) Ribbon representation in the same orientation as panel A.
3SBWW-2 β-sheet structure lacks the Trp–Pro interaction present in WW domains
In contrast to what was expected according to design criteria where Pro 22 should interact with Trp 4 and Tyr 11 (Fig. 1), the Pro residue in the β-sheet structure calculated for peptide 3SBWW-2 is located at the disordered C-terminal tail and not close to the aromatic residues Trp 4 and Tyr 11.
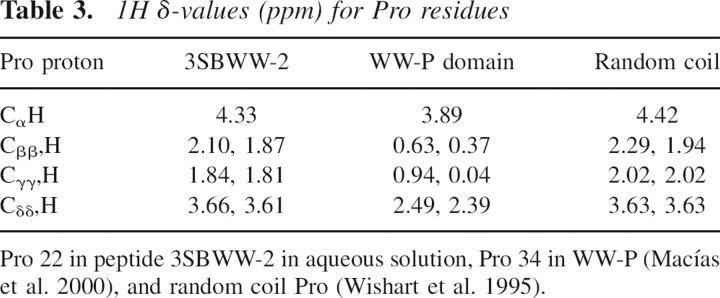
Analysis of the Pro 1H chemical shifts also indicates the absence of the Trp–Pro interaction in peptide 3SBWW-2. In WW domains, δ-values of Pro protons are strongly affected by the anisotropic effect of the aromatic rings of Trp and Tyr (Table 3). In contrast, the 1H δ-values for Pro 22 in peptide 3SBWW-2 are very close to the Pro random coil δ-values (Table 3). The absence of anisotropy effects on the protons of Pro 22 suggests that the side chains of the aromatic residues, Trp 4 and Tyr 11, are far from the Pro residue, as seen in the calculated structures (Fig. 7).
Table 3.
1H δ-values (ppm) for Pro residues
β-Sheet population in 3SBWW-2 is higher than in Betanova-LLM and lower than in WW-P
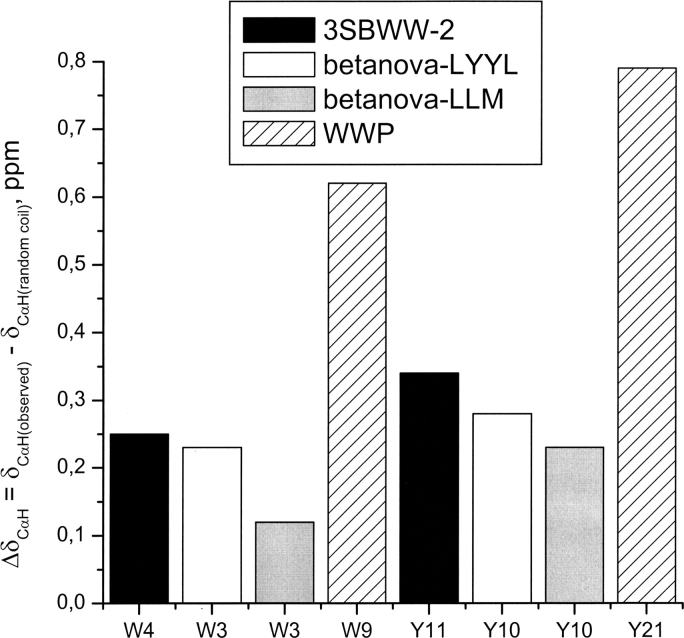
A qualitative examination of those chemical shifts that deviates strongly from their random coil values upon β-sheet formation provides a suitable way to compare the stability of the β-sheet structure adopted by the designed peptide 3SBWW-2 with those of Betanova-LLM and WWP domain. W4 and Y11 are the residues most ap-propriate for such comparison, since they are identical in the three peptides under consideration (see Table 1). Considering that larger deviations of the 1H δ-values correlate with higher β-sheet populations, the ΔδCαH values measured for W4 and Y11 (Fig. 8) indicate that the β-sheet formed by the designed peptide 3SBWW-2 is more populated than that of Betanova-LLM, which is in good agreement with the CD analysis. By the same criterion, β-sheet structure is less populated in peptide 3SBWW-2 than in WW-P domain.
Figure 8.
Histogram showing the ΔδCαH (δCαH observed − δCαH random coil, ppm) for Trp and Tyr residues at equivalent positions in peptides 3SBWW-2 (filled bars) and Betanova-LYYL (open bars) in H2O/D2O 9:1 (v/v) at pH 5.5 and 10°C, in Betanova-LLM (López de la Paz et al. 2001) in aqueous solution (gray bars), and for the WWP domain (striped bars) (Macías et al. 2000).
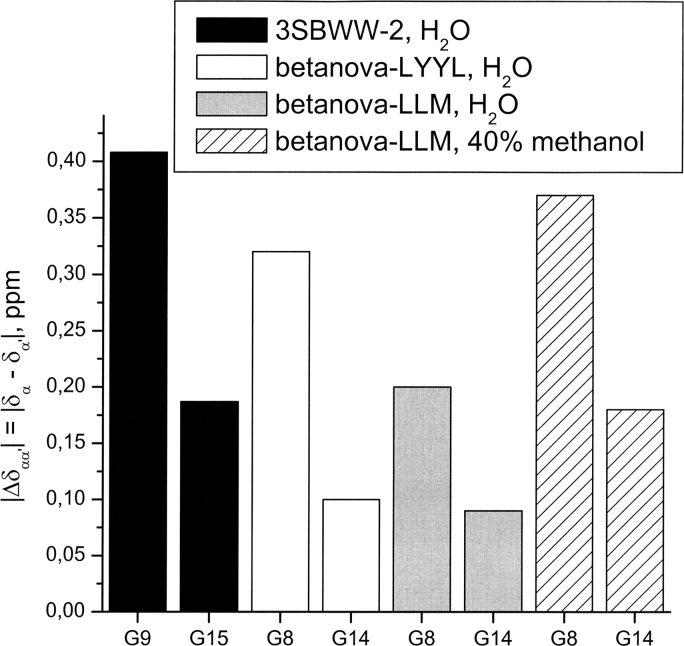
The splitting between CαH and Cα′H protons of Gly residues in β-turns (Δδαα′ = δα−δα′ ppm) has also been taken as a diagnostic of β-hairpin formation (Griffiths-Jones and Searle 2000), and it is useful to compare β-sheet stability in peptides with the same β-turn sequences, as are peptides 3SBWW-2 and Betanova-LLM, but not in peptides with different β-turn sequences, such as the WW-P domain (Table 1). The Δδαα′ values (Fig. 9) in aqueous solution for G9 and G15 residues of 3SBWW-2 are higher than those of equivalent residues (G8 and G14) of Betanova-LLM in either aqueous solution or in 40% (v/v) aqueous methanol solution where β-sheet stability increases in Betanova-LLM (López de la Paz et al. 2001). This result confirms that β-sheet population is higher for 3SBWW-2 than for Betanova-LLM.
Figure 9.
Histogram showing the splitting of CαH and Cα′H protons (ΔδGly = δCαH − δCα′H, ppm) for G9 and G15 residues in peptide 3SBWW-2 in H2O/D2O 9:1 (v/v) at pH 5.5 and 10°C (filled bars) and for G8 and G14 in Betanova-LYYL (open bars), Betanova-LLM (López de la Paz et al. 2001) in aqueous solution (gray bars), and in 40% methanol (striped bars).
The C-terminal extension makes a contribution to β-sheet stability in 3SBWW-2
The fact that the β-sheet structure adopted by peptide 3SBWW-2 is more stable than that formed by Betanova-LLM, while the C-terminal region of peptide 3SBWW-2 is disordered, raises the question of whether the C-terminal extension contributes or not to β-sheet stability. To clarify this point, we have designed a new peptide, named Betanova-LYYL (Table 1), that consists of the Betanova template and the same strand residues as peptide 3SBWW-2, and that lacks the C-terminal T–D–P sequence, characteristic of WW domains (see above).
The aromatic signature at 230 nm, which is characteristic of WW domains, is present in the CD spectrum of 3SBWW-2 and could also be detected in that of Betanova-LYYL (Fig. 2). However, based on the ratios between the ellipticity at 217 nm and 204 nm (Table 2), β-sheet content in Betanova-LYYL is smaller than in 3SBWW-2, though clearly higher than in Betanova-LLM.
Based on the profiles of ΔδCαH, ΔδNH, ΔδCα, and ΔδCβ conformational shift values (Fig. 4) and the set of observed NOEs, peptide Betanova-LYYL adopts the same three-stranded β-sheet structure as peptide 3SBWW-2 (Fig. 5) and as the previously reported Betanova peptides (López de la Paz et al. 2001). According to the ΔδCαH values measured for W4 and Y11 (Fig. 8) and the Δδαα′ splitting values of Gly residues (Fig. 9), the β-sheet adopted by Betanova-LYYL is less populated than peptide 3SBWW-2, but more than Betanova-LLM, in concordance with the CD analysis. The fact that the magnitudes of the conformational shifts observed for Betanova-LYYL are smaller than in the case of peptide 3SBWW-2 (Fig. 5) reinforces the conclusion that the β-sheet in Betanova-LYYL is less populated than that in 3SBWW-2. This indicates that the disordered C-terminal extension present in peptide 3SBWW-2 and absent in Betanova-LYYL contributes to β-sheet stability.
Discussion
The main purpose of the current work was to design a minimalist three-stranded β-sheet that was as stable as WW domains. To that end, we designed a series of peptides that are hybrids of Betanova-LLM (López de la Paz et al. 2001) and WW-P, a prototype WW domain (Macías et al. 2000). These peptides were cloned and expressed in E. coli. The ability of the purified peptides to form the β-sheet structure was evaluated by CD on the basis of the R217/204 ratio. According to the CD criterion, 3SBWW-2 has the highest β-sheet population. Considering that peptides 3SBWW-2 and 3SBWW-4 only differ by the presence of an N-terminal extension (Table 1), it is surprising that, according to the R217/204 ratio, the β-sheet seems to be more stable in peptide 3SBWW-2. It is quite possible that the additional N-terminal extension present in 3SBWW-4 is unstructured, as it is the C-terminal extension in 3SBWW-2 (see Results and below). In this case, the random coil contribution to CD spectra will increase and thus the spectra will look less structured. The R217/204 ratio is useful as an indicator of β-sheet formation and is very suitable for fast screening of a series of β-sheet-forming peptides, though sometimes it might not reflect the precise ranking of β-sheet population. In any case, we focused on peptide 3SBWW-2, the best β-sheet-forming peptide according to CD, to perform a complete NMR investigation.
Based on the NMR data, we showed that, as intended, 3SBWW-2 in aqueous solution adopts a three-stranded antiparallel β-sheet that is more stable than the parent Betanova-LLM and the control Betanova-LYYL, but is still not as stable as that of WW domains (see Results), and that the control Betanova-LYYL β-sheet is significantly more stable than that of the parent Betanova-LLM. We can get insights into the contributions to β-sheet stability by pairwise comparison of the structural characteristics of these four peptides.
First, we consider the last two peptides whose sequences differ only on three-strand residues, Y7–Y12–T17 in Betanova-LYYL versus Q6–T11–M16 in Betanova-LLM (Table 1). The differences in the intrinsic β-sheet propensities (Minor and Kim 1994a, b; Smith et al. 1994) of these residues as well as in their cross-strand side chain–side chain pairwise interactions, ranked according to the reported statistically derived probabilities (Wouters and Curmi 1995; Hutchinson et al. 1998; Pantoja-Uceda et al. 2006), do not suffice to explain the higher stability of the β-sheet adopted by peptide Betanova-LYYL relative to Betanova-LLM. As reported for other β-sheet peptides (Santiveri et al. 2003, 2005), the differences in β-sheet stability between Betanova-LYYL and Betanova-LLM most probably lies on the differences in side chain–side chain packing. Since peptide 3SBWW-2 has strand residues identical to Betanova-LYYL, the same favorable side chain packing is contributing to its β-sheet stability.
Concerning the C-terminal extension present in 3SBWW-2, it was expected to contribute to β-sheet stability by the Pro residue interacting with the side chains of two aromatic residues, a Trp at the N-terminal strand and Tyr in the middle one, forming a small hydrophobic cluster, as occurs in WW-P (Fig. 1). Surprisingly, the C-terminal extension contributes to β-sheet stability, as indicated by the higher β-sheet stability of 3SBWW-2 relative to Betanova-LYYL, but it is disordered. Although peptide 3SBWW-2 contains Trp, Tyr, and Pro residues at positions equivalent to those in WW-P (Table 1), that interaction does not exist (Fig. 7). Thus, one explanation for the lower β-sheet stability in peptide 3SBWW-2 relative to WW-P lies in the absence of the small hydrophobic core formed by these Trp, Tyr, and Pro residues. A plausible explanation for the stabilization provided by the C-terminal tail is by decreasing the loss of entropy upon folding.
Another noticeable difference between 3SBWW-2 and WW-P structures is the significant differences in the length of the loops, in particular, in the first one where a seven-residue segment was shortened to only two residues (Table 1). The shortest β-sheet backbone of the 3SBWW peptides is probably more rigid than that of WW domains. In 3SBWW peptides, the two tight turns together with the very short β-strands, only four residues long, might originate some strain. This strain might be counterbalanced at least partially by having a disordered tail (see above). It should be noted that Betanova and 3SBWW β-sheet backbones are the shortest ones reported. Substitutions of the two central residues of the first loop in the Pin1 WW domain without modifying its length did not alter β-sheet structure (Kaul et al. 2001). Taken all together, for β-sheet design one should keep in mind that the combination of tight turns and very short strands might destabilize the structure.
On the whole, from these results we can gather some lessons for protein design: (1) Adequate side chain packing contributes greatly to β-sheet stability; (2) residues at the peptide ends not taking part in the β-sheet structure might contribute to its stability by decreasing the loss of entropy upon folding; and (3) very short strands and tight turns might lead to some structural strain and, hence, be destabilizing or less stabilizing than expected.
Materials and methods
Construction of GST–3SBWW fusion proteins
To construct GST fusion proteins, DNA sequences corresponding to the different 3SBWW forms were synthetically synthesized. Complementary single-strand oligonucleotides coding for the desired sequences were hybridized. The products were designed to generate preformed BamHI and HindIII cohesive ends (sequences of all oligonucleotides are available from the authors upon request). The double-stranded DNAs were subsequently cloned into BamHI and HindIII restriction sites of pGAT2 expression plasmid (EMBL) downstream from the glutathione-S-transferase gene and the thrombin cleavage site. Each construct was confirmed by DNA sequencing.
Expression and purification of 3SBWW peptides
E. coli BL21 cells carrying pGAT2–3SBWW plasmids expressed fusion protein GST linked to the 3SBWW constructs. Briefly, E. coli were grown at 37°C in Luria Broth containing 100 μg/mL ampicillin until absorbance at 600 nm was 0.4–0.6. The expression of fusion proteins was induced by the addition of 0.5–1.0 mM IPTG for 4–6 h. Cells were collected by centrifugation at 5000g. The E. coli pellets (from 1 L) were resuspended in 15 mL PBS containing a mixture of a protease inhibitors. The cells were disrupted in a French press and insoluble material was removed by ultracentrifugation at 38,000g for 1 h. The soluble cell lysate containing the expressed fusion protein was mixed with 1 mL of a 50% slurry of glutathione-agarose, preequilibrated in PBS. After several washes with PBS, the bound GST–DBP was eluted with 20 mM oxidized glutathione in 10 mM Tris Buffer (pH 8.0). The fusion protein was cleaved with thrombin and dialyzed against PBS to remove oxidized glutathione. GST was separated from recombinant 3SBWW proteins using a HiLoad 26/60 Superdex Prep Grade gel filtration column in a FPLC system (Pharmacia). Sequence fidelities of the re3SBWW peptides were confirmed by mass spectrometry. The re3SBWW peptides were dialyzed against H2O and lyophilized.
Peptide synthesis
Peptide Betanova-LYYL was synthesized by Thermo Electron Corporation.
Far-UV circular dichroism
The 3SBWW designed peptides were dissolved in 50 mM sodium phosphate (pH 7.0) at 50 μM and 10 μM concentrations for CD experiments. The concentration of the peptide samples was determined by measurements of ultraviolet absorbance. CD spectra were acquired on a Jasco-710 instrument calibrated with (1S)-(+)-10-camphorsulphonic acid. Usually 20 accumulations were averaged to obtain each spectrum in the range 190–250 nm at a temperature of 293 K by taking points every 0.2 nm, with a 100 nm/min scan rate, an integration time of 1 sec, and a 1-nm bandwidth. Cells with pathlengths of 0.1 cm and 0.5 cm were used.
NMR spectroscopy
3SBWW-2 and Betanova-LYYL peptide samples for NMR experiments were prepared by dissolving the lyophilized peptide in H2O/D2O 9:1 (v/v) or in pure D2O (pH 5.5). Peptide concentrations were 1–2 mM. The pH of 3SBWW-2 samples was checked with a glass microelectrode and was not corrected for isotope effects. NMR temperature probe was calibrated using a methanol sample. Sodium 2,2-dimethyl-2-silapentane-5-sulphonate (DSS) was used as an internal reference for 1H δ-values. The 13C δ-values were indirectly referenced multiplying the spectrometer frequency that corresponds to 0 ppm in 1H spectrum, assigned to internal DSS by 0.25144954 (Bax and Subramanian 1986; Spera and Bax 1991). NMR spectra were acquired on AV-600 and AV-800 US2 Bruker pulse spectrometers operating at proton frequency of 600.13 and 800.2 MHz, respectively. 2D homonuclear correlated spectroscopy (COSY; Aue et al. 1976), total correlated spectroscopy (TOCSY; Rance 1987), and nuclear Overhauser enhancement spectroscopy (NOESY; Jeener et al. 1979; Kumar et al. 1980) spectra were recorded at 10°C and acquired in the phase-sensitive mode using the time-proportional phase incrementation mode (Redfield and Kuntz 1975). A mixing time of 120 msec was used for NOESY spectra. 2D 1H–13C HSQC spectra (Bodenhausen and Ruben 1980) at 10°C in natural 13C abundance were recorded in D2O non-labeled protein samples. Water suppression was achieved either by selective presaturation or including a WATERGATE module (Piotto et al. 1992) in the original pulse sequences prior to acquisition. 2D acquisition data matrices were defined by 2048 × 512 points in t2 and t1, respectively. Data were processed using the standard XWIN-NMR Bruker program on a Silicon Graphics computer. The 2D data matrix was multiplied by a square-sine-bell window function with the corresponding shift optimized for every spectrum and zero-filled to a 4 K × 2 K complex matrix prior to Fourier transformation. Baseline correction was applied in both dimensions.
NMR assignment
1H NMR signals of peptides 3SBWW-2 and Betanova-LYYL were assigned by standard sequential assignment methods (Wüthrich et al. 1984; Wüthrich 1986). 13C resonances were assigned on the basis on the cross-correlations observed in HSQC spectra between the proton and the carbon to which it is bonded. The 1H and 13C δ-values are available as Supplemental Material.
Structure calculation
Distance constraints for structure calculations were derived from the 2D 120-msec mixing time NOESY spectra recorded either in H2O or in D2O. The NOE cross-peaks were integrated by using the automatic integration subroutine of the SPARKY program (T.D. Goddard and D.G. Kneller) and then calibrated and converted to upper limit distance constraints within the DYANA program (Guntert et al. 1997). ϕ and ψ angle restraints were derived from 1Hα, 13Cα, and 13Cβ chemical shifts by using the TALOS program (Cornilescu et al. 1999). ϕ angles for those residues for which the derived angle restraints were ambiguous were constrained to the range −180° to 0°, except for Asn and Gly residues. Structures were calculated using the DYANA program (Guntert et al. 1997) and an annealing strategy.
Electronic supplemental material
Supplemental material includes two tables listing the 1H and 13C chemical shifts of peptides 3SBWW-2 and Betanova-LYYL in aqueous solution.
Acknowledgments
We thank Dr. M. López de la Paz for providing us with unreported data on Betanova peptides. A.M. Fernández-Escamilla was a recipient of a Marie Curie postdoctoral fellowship from the European Union. We are grateful to Dr. D. Laurents for English grammar revision.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: M. Angeles Jiménez, Instituto de Química-Física Rocasolano, Consejo Superior de Investigaciones Científicas, Serrano 119, Madrid 28006, Spain; e-mail: majimenez@iqfr.csic.es; fax: 34-1-5642431.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062186506.
References
- Aue, W.P., Bertholdi, E., Ernst, R.R. 1976. Two-dimensional spectroscopy. application to NMR. J. Chem. Phys. 64: 2229–2246. [Google Scholar]
- Bax, A. and Subramanian, J. 1986. Sensitivity-enhanced two-dimensional heteronuclear shift correlation NMR spectroscopy. J. Magn. Reson. 67: 565–570. [Google Scholar]
- Bodenhausen, G. and Ruben, D.J. 1980. Natural abundance 15N NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 69: 185–189. [Google Scholar]
- Cornilescu, G., Delaglio, F., Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- Crane, J.C., Koepf, E.K., Kelly, J.W., Gruebele, M. 2000. Mapping the transition state of the WW domain β-sheet. J. Mol. Biol. 298: 283–292. [DOI] [PubMed] [Google Scholar]
- Das, C., Raghothama, S., Balaram, P. 1998. A designed three stranded β-sheet peptide as a multiple β-hairpin model. J. Am. Chem. Soc. 120: 5812–5813. [Google Scholar]
- de Alba, E., Santoro, J., Rico, M., Jiménez, M.A. 1999. De novo design of a monomeric three-stranded antiparallel β-sheet. Protein Sci. 8: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deechongkit, S. and Kelly, J.W. 2002. The effect of backbone cyclization on the thermodynamics of β-sheet unfolding: Stability optimization of the PIN WW domain. J. Am. Chem. Soc. 124: 4980–4986. [DOI] [PubMed] [Google Scholar]
- Deechongkit, S., Nguyen, H., Powers, E.T., Dawson, P.E., Gruebele, M., Kelly, J.W. 2004. Context-dependent contributions of backbone hydrogen bonding to β-sheet folding energetics. Nature 430: 101–105. [DOI] [PubMed] [Google Scholar]
- Deechongkit, S., Nguyen, H., Jager, M., Powers, E.T., Gruebele, M., Kelly, J.W. 2006. β-Sheet folding mechanisms from perturbation energetics. Curr. Opin. Struct. Biol. 16: 94–101. [DOI] [PubMed] [Google Scholar]
- Fesinmeyer, R.M., Hudson, F.M., Olsen, K.A., White, G.W., Euser, A., Andersen, N.H. 2005. Chemical shifts provide fold populations and register of β-hairpins and β-sheets. J. Biomol. NMR 33: 213–231. [DOI] [PubMed] [Google Scholar]
- Fisinger, S., Serrano, L., Lacroix, E. 2001. Computational estimation of specific side chain interaction energies in α helices. Protein Sci. 10: 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S.R. and Searle, M.S. 2000. Structure, folding, and energetics of cooperative interactions between β-strands of a de novo designed three-stranded antiparallel β-sheet peptide. J. Am. Chem. Soc. 122: 8350–8356. [Google Scholar]
- Guntert, P., Mumenthaler, C., Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273: 283–298. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G., Sessions, R.B., Thornton, J.M., Woolfson, D.N. 1998. Determinants of strand register in antiparallel β-sheets of proteins. Protein Sci. 7: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager, M., Nguyen, H., Crane, J.C., Kelly, J.W., Gruebele, M. 2001. The folding mechanism of a β-sheet: The WW domain. J. Mol. Biol. 311: 373–393. [DOI] [PubMed] [Google Scholar]
- Jeener, J., Meier, B.H., Bachmann, P., Ernst, R.R. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71: 4546–4553. [Google Scholar]
- Jiang, X., Kowalski, J., Kelly, J.W. 2001. Increasing protein stability using a rational approach combining sequence homology and structural alignment: Stabilizing the WW domain. Protein Sci. 10: 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, R., Angeles, A.R., Jager, M., Powers, E.T., Kelly, J.W. 2001. Incorporating beta-turns and a turn mimetic out of context in loop 1 of the WW domain affords cooperatively folded β-sheets. J. Am. Chem. Soc. 123: 5206–5212. [DOI] [PubMed] [Google Scholar]
- Koepf, E.K., Petrassi, H.M., Sudol, M., Kelly, J.W. 1999. WW: An isolated three-stranded antiparallel β-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 8: 841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme, T., Ramírez-Alvarado, M., Serrano, L. 1998. Design of a 20-amino acid, three-stranded β-sheet protein. Science 281: 253–256. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Ernst, R.R., Wüthrich, K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton–proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 95: 1–6. [DOI] [PubMed] [Google Scholar]
- López de la Paz, M., Lacroix, E., Ramírez-Alvarado, M., Serrano, L. 2001. Computer-aided design of β-sheet peptides. J. Mol. Biol. 312: 229–246. [DOI] [PubMed] [Google Scholar]
- Macías, M.J., Gervais, V., Civera, C., Oschkinat, H. 2000. Structural analysis of WW domains and design of a WW prototype. Nat. Struct. Biol. 7: 375–379. [DOI] [PubMed] [Google Scholar]
- Minor, D.L. Jr. and Kim, P.S. 1994a. Context is a major determinant of β-sheet propensity. Nature 371: 264–267. [DOI] [PubMed] [Google Scholar]
- ——— 1994b. Measurement of the β-sheet-forming propensities of amino acids. Nature 367: 660–663. [DOI] [PubMed] [Google Scholar]
- Nguyen, H., Jager, M., Moretto, A., Gruebele, M., Kelly, J.W. 2003. Tuning the free-energy landscape of a WW domain by temperature, mutation, and truncation. Proc. Natl. Acad. Sci. 100: 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja-Uceda, D., Santiveri, C.M., Jimenez, M.A. 2006. De novo design of monomeric β-hairpin and β-sheet peptides. In Protein design: Methods and applications (eds. Guerois R. and López de la Paz, M.) . pp. 27–51. Humana Press, Totowa, NJ Vol. 340:. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., Sklenar, V. 1992. Gradient-tailored excitation for single quantum NMR spectroscopy in aqueous solution. J. Biomol. NMR 6: 661–665. [DOI] [PubMed] [Google Scholar]
- Rance, M. 1987. Improved techniques for homonuclear rotating-frame and isotropic mixing experiments. J. Magn. Reson. 74: 557–564. [Google Scholar]
- Redfield, A.G. and Kuntz, S.D. 1975. Quadrature Fourier detection: Simple multiplex for dual detection. J. Magn. Reson. 19: 250–259. [Google Scholar]
- Santiveri, C.M., Rico, M., Jiménez, M.A. 2001. 13Cα and 13Cβ chemical shifts as a tool to delineate β-hairpin structures in peptides. J. Biomol. NMR 19: 331–345. [DOI] [PubMed] [Google Scholar]
- Santiveri, C.M., Rico, M., Jiménez, M.A., Pastor, M.T., Pérez-Payá, E. 2003. Insights into the determinants of β-sheet stability: 1H and 13C NMR conformational investigation of three-stranded antiparallel β-sheet-forming peptides. J. Pept. Res. 61: 177–188. [DOI] [PubMed] [Google Scholar]
- Santiveri, C.M., Santoro, J., Rico, M., Jiménez, M.A. 2004. Factors involved in the stability of isolated β-sheets: Turn sequence, β-sheet twisting, and hydrophobic surface burial. Protein Sci. 13: 1134–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiveri, C.M., Pantoja-Uceda, D., Rico, M., Jiménez, M.A. 2005. β-hairpin formation in aqueous solution and in the presence of trifluoroethanol: A 1H and 13C nuclear magnetic resonance conformational study of designed peptides. Biopolymers 79: 150–162. [DOI] [PubMed] [Google Scholar]
- Schenck, H.L. and Gellman, S.H. 1998. Use of a designed triple-stranded antiparallel β-sheet to probe β-sheet cooperativity in aqueous solution. J. Am. Chem. Soc. 120: 4869–4870. [Google Scholar]
- Schwarzinger, S., Kroon, G.J., Foss, T.R., Chung, J., Wright, P.E., Dyson, H.J. 2001. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 123: 2970–2978. [DOI] [PubMed] [Google Scholar]
- Sharman, G.J. and Searle, M.S. 1998. Cooperative interaction between the three strands of a designed antiparallel β-sheet. J. Am. Chem. Soc. 120: 5291–5300. [Google Scholar]
- Smith, C.K., Withka, J.M., Regan, L. 1994. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry 33: 5510–5517. [DOI] [PubMed] [Google Scholar]
- Spera, S. and Bax, A. 1991. Empirical correlation between protein backbone conformation and Cα and Cβ 13C NMR chemical shifts. J. Am. Chem. Soc. 113: 5490–5492. [Google Scholar]
- Sudol, M. 1996. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65: 113–132. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Sykes, B.D., Richards, F.M. 1991. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222: 311–333. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Bigam, C.G., Holm, A., Hodges, R.S., Sykes, B.D. 1995. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5: 67–81. [DOI] [PubMed] [Google Scholar]
- Wouters, M.A. and Curmi, P.M. 1995. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins 22: 119–131. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K. In NMR of proteins and nucleic acids .1986. John Wiley & Sons, New York.
- Wüthrich, K., Billeter, M., Braun, W. 1984. Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton–proton distances. J. Mol. Biol. 180: 715–740. [DOI] [PubMed] [Google Scholar]