Abstract
The extracellular part of the fibroblast growth factor (FGF) receptor (FGFR) consists of up to three Ig modules (Ig1–Ig3), in which the Ig2 and Ig3 modules determine affinity and specificity for FGF and heparin. The FGFR isoforms lacking the Ig1 module have higher affinity for FGF and heparin than the triple Ig-module isoforms, suggesting that the Ig1 module is involved in the regulation of the FGFR–ligand interaction. We show here by surface plasmon resonance and NMR analyses that the Ig1 module binds to the Ig2 module, and identify by NMR the binding sites involved in the Ig1–Ig2 interaction. The identified binding site in the Ig2 module was found to be in the area of the FGF–Ig2 and Ig2–heparin contact sites, thus providing direct structural evidence that the Ig1 module functions as a competitive autoinhibitor of the FGFR–ligand interaction. Furthermore, the Ig1 binding site of the Ig2 module overlaps the Ig2–Ig2 contact site. This suggests that the function of the Ig1 module is not only regulation of the FGFR–ligand binding affinity but also prevention of spontaneous FGFR dimerization (through a direct Ig2–Ig2 interaction) in the absence of FGF.
Keywords: FGFR Ig module 1 function, NMR, SPR
Fibroblast growth factor receptors (FGFR1–FGFR4) regulate a multitude of cellular processes via interactions with fibroblast growth factors (FGF1–FGF23) (McKeehan et al. 1998; Itoh and Ornitz 2004) and cell adhesion molecules (Doherty and Walsh 1996; Kiselyov et al. 2003, 2005). FGFR consists of up to three Ig modules (Ig1–Ig3), a trans-membrane domain and a cytoplasmic tyrosine kinase domain. The Ig1–Ig2 linker is very long, consisting of 20–30 amino acid residues. FGFRs also bind heparin/heparan sulphate, which is required for the high-affinity FGF–FGFR interaction (Yayon et al. 1991; Ornitz et al. 1992). FGF–FGFR binding results in dimerization of FGFR leading to autophosphorylation of the receptor tyrosine kinase domains. Based on crystal structures of the ternary FGF–FGFR–heparin complex, two models, a symmetric and an asymmetric, of FGFR dimerization have been proposed. In the symmetric model (Plotnikov et al. 1999, 2000; Schlessinger et al. 2000), dimerization of the two FGF–FGFR complexes is stabilized by the FGF–FGFR interactions through a primary and a secondary interaction site (involving Ig2, Ig3, and Ig2–3 linker), a direct FGFR–FGFR interaction (Ig2–Ig2 binding), and heparin–FGF and heparin–FGFR (involving Ig2) interactions. In the asymmetric model (Pellegrini et al. 2000), the secondary FGF–FGFR interaction site and the direct FGFR–FGFR interaction are absent. Regulation of the FGFR–ligand binding is primarily achieved by alternative splicing of FGFRs. There are FGFR isoforms lacking the Ig1 module (FGFR1 and 2), the Ig1 module combined with the Ig1–Ig2 linker sequence (FGFR2), or the Ig1–Ig2 linker alone (in FGFR3) (McKeehan et al. 1998; Shimizu et al. 2001).
The physiological significance of the Ig1 module is not well elucidated. The triple Ig-module form of FGFR1 and FGFR3 has lower affinity for FGF and heparin compared to the double Ig-module form (Wang et al. 1995; Olsen et al. 2004), and the Ig1 module has been shown to bind an FGFR3 Ig2–Ig3 fragment with a dissociation constant (K d) of 20 μM (Olsen et al. 2004). However, since the residues (as well as the module) of the FGFR3 fragment involved in this interaction have not been identified, the mechanism by which the Ig1 module affects the FGFR–ligand interaction is not known. The Ig1 module may be presumed to reduce the affinity of the FGFR–ligand interaction in several ways: by an allosteric mechanism, a competitive inhibition, or a combination of the two effects. It is also possible that the FGFR–FGF interaction is affected by the Ig1 module by one mechanism, whereas the FGFR–heparin interaction is affected by a different mechanism. Thus, this subject requires further analysis.
Results and Discussion
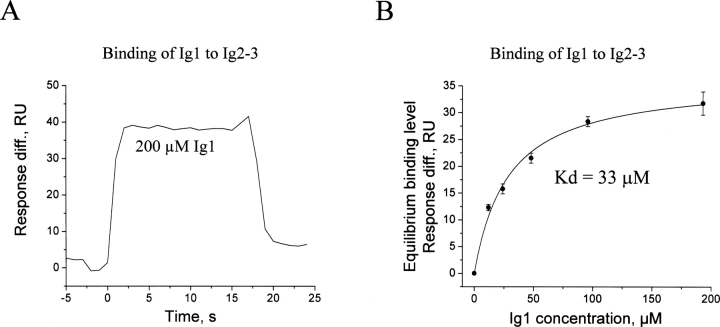
To study the function of the FGFR1 Ig1 module, we have recently determined the structure of the module by nuclear magnetic resonance (NMR) analysis (Kiselyov et al. 2006a). Since Ig1 in FGFR3 binds to Ig2–3 with a K d value of 20 μM (Olsen et al. 2004), it was of interest to determine this binding for FGFR1. Therefore, binding of soluble Ig1 to immobilized Ig2–3 modules of FGFR1 was studied by surface plasmon resonance (SPR) analysis. The time course of the binding, similar to that for FGFR3, is characterized by very fast association and dissociation phases (Fig. 1A). A plot of the equilibrium binding response versus the concentration of Ig1 is shown in Figure 1B. The calculated K d value for the binding was 33±6 μM, which is very close to the 20 μM K d value determined for FGFR3. It should be noted that the maximum binding level of the Ig1 module (∼30 RU) may seem low, however, the maximum binding level of FGF1 at a saturating concentration of 100 nM was ∼100 RU (data not shown). The calculated K d value for the FGF1 binding was ∼5 nM. Thus, the maximum binding level of the Ig1 module when compared to that of FGF1 is in line with the expected value.
Figure 1.
Binding of the FGFR1 Ig1 module to the combined FGFR Ig2–Ig3 modules. (A) The association and dissociation phases of the Ig1 binding at a concentration of 200 μM. No unspecific binding of the Ig1 module to the control surface has been detected. (B) Plot of the equilibrium binding level of the Ig1 module to the Ig2–3 modules versus the concentration of the Ig1 module. The binding is given as an average of six replicates, with the error bar showing standard deviations. The data were fitted with the theoretical curve in order to calculate the dissociation constant (K d).
In order to identify the residues involved in the interaction between Ig1 and Ig2–3, NMR analysis was employed. This method requires production of 15N labeled proteins and assignment of their 15N, 1H resonance frequencies for the backbone atoms. The resonance assignment of the Ig1 module of mouse FGFR1 has previously been reported (Kiselyov et al. 2006b). Since resonance assignment of the 25-kDa Ig2–3 construct of FGFR1 is a very time-consuming process, we decided to perform assignment of only the Ig2 module of mouse FGFR1, because the Ig2 module contains binding sites for FGF (both primary and secondary), heparin, and the Ig2 module itself (whereas the Ig3 module contains the binding site only for FGF) and because the Ig1 module inhibits both the FGF–FGFR1 and heparin–FGFR1 interactions (Wang et al. 1995).
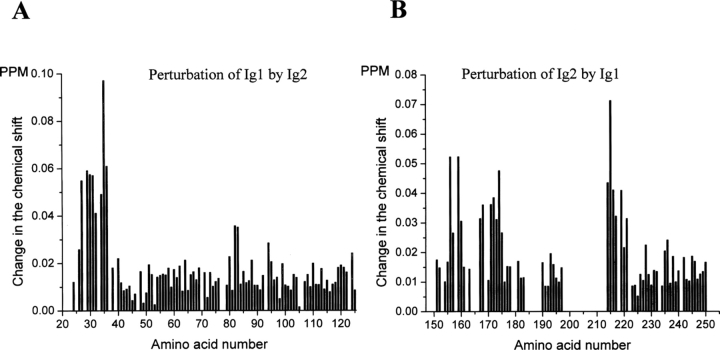
In a 15N-HSQC spectrum of a 15N-labeled protein, a signal for all amino acids with both a nitrogen and a proton can be observed. The changes in chemical shifts of the signals provide a method for identification in a protein of amino acid residues that are perturbed by the binding of another molecule. A 2-mM unlabeled Ig1 module was added to a 0.5 mM 15N-labeled sample of the Ig2 module, and vice versa, a 2 mM unlabeled Ig2 module was added to a 0.5 mM 15N-labeled sample of the Ig1 module. The recorded changes of chemical shifts are shown in Figure 2A–B. The residues of the Ig1 module that exhibited significant perturbation (higher than 0.04 ppm) by the Ig2 module were L27, E29, Q30, A31, Q32, W34, G35, and V36 (Fig. 2A), and the residues of the Ig2 module that exhibited significant perturbation (higher than 0.025 ppm) by the Ig1 module were T156, S157, E159, K160, A167, V168, A171, K172, T173, V174, K175, S214, I215, I216, M217, and S219 (Fig. 2B). The changes of the chemical shifts of these residues demonstrate that the presence of one module close to the other module alters the chemical environment at the perturbed residues, indicating that the perturbed residues are either a part or in the vicinity of the binding site for the interaction between the two modules. Mapping of the perturbed residues onto the structures of the Ig1 and Ig2 modules is shown in Figure 3A. Since the structure of the mouse Ig2 module is not known, the structure of the human Ig2 module (Plotnikov et al. 1999) was used for mapping. The perturbed residues in the Ig1 module are located in the A/A′ loop region of the module, which is noteworthy because this loop in the Ig1 module is much longer than that of the Ig2 and Ig3 modules (Kiselyov et al. 2006a), and form a single patch, whereas the perturbed residues in the Ig2 module are located in two patches: a larger patch consisting of 12 residues (A167, V168, A171, K172, T173, V174, K175, S214, I215, I216, M217, S219) and a smaller patch consisting of four residues (T156, S157, E159, K160). The two patches are located very close to each other. Thus, these data indicate that there is a specific interaction between the soluble Ig1 and Ig2 modules of FGFR1. Due to the fact that the Ig1–Ig2 linker is unusually long (30 amino acids), we presume that the Ig1 module may be involved in the intramolecular binding to the Ig2 module of the triple-Ig form of FGFR1. It should be noted that since the Ig1 module binds to the C-terminal part of the Ig2 module, it is possible that the Ig1 module may also bind the N-terminal part of the Ig3 module and/or the Ig2–Ig3 linker, which presumably could stabilize the intramolecular binding of the Ig1 module.
Figure 2.
NMR titration of the interaction between the Ig1 and Ig2 modules of FGFR1. (A) Changes in chemical shifts of 0.5 mM 15N-labeled Ig1 module after addition of 2 mM unlabeled Ig2 module. (B) Changes in chemical shifts of 0.5 mM 15N-labeled Ig2 module after addition of 2 mM unlabeled Ig1 module. The data are given as averages from two independent experiments. The change of the chemical shift was calculated using the following expression: [(5 * ΔH)2 + (ΔN)2]0.5, where ΔH is the change of the 1H chemical shift and ΔN is the change of the 15N chemical shift.
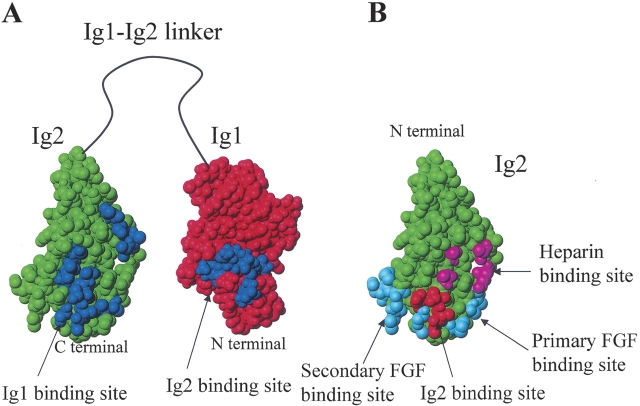
Figure 3.
Mapping of the various FGFR1–ligand binding sites onto the structures of the FGFR1 Ig1 and Ig2 modules. (A) Mapping of the residues of the Ig2 module perturbed by the interaction with the Ig1 module (blue) onto the structure of the Ig2 module (green); and mapping of the residues of the Ig1 module perturbed by the interaction with the Ig2 module (blue) onto the structure of the Ig1 module (red). (B) Mapping of the residues of the Ig2 module involved in binding to the Ig2 module (red), heparin (magenta), FGF through the primary interaction site (cyan) (according to both the symmetrical and asymmetrical model) and secondary interaction site (cyan) (according to the symmetrical model).
According to the symmetrical model of the FGFR dimerization, the primary Ig2 site binding to FGF consists of L165, A167, P169, and V248; the secondary Ig2–FGF binding site consists of P199, D200, I203, G204, G205, S219, and V221; the Ig2–heparin binding site consists of K160, K163, K175, and K177; and the Ig2–Ig2 binding site consists of A171, K172, T173, and D218. Mapping of these binding sites onto the structure of the Ig2 module is shown in Figure 3B. A167 from the primary and S219 from the secondary Ig2–FGF binding sites, K175 from the Ig2–heparin binding site, and A171, K172, T173 from the Ig2–Ig2 binding site are among the residues in the Ig2 module which are perturbed by Ig1 binding. As can be seen from Figure 3A–B, both the primary and secondary Ig2–FGF and the Ig2–heparin binding sites are adjacent to the larger Ig2 patch of perturbed residues, whereas three out of four residues of the Ig2–Ig2 binding site are located within this patch. Based on this, we surmise that the Ig1 binding to Ig2 may compete with the binding of FGFR1 to FGF and heparin/heparan sulfate and, thus, affect the affinity of the FGFR1–ligand interaction, which is supported by the fact that the triple-Ig form of FGFR1 has a lower affinity for FGF and heparin/heparan sulfate than the double-Ig form (Wang et al. 1995). It should also be noted that the acid box located in the Ig1–Ig2 linker has been hypothesized (Plotnikov et al. 1999) to directly interact with the heparin binding site of the Ig2 module, and in this way, on the one hand to stabilize the Ig1 binding to Ig2/Ig3, and on the other hand to inhibit the binding of heparin to the Ig2 module. As can be seen from Figure 3, the Ig2 site involved in binding to Ig1 is immediately adjacent to the primary and secondary sites of Ig2 for FGF, whereas the Ig2 site for heparin is somewhat further away (although also very close). Thus, it is possible that when Ig1 binds to Ig2, there is enough room for the acid box to bind to Ig2 as well. However, whether or not this is true cannot be deduced from our data and should be addressed in a separate study.
Furthermore, since the Ig2 site binding to Ig1 overlaps the Ig2–Ig2 binding site, the Ig1 module can be presumed to be an inhibitor of spontaneous FGFR1 dimerization. The significance of the tighter activation control due to the presence of Ig1 can be demonstrated by the fact that a switch in expression from the triple-Ig form of FGFR to the double-Ig form correlates with an increase in glioma malignancy (Yamaguchi et al. 1994). Thus, expression of the double-Ig form of FGFR, which is expected to be under less tight control of activation due to the lack of Ig1, probably leads to a growth advantage of tumor cells.
The presented results provide structural basis for understanding the autoinhibitory function of the Ig1 module of FGFR.
Materials and methods
Production of recombinant proteins
The Ig1 and Ig2 modules of mouse FGFR1 consist of a His-tag, AGHHHHHH, and amino acids 23–119 and 140–251, respectively (swissprot p16092). The combined Ig2–3 modules of mouse FGFR1 (3c isoform) consist of a His-tag, RSHHHHHH, and amino acids 141–365 (swissprot p16092). The Ig1 and Ig2 modules as well as the combined Ig2–3 modules of mouse FGFR1 (3c isoform) were produced as previously described (Kiselyov et al. 2003, 2006b). Briefly, the Ig1 and Ig2 modules were expressed in the KM71 strain of yeast Pichia pastoris (Invitrogen), and the Ig2–3 modules, in Drosophila S2 cells (Invitrogen) according to the manufacturer's instructions. All the proteins were purified by affinity chromatography using Ni2+-NTA resin (Qiagen) and/or ion exchange chromatography and gel filtration.
SPR analysis
Binding analysis was performed using a BIAcoreX instrument (Biosensor AB) at 25°C using 10 mM sodium phosphate (pH 7.4), 150 mM NaCl as a running buffer. The flow rate was 5 μL/min. Approximately 1500 RU of the Ig2–3 modules of FGFR2 were immobilized on the sensor chip CM5 (Biosensor AB) as previously described (Kiselyov et al. 2003). Binding of the FGFR1 Ig1 module to the immobilized receptor Ig2–3 modules was studied in the following way: the Ig1 module was injected at a specified concentration simultaneously into a flow-cell with the immobilized Ig2–3 modules (Fc1-cell) and a control flow-cell with nothing immobilized (Fc2-cell). The control Fc2-cell has been activated and blocked in the same way as the Fc1-cell. The curve representing a possible unspecific binding of the Ig1 module to the surface of the Fc2-cell was subtracted from the curve representing binding of the Ig1 module to the immobilized Ig2–3 modules and the surface of the Fc1-cell. The resulting curve was used for analysis.
NMR measurements
The following samples were used for recording of NMR spectra: 2 mM Ig2 module, 2 mM 15N-labeled Ig2 module, 0.5 mM 15N, 13C(50%)-labeled Ig2 module. The buffer was 10 mM sodium phosphate (pH 7.4), 150 mM NaCl, except for the double-labeled sample, where 10 mM sodium phosphate (pH 7.4), 30 mM NaCl was used. The following NMR spectra were recorded and used for assignment of the Ig2 module: TOCSY (45 msec and 70 msec mixing time), NOESY (80 msec and 200 msec mixing time), DQFCOSY, 15N-HSQC, 15N-TOCSY-HSQC (70 msec mixing time), 15N-NOESY-HSQC (125 msec mixing time), HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HNCA, and HN(CO)CA. All spectra were recorded using the standard setup provided by ProteinPack. The spectra were processed by NMRPipe (Delaglio et al. 1995) and analyzed by Pronto3D (Kjær et al. 1994). The NMR experiments were performed using Varian Unity Inova 750 and 800 MHz spectrometers. All spectra were recorded at 298 K.
Acknowledgments
This work was supported by grants from Købmand i Odense Johann og Hanne Weimann, f. Seedorffs Legat (V.V.K.), The John and Birthe Meyer Foundation (F.M.P.), The Danish Medical Research Council (E.B. and V.B.), The Danish Cancer Society (E.B.), the Lundbeck Foundation (E.B. and V.B.), and the EU integrated project PROMEMORIA.
Footnotes
Reprint requests to: Flemming M. Poulsen, Institute of Molecular Biology, Øster Farimagsgade 2A, Copenhagen DK-1353, Denmark; e-mail: fmp@apk.molbio.ku.dk; fax: 45-353-22075.
Abbreviations: FGFR, fibroblast growth factor receptor; FGF, fibroblast growth factor; Ig, immunoglobulin; NMR, nuclear magnetic resonance; SPR, surface plasmon resonance.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062206106.
References
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Doherty, P. and Walsh, F.S. 1996. CAM-FGF receptor interactions: A model for axonal growth. Mol. Cell. Neurosci. 8: 99–111. [DOI] [PubMed] [Google Scholar]
- Itoh, N. and Ornitz, D.M. 2004. Evolution of the Fgf and Fgfr gene families. Trends Genet. 20: 563–569. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Skladchikova, G., Hinsby, A.M., Jensen, P.H., Kulahin, N., Soroka, V., Pedersen, N., Tsetlin, V., Poulsen, F.M., Berezin, V. et al. 2003. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure 11: 691–701. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Soroka, V., Berezin, V., Bock, E. 2005. Structural biology of NCAM homophilic binding and activation of FGFR. J. Neurochem 94: 1169–1179. [DOI] [PubMed] [Google Scholar]
- Kiselyov, V.V., Bock, E., Berezin, V., Poulsen, F.M. 2006a. NMR structure of the first Ig module of mouse FGFR1. Protein Sci. 15: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov, V.V., Berezin, V., Bock, E., Poulsen, F.M. 2006b. 1H and 15N resonance assignment of the first module of FGFR1. J. Biomol. NMR (in press). [DOI] [PubMed]
- Kjær, M., Andersen, K.V., Poulsen, F.M. 1994. Automated and semiautomated analysis of homo- and heteronuclear multidimensional nuclear magnetic resonance spectra of proteins: The program Pronto. Methods Enzymol. 239: 288–307. [DOI] [PubMed] [Google Scholar]
- McKeehan, W.L., Wang, F., Kan, M. 1998. The heparan sulfate-fibroblast growth factor family: Diversity of structure and function. Prog. Nucleic Acid Res. Mol. Biol. 59: 135–176. [DOI] [PubMed] [Google Scholar]
- Olsen, S.K., Ibrahimi, O.A., Raucci, A., Zhang, F., Eliseenkova, A.V., Yayon, A., Basilico, C., Linhardt, R.J., Schlessinger, J., Mohammadi, M. 2004. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. 101: 935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz, D.M., Yayon, A., Flanagan, J.G., Svahn, C.M., Levi, E., Leder, P. 1992. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell. Biol. 12: 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, L., Burke, D.F., von Delft, F., Mulloy, B., Blundell, T.L. 2000. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407: 1029–1034. [DOI] [PubMed] [Google Scholar]
- Plotnikov, A.N., Schlessinger, J., Hubbard, S.R., Mohammadi, M. 1999. Structural basis for FGF receptor dimerization and activation. Cell 98: 641–650. [DOI] [PubMed] [Google Scholar]
- Plotnikov, A.N., Hubbard, S.R., Schlessinger, J., Mohammadi, M. 2000. Crystal structures of two FGF–FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101: 413–424. [DOI] [PubMed] [Google Scholar]
- Schlessinger, J., Plotnikov, A.N., Ibrahimi, O.A., Eliseenkova, A.V., Yeh, B.K., Yayon, A., Linhardt, R.J., Mohammadi, M. 2000. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6: 743–750. [DOI] [PubMed] [Google Scholar]
- Shimizu, A., Tada, K., Shukunami, C., Hiraki, Y., Kurokawa, T., Magane, N., Kurokawa-Seo, M. 2001. A novel alternatively spliced fibroblast growth factor receptor 3 isoform lacking the acid box domain is expressed during chondrogenic differentiation of ATDC5 cells. J. Biol. Chem. 276: 11031–11040. [DOI] [PubMed] [Google Scholar]
- Wang, F., Kan, M., Yan, G., Xu, J., McKeehan, W.L. 1995. Alternately spliced NH2-terminal immunoglobulin-like Loop I in the ectodomain of the fibroblast growth factor (FGF) receptor 1 lowers affinity for both heparin and FGF-1. J. Biol. Chem. 270: 10231–10235. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T.P., Harpal, K., Henkemeyer, M., Rossant, J. 1994. Fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Dev. 8: 3032–3044. [DOI] [PubMed] [Google Scholar]
- Yayon, A., Klagsbrun, M., Esko, J.D., Leder, P., Ornitz, D.M. 1991. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64: 841–848. [DOI] [PubMed] [Google Scholar]