Abstract
The crystal structure of the active-site mutant Cys25 → Ala of glycosylated human procathepsin S is reported. It was determined by molecular replacement and refined to 2.1 Å resolution, with an R-factor of 0.198. The overall structure is very similar to other cathepsin L-like zymogens of the C1A clan. The peptidase unit comprises two globular domains, and a small third domain is formed by the N-terminal part of the prosequence. It is anchored to the prosegment binding loop of the enzyme. Prosegment residues beyond the prodomain dock to the substrate binding cleft in a nonproductive orientation. Structural comparison with published data for mature cathepsin S revealed that procathepsin S residues Phe146, Phe70, and Phe211 adopt different orientations. Being part of the S1′ and S2 pockets, they may contribute to the selectivity of ligand binding. Regarding the prosequence, length, orientation and anchoring of helix α3p differ from related zymogens, thereby possibly contributing to the specificity of propeptide–enzyme interaction in the papain family. The discussion focuses on the functional importance of the most conserved residues in the prosequence for structural integrity, inhibition and folding assistance, considering scanning mutagenesis data published for procathepsin S and for its isolated propeptide.
Keywords: crystal structure, cysteine proteinase, procathepsin S, propeptide
Cathepsin S (EC 3.4.22.27) is a cysteine protease of the papain family (clan C1A) (Kirschke et al. 1995). In mammals, papain-like proteases are mainly involved in lysosomal protein degradation. Cathepsin S, first mentioned by Turnsek et al. (1975), has been described in detail by Kirschke et al. (1986). Like the related cathepsins L and K, it is an endopeptidase, but, in contrast to them, it is stable at neutral pH. Cathepsin S has been found almost exclusively in antigen-presenting cells, B lymphocytes, and dendritic cells, where it is involved in processing of the invariant chain of the MHC II complex (Riese et al. 1996). Selective inhibition of cathepsin S results in a delayed immune response (Riese et al. 1998). The enzyme is probably also necessary for normal retinal function, as suggested by adenovirus-mediated antisense gene transfer experiments in rats (Lai et al. 2000).
The structure of mature cathepsin S is known from X-ray data obtained with a Cys25 → Ser mutant (Turkenburg et al. 2002). It is a monomer with two domains (▶, L and R, left and right in the standard view). The interface of the two domains provides the structure for the active-site cleft, comprising the substrate-binding subsites (Schechter and Berger 1967) and the catalytic triad. The catalytic His164 and the stabilizing Asn184 are part of the polar surface of the R domain, whereas the L domain contributes the catalytic Cys25. The cathepsin S structure is not only very similar to that of the related mammalian enzymes, e.g., cathepsins K, L, and H, but also to the prototype of clan C1A, the plant enzyme papain (Turkenburg et al. 2002). As most other peptidases, cathepsin S derives from an inactive precursor by auto- or heterocatalytic processing. In the zymogen, the N terminus of the catalytic unit bears a 99 amino acid extension. Procathepsin S has been classified as a cathepsin L-like endopeptidase, one of three subfamilies in the papain clan (Karrer et al. 1993; Kirschke et al. 1995; Wex et al. 2000). The structures of two zymogens of this subfamily, procathepsins L (Coulombe et al. 1996) and K (LaLonde et al. 1999; Sivaraman et al. 1999), have been determined by X-ray analysis. It was found that there is no difference in the structure of the catalytic units between enzymes and zymogens, i.e., the catalytic power is already present in the zymogen, but nearly completely masked by the prosegment. Part of this prosequence occupies the active site cleft in the reverse nonproductive orientation. This inhibitory mechanism is not restricted to the cathepsin L-like subfamily, but was also found for cathepsins B and X, which feature much shorter prosequences (Sivaraman et al. 2000; Quraishi and Storer 2001). The prosegment structures of the two cathepsin L-like subfamily members known so far are similar. The N-terminal part forms a small domain (P-domain), whose backbone is built by two crossed helices, α1p and α2p. The P-domain anchors in the corresponding binding loop of the R-domain and forces the adjacent extended part of the prosequence into the active site cleft (Coulombe et al. 1996; LaLonde et al. 1999; Sivaraman et al. 1999).
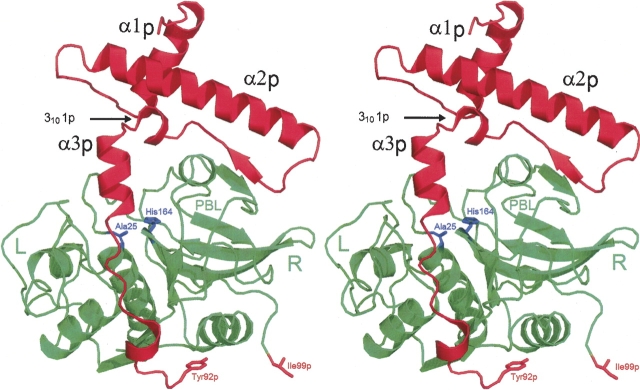
Figure 1.
Overall structure of the human procathepsin S Cys25 → Ala mutant. The amino acid sequence of the molecule comprises 316 residues (including the N-terminal methionine), of which 308 were visible in the electron density, 91 for the prosegment (in red, suffix “p”) and 217 for the catalytic unit (in green). The gap at the C-terminal end of the prosegment (Tyr92p–Ile99p) results from poorly defined electron density in this region. For clarity, only some of the secondary structure elements (α1p, α2p, 3101p, and α3p) of the prosequence are labeled. “L” and “R” specify the left and right domain of the catalytic unit. “PBL” is the prosegment-binding loop. The C terminus (Ile217) and the active site residues (Ala25 and His164) are indicated. Stereo picture prepared with PyMOL (DeLano 2002).
The inhibitory function of the prosegment has been intensely studied in vitro with isolated propeptides, revealing that they are competitive tight binding inhibitors of their mature enzymes (Fox et al. 1992; Taylor et al. 1995; Carmona et al. 1996; Volkel et al. 1996; Maubach et al. 1997). Inhibition studies with cognate, noncognate, and even chimeric propeptides of the closely related cathepsins S, L, and K, revealed that there is some selectivity, but not an absolute specificity (Carmona et al. 1996; Maubach et al. 1997; Guo et al. 2000). The crystallization of a cathepsin–propeptide complex has not yet been reported. Therefore, the orientation of an isolated inhibitory propeptide in the active site can only be suggested. As achieving selectivity is a challenge in the design of cysteine protease inhibitors with pharmaceutical significance, structural data on prosequence–enzyme interactions are of significant value (Chowdhury et al. 2002).
Further interest came from the discovery of prosegment-dependent folding of cathepsin L-like subfamily members, as discussed for the first time by Smith and Gottesman (1989), but proved only a decade later (Yamamoto et al. 1999; Wiederanders 2000). Identification of a missense mutation in the prosequence of cathepsin K in a patient with pycnodysostosis, resulting in a misfolded, inactive protein (Hou et al. 1999), illustrates the clinical relevance. One can assume that the prosequence forms a well-structured template, stabilizing a critical transition state of the catalytic unit in an enzyme-like manner. This lowers the energy barrier along the folding-reaction coordinate. Considering such prosequences as single-turnover catalysts, they have been termed foldases (Zhu et al. 1989).
The consequences of point mutations of highly conserved cathepsin S prosequence residues on refolding and inhibition have been studied systematically (Schilling et al. 2001). These results will be discussed in the light of the 2.1 Å X-ray structure of the Cys25 → Ala procathepsin S mutant presented here.
Results
Quality of the structural model and overall structure
The asymmetric unit of the crystals contains one molecule of the Cys25 → Ala procathepsin S mutant and 242 water molecules. The RMSD of bond lengths and angles of the refined structural model are 0.009 Å and 1.35°, respectively, indicative of good steric geometry. A Ramachandran plot (Ramachandran and Sasisekharan 1968) had 86.1% of the residues in the most favored region and 13.5% of the residues in the additionally allowed regions. No residues were found in the disallowed region, and only one residue (Tyr92p) adopted conformational angles in the generously allowed region. The published amino acid sequence (99 residues in the prosegment, marked with suffix p, and 217 in the catalytic domain) (Shi et al. 1992; Wiederanders et al. 1992) fits well into the electron density, with the exception of a few solvent-exposed residues at both ends of the prosequence, Ala1p–Gln2p and Lys93p–Arg98p at the N and C terminus, respectively. As electron density was poorly defined, these residues are very likely disordered in the crystal and, consequently, were omitted from the structural model (▶). Procathepsin S was crystallized in the Asn89p-glycosylated form, as indicated by the shape of the electron density extending from this side chain, but the difference electron density map did not provide insight into details of the glycosidic moiety. Structural integrity of the mutant zymogen was proven by SDS-PAGE; additional bands due to autocatalytic cleavage have not been observed.
As can be seen in ▶, the overall fold of procathepsin S is very similar to that of the human cathepsin L-like subfamily members elucidated so far, procathepsins K and L (PDB 1BY8 and PDB 1CS8, respectively) (Coulombe et al. 1996; LaLonde et al. 1999; data not shown) to procaricain from Carica papaya (PDB 1PCI) (Groves et al. 1996). Regarding the overall structure of the catalytic unit (McGrath et al. 1998; Turkenburg et al. 2002; Ward et al. 2002; Pauly et al. 2003), there is no difference between the zymogen and the mature enzyme, just as observed previously for cathepsins L and K when compared to their proenzymes (Coulombe et al. 1996; LaLonde et al. 1999; Sivaraman et al. 1999). Superpositon of mature cathepsin S (PDB 1GLO), (Turkenburg et al. 2002) onto the catalytic unit of procathepsin S (this study) results in an RMSD on Cαs of only 0.36 Å with no outliers (largest deviation 1.36 Å).
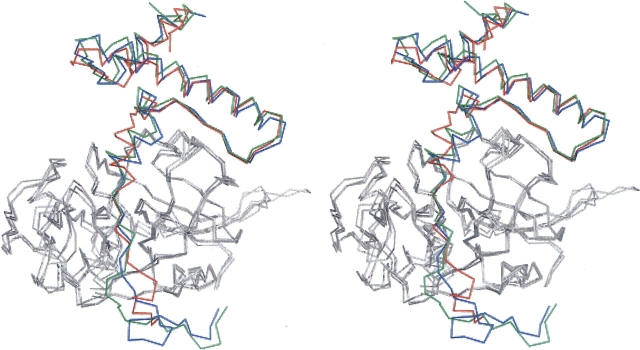
Figure 2.
Superposition of the Cα tracing of procathepsin S (red), K (blue), and L (green), as taken from PDB entries 2C0Y, 1BY8, and 1CS8, respectively. Only the prosequences are drawn in colors, the catalytic units are gray. The superposition underlines the high similarity for procathepsin S with procathepsin K (RMSD 0.79 Å) and procathepsin L (RMSD 0.88 Å). Stereo picture prepared with PyMOL (DeLano 2002).
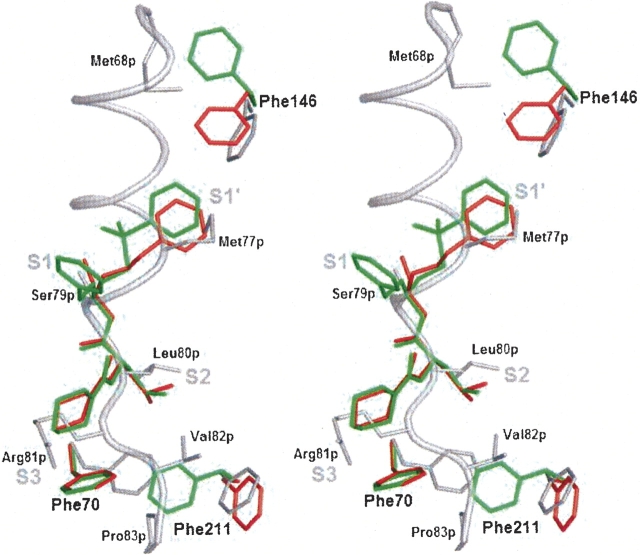
Due to occupation by part of the prosegment, subsite structures in the proenzyme are different in some details from those in mature cathepsin S in complex with various inhibitors (PDB entries 1MS6 and 1NPZ) (Ward et al. 2002; Pauly et al. 2003). As shown in ▶, the superposition of the structures reveals different orientations of the side chains of phenylalanines 70, 146, and 211.
Figure 3.
Flexibility of Phe146, Phe70, and Phe211 lining the substrate binding cleft, illustrated by superposition of procathepsin S and two cathepsin S/inhibitor complexes. (Gray) procathepsin S (this work); (red) cathepsin S/morpholine-4-carboxylic acid {1S-(2-benzyloxy-1R-cyanoethylcarbamoyl)-3-methyl butyl} amide complex (PDB entry 1MS6); (green) cathepsin S/4-morpholinecarbonyl-Phe-(S-benzyl)Cys-ψ (CH=O) complex (PDB entry 1NPZ). Stereo picture of superposition prepared with PyMOL (DeLano 2002).
Structure of the prosegment
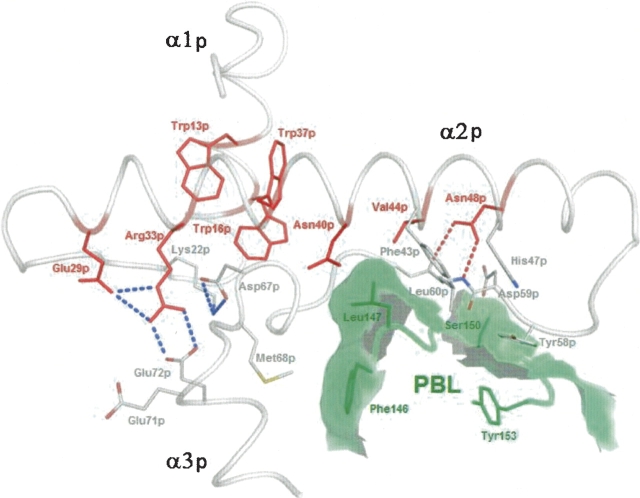
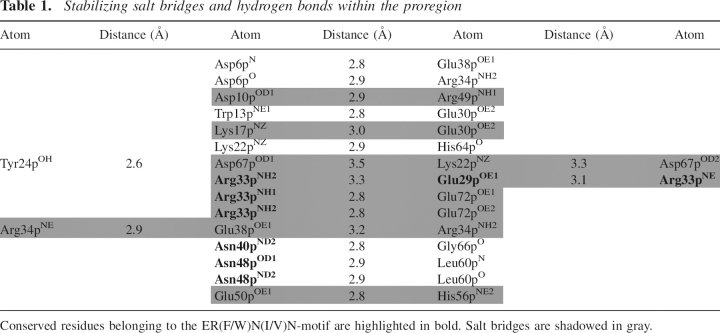
The N-terminal part of the prosegment forms the small P-domain, comprising three α-helices (α1p = 10p–20p, α2p = 29p–52p, α3p = 70p–76p) and one β-strand (β1p = 57p–60p) (▶). α2p and β1p are part of a hairpin structure. The side chains of Leu9p, Trp13p, Trp16p, Ile36p, Trp37p, Leu41p, Met62p, and the aliphatic part of Glu38p contribute to the hydrophobic core of the P-domain. Helix α1p, bearing Trp13p and Trp16p, and helix α2p with Trp37p are connected by a seven-residue loop. They intersect at an angle of ∼65°, so that in the center of the core, the aromatic rings of the specified tryptophans are positioned perpendicular to each other (▶). Outside the core, hydrogen bonds and salt bridges, listed in ▶, contribute to the stabilization of the P-domain. The salt bridges Lys22p–Asp67p and Arg33p–Glu72p are fixing the α3p-helix to the scaffold of the P-domain (▶). Amino acids contributing to the P-domain's stability are well conserved; sequence identity of the propeptides in the cathepsin L-like subfamily is between 26% and 58% (Karrer et al. 1993; Coulombe et al. 1996; Groves et al. 1998). Not only the primary, but also the tertiary structure of the prosegments of the cathepsin L-like subfamily members is very similar, as illustrated in ▶ by main chain superposition of procathepsins S, K, and L. In all these zymogens, the prosegments embrace the catalytic domains like a clamp. However, ▶ also reveals differences. The α3p-helix in procathepsin S is rotated within the paper plane by ∼20° counterclockwise compared to procathepsins K and L (and ∼10° clockwise to procathepsin B; data not shown). The consequence is an additional 310-helix (3101p = 64p–67p) between β1p and α3p, and larger distances between some α3p residues (Val73p, Ser75p, Leu76p, and Met77p) and the primed subsites in procathepsin S than in the related zymogens (▶,▶,▶). Beyond the C terminus of helix α3p, the prosegment adopts an extended conformation and follows the substrate-binding cleft, in a direction reverse to substrate orientation. A water molecule fills the cavity created by the catalytic site mutation (Cys 25 → Ala), forming hydrogen bonds with the side chains of Gln19 and His164 of the catalytic unit and with Ser79p, and α3p residue (▶).
Figure 4.
Conserved residues involved in stabilization of the procathepsin S P-domain. Members of the ER(F/W)N(I/V)N motif are labeled in red. Dashed red lines symbolize hydrogen bonds, and blue lines symbolize salt bridges. Residues of the prosegment binding loop (PBL) of the enzyme are printed in dark green, the calculated contact area of PBL with the prosequence is shadowed in light green. Picture prepared with PyMOL (DeLano 2002).
Table 1.
Stabilizing salt bridges and hydrogen bonds within the proregion
Figure 5.
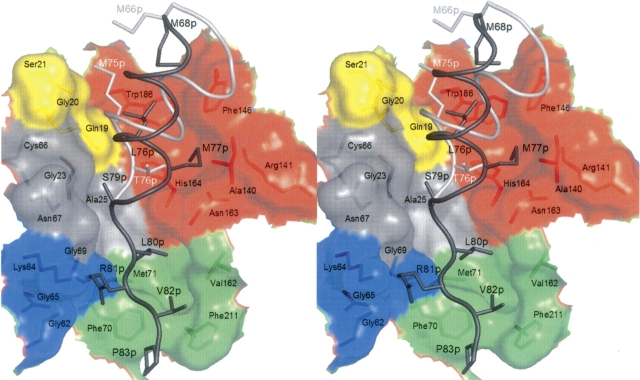
Interaction of the prosegment (cathepsin S, dark gray; cathepsin K, light gray; color code for numbering and chain) with the active site cleft of cathepsin S. The subsites are colored as follows: S2′, yellow; S1′, red; S1, gray; S2, blue; and S3, green. Stereo picture prepared with PyMOL (DeLano 2002).
Figure 6.
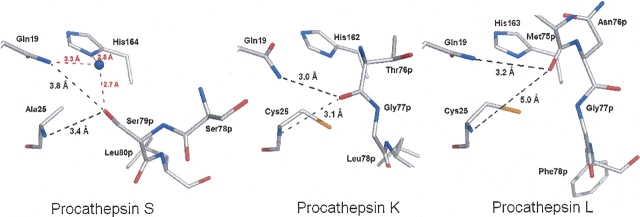
Prosegment interaction with the active site. (Black dashed lines) Coordination with main chain nitrogen of residue 25 and side chain nitrogen of Gln19 of the oxyanion hole. Procathepsin S, (red dashed lines) coordination of an additional water molecule (blue ball) filling the void created by the active site mutation Cys25 → Ala. The side chain of Ser79p projects into the oxyanion hole. Procathepsin K, the main chain oxygen of Thr76p reaches the oxyanion hole. Procathepsin L, the main chain oxygen of Met75p is closest to the oxyanion hole. Picture prepared with PyMOL (DeLano 2002).
Interactions between the prosegment and the catalytic unit
At first glance, the orientation of the cathepsin S prosegment relative to the catalytic unit seems to be very similar to what is seen in the procathepsins L, K, and B. In all these papain family members, the catalytic unit contacts the prosegment via two regions, i.e., the prosegment binding loop (PBL) and the substrate binding cleft (S′ and S subsites). Nevertheless, detailed analysis reveals some peculiarities of these two regions in procathepsin S. The following interactions are observed:
Residues of the α2p helix and the β1p strand pack against the platform of the PBL, a large omega loop comprising residues Pro143–Glu155. On one side of this loop, three residues of the β1p strand are involved in a conserved hydrophobic–hydrophilic–hydrophobic interaction profile: the side chains of Tyr58p, Asp59p, and Leu60p contact the side chains of Tyr153, Ser150, and Phe43p/His47p/Tyr58p, respectively (▶). This is a structural feature conserved in the subfamily, as in procathepsin K Tyr56p, Glu57p and Leu58p contact Tyr150, Lys147, and Tyr41p/His45p/Tyr56p and in procathepsin L Phe56p, Thr57p, and Met58p contact Tyr151, Glu148 (main chain), and Met41p/His45p/Phe56p. On the other side of the PBL, Phe146 makes hydrophobic contacts to a cluster of residues mainly from helix α3p: Leu65p, Met68p, Val73p, Leu76p, and Met77p. Helix α3p is differently oriented in procathepsin S compared to other procathepsins. This cannot be explained by altered hydrophobicity, as the hydrophobic character of this region is conserved for all residues except the last (procathepsin K: Leu63p, Met66p, Val71p, Met75p, and Thr76p; procathepsin L: Phe63p, Met66p, Phe71p, Met75p, and Asn76p).
The S1′ pocket, occupied by the last residue of α3p, Met77p, is formed by the hydrophobic moieties of Ala140, Arg141, Phe146, His164, and Trp186 (▶). Phe146 does not only border the S1′ pocket, but also contributes to an adjacent poorly defined pocket, distal from S1′. McGrath et al. (1998) assume that Phe146 could bind a P2′ residue, but the inhibitor in their structure does not provide an equivalent moiety. This may also be the reason for Turkenberg et al. (2002) not to predict more than one primed subsite in mature cathepsin S. However, the zymogen structure permits differentiation between S1′, faced by Met77p, and S2′, faced by Leu76p (▶). Crystal structures of procathepsins L and K revealed S2′ pockets, occupied by Met75p, as extensions of the crevice formed by the tryptophans 189 and 184, respectively, occupied by Met75p (Coulombe et al. 1996; LaLonde et al. 1999; Sivaraman et al. 1999). In procathepsin S, the shorter side chain of Leu76p, which substitutes for Met75p, is in van der Waals contact with Trp186, but does not completely fill S2′ (▶). Thus, not all sides of the S2′ pocket are well defined.
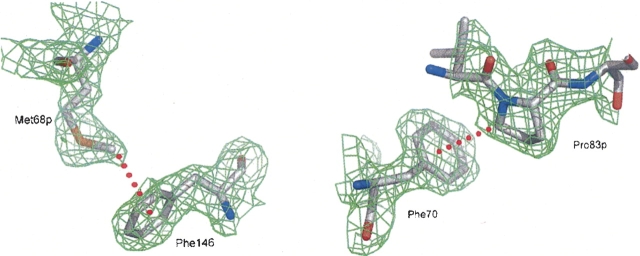
Another relevant contact on the S′ side is the C—H…π interaction of Met68pCE as the H-donor with the aromatic ring of Phe146 as the acceptor (▶, ▶) (Brandl et al. 2001). This interaction is unique within the cathepsin L-like subfamily, as the position of Phe146 is occupied by Leu144 in cathepsin L and by Gln143 in cathepsin K. Therefore, methionines 66p in the procathepsins L and K, corresponding to Met68p in procathepsin S, rotate away and are solvent exposed.
The last residue of helix α3p and the extended part of the prosegment (Ser78p–Pro83p) interact with the nonprimed subsites. As detailed in ▶, side chain and main chain atoms of Ser78p and Ser79p are in van der Waals contact with the S1 pocket. Compared to procathepsins K and L, helix α3p of procathepsin S is extended by Ser78p. The side chain hydroxyl group of Ser79p fills the oxyanion hole, formed by Ala25N and Gln19NE2. This mode of interaction is completely different from the situation in the procathepsins K and L, where the highly conserved oxyanion hole is occupied by the carbonyl oxygens of Thr76p and Met75p, respectively. This unique structural feature of procathepsin S is of local nature, as the adjacent Leu80p is well aligned with Leu78p and Phe78p in the procathepsins K and L, respectively (▶).
Analogous to the other procathepsins, Leu80p projects into the S2 pocket and Arg81p into the S3 pocket. Beyond this, Pro83pCD, formally in position P5, forms a C–H…π bond (Brandl et al. 2001) with the solvent-exposed Phe70 (▶) at the distal end of the P2 pocket. The position of Phe70 required for this interaction is only found in the proenzyme and forces Phe211 to swing out toward the bulk solvent.
Figure 7.
C–H…π interactions between prosequence and active-site cleft residues of cathepsin S. Prosequence residues are marked with “p.” The 2Fo−Fc electron density is contoured in green at a level of 1σ. Red dashed lines symbolize C–H…π interactions: Met68pCE…Phe146π (distance = 4.02 Å; left) and Pro83pCD…Phe70π (distance = 3.54 Å; right). Pictures prepared with PyMOL (DeLano 2002).
Discussion
The overall structure of procathepsin S is very similar to that of the two other cathepsin L- like subfamily members investigated so far, procathepsins L and K (▶) (Coulombe et al. 1996; LaLonde et al. 1999; Sivaraman et al. 1999). The active-site cleft between the two domains of the catalytic unit is already preformed in the precursor, but blocked by the C-terminal part of the proregion, which covers the catalytic center in a nonproductive orientation and anchors via residues Ser78p–Pro 83p in the nonprimed subsites. The larger part of the prosequence forms a small domain with the crossed helices α1p and α2p as scaffold. The P-domain contacts the prosegment-binding loop and the primed subsites of the catalytic unit via residues Tyr58p–Leu60p and the hydrophobic cluster Leu65p, Met68p, Val73p, Leu76p, and Met77p, respectively.
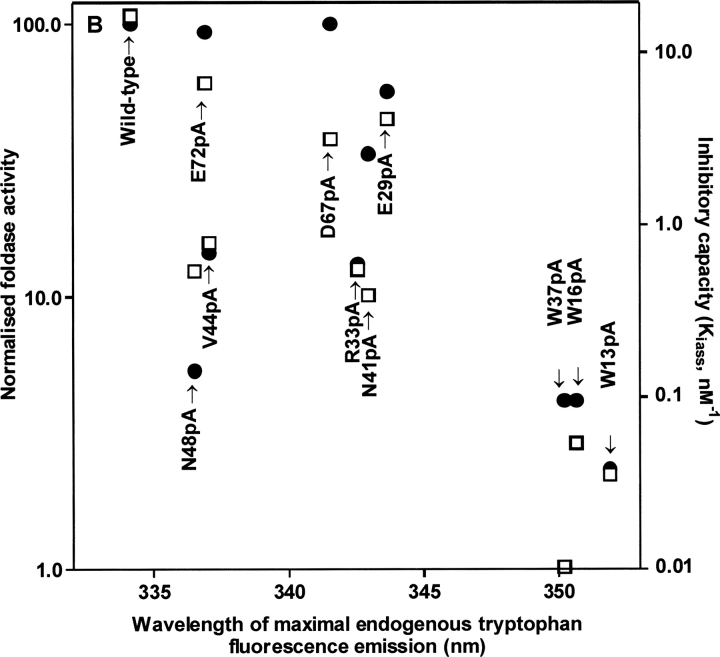
About one decade ago, motifs of conserved amino acids, e.g., ER(F/W)N(I/V)N (Karrer et al. 1993) and GNFD (Vernet et al. 1995), were identified in the prosequences of papain family members; in fact, the first of these is the basis for the recent subfamily classification (Wex et al. 2000). Conservation suggests functional and/or structural importance. The role of 10 conserved prosegment residues in procathepsin S has been probed by alanine scanning, both in vitro (Schilling et al. 2001; Pietschmann et al. 2002) and in vivo (Kreusch et al. 2000). Pietschmann et al. (2002) correlated the structural integrity of recombinant propeptide mutants, scaled by the red shift of the endogenous tryptophan fluorescence, with two basic functions, inhibition and folding, of the parental enzyme. These results are re-presented in ▶ because they are essential for the discussion of the functional importance of some structural features described in this paper. It can be seen that there is a rough reciprocal correlation between structural break down of the mutant P-domain and the functional parameters. Pulse-chase experiments in HEK-cells with zymogens, bearing the same mutations, confirmed this trend (Kreusch et al. 2000). This favors the hypothesis, based on physicochemical studies, that isolated propeptides, at least in complex with the corresponding enzymes, have the same fold as the proregions in the maternal zymogens (Maubach et al. 1997; Jerala et al. 1998; Ogino et al. 1999). Autonomous folding of the isolated cathepsin S propeptide was also shown by crystallization (Kaulmann et al. 2003). Consequently, the in vitro results with isolated cathepsin S propeptide mutants can be interpreted in the light of the zymogen structure.
Figure 8.
Effect of scanning mutagenesis of highly conserved prosequence residues on human cathepsin S propeptide function; “XnpA” means that amino acid X in position n of the propeptide is substituted by Ala. Propeptide mutants have been expressed, purified, and characterized as published by Kreusch et al. (2000). Renaturation rates (left ordinate, •) are from Pietschmann et al. (2002). Inhibiton constants (right ordinate, □) and red shift of the endogenous tryptophan fluorescence, standing for compactness of tertiary structure (abscissa), are from Schilling et al. (2001).
Tryptophans 13p, 16p, and 37p stabilize the core of the prodomain scaffold
The tryptophans 13p, 16p, and 37p form a cluster at the intersection point of the helices α1p and α2p (▶), thereby stabilizing the core of the prodomain through hydrophobic interactions. The integrity of this cluster is of outstanding importance for the P-domain structure. This is deduced from two experimental series, where each of these tryptophan residues was alanine-substituted by site-directed mutagenesis. ▶ illustrates that tryptophan fluorescence emission spectra of only three propeptide mutants, Trp13p → Ala, Trp16p → Ala, and Trp37p → Ala, show a strong red shift (Schilling et al. 2001). The three analogous zymogen mutants, when expressed in HEK cells, were degraded by the quality control machinery of the cell (Kreusch et al. 2000). All the other mutants specified in ▶, including those involved in salt bridges and hydrogen bonds (e.g., Glu29p, Arg33p, Asn48p, Asp67p, and Glu72p; cf. ▶), behaved more similarly to the wild-type control, in vitro as well as in vivo. Thus, the contribution of hydrophobic interactions between the three aromatic core residues to P-domain stability significantly exceeds that of salt bridges and hydrogen bonds, as discussed for procathepsin L by Coulombe et al. (1996).
Val44p and Asn48p strengthen P-domain binding to the enzyme
As shown in ▶, outside of the core of the prodomain the hairpin between the α2p-helix and the β1p-strand is stabilized by Val 44p interdigitating between Leu60p and Met62p. Furthermore, the Asn48p side chain is involved in two hydrogen bonds to main chain atoms of Leu60p, one of the residues anchored to the prosegment-binding loop of the enzyme. Val44p also forms van der Waals contacts to the side chain of Leu147 in the PBL. Alanine substitution of Val44p and Asn48p did not considerably shift the fluorescence emission to longer wavelengths (▶). The same mutations in the zymogen affected neither targeting nor maturation and secretion in transfected HEK cells (Kreusch et al. 2000). Although the P-domain structure of these mutants remained intact, the inhibitory capacity was shown to be reduced by about one order of magnitude (▶). The kinetics of complex formation of the propeptide mutants Val44 → Ala and Asn48 → Ala with the maternal enzyme revealed the reason for this: dissociation of the complex is much faster (0.02 sec−1 and 0.03 sec−1, respectively) than with other mutants and the wild-type propeptide (0.001 sec−1; all data from ▶ in Schilling et al. 2001). Thus, these two residues, members of the ER(F/W)N(I/V)N motif, contribute to the tight binding of the P-domain to the enzyme by stabilization of the hairpin-anchor as well as by direct interaction with the harboring PBL.
Autoprocessing may result from loss of the charge of Asp67p and Glu72p
Ionic interaction of two highly conserved ionizable groups (Asp65p, Glu70p) in procathepsin L has been discussed to be involved in pH-dependent modulation of prosegment–enzyme interaction, i.e., autoprocessing (Coulombe et al. 1996). The homologous acidic residues in procathepsin S participate in the salt bridges Asp67p–Lys22p and Glu72p–Arg33p (▶; ▶). The latter salt bridge connects the α3p helix to the N terminus of the α2p helix and may contribute to proper orientation of the α3 helix toward the active site cleft. The loss of the negative charge in position 67p was discussed to be involved in the pH-dependent activation of the papain precursor (Vernet et al. 1995). Substition of each of these carboxylic residues by alanine (in separate experiments) did not considerably alter the structure and function of the cathepsin S propeptide (▶) (Schilling et al. 2001). These data suggest that dissociation of the proregion from the active-site cleft results from the loss of the negative charge of the two acidic residues, Asp 67p and Glu72p, by protonation. The functional consequence is that lowering the pH triggers the zymogen activation in a cooperative manner.
Materials and methods
Cloning and expression of the human procathepsin S (Cys25 → Ala) mutant in baculovirus-infected insect cells
The complete sequence of human pre-procathepsin S, with the only potential glycosylation site in the proregion at Asn89p (HCS1/4) (Wiederanders et al. 1992), was cloned into the plasmid pcDNA3.1 (Invitrogen) as described previously (Kreusch et al. 2000). The Cys25 → Ala mutation, introduced to prevent autocatalytic processing of the zymogen, was created by PCR-based site-directed mutagenesis according to the ExSite protocol (Stratagene) with the following oligonucleotide primers (the altered bases are underlined):
sense 5′-TTGTGGTGCTGCCTGGGCTTTCAGTGCTGTGGG and antisense 5′-AAAGCCCAGGCAGCACCACAAGAACCTTGATATTTCAC.
The mutant pre-procathepsin S gene was cloned into the vector pBlueBac4.5 (Invitrogen). The proenzyme DNA sequence was amplified using the primers 5′-TCACAGGATCCATGAAACGGCTGGTTTG (sense) and 5′-ACGGGGAATTCCTAGATTTCTGGGTAAGAGG (antisense), including BamHI and EcoRI restriction sites (italic); the sense primer also contained the triplet for the leading methionine (underlined). The insert was sequenced to check correct amplification and ligation. Recombinant baculovirus was generated by homologous recombination between the transfer vector and linearized Bac-N-Blue AcMNPV DNA in Spodoptera frugiperda 9 (Sf9) cells (Invitrogen). Insectin liposomes were used to optimize the transfection. Recombinant viruses were plaque-purified, amplified, and tested for their ability to express the human procathepsin S mutant. Large-scale expression of the recombinant protein was performed by G. Schmid (Hoffmann LaRoche, Basel, Switzerland). After centrifugation at 10,000 rpm, the supernatant of the culture medium was concentrated 10-fold, and stored in aliquots at –20°C for subsequent purification.
Purification of recombinant human procathepsin S (Cys25 → Ala) mutant
Capturing was performed with affinity chromatography using 50 mL concanavalin A sepharose (Amersham Pharmacia Biotech) packed in an XK 16/20 column (Amersham Pharmacia Biotech) at a flow rate of 2 mL/min. The gel was equilibrated with 0.1 M Tris (pH 7.4), 0.5 M NaCl, 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2. After loading and washing (with three column volumes 0.1 M Tris at pH 7.4, 0.5 M NaCl), the protein was eluted with 0.25 M methyl-α-D-mannopyranoside, added to the washing buffer. Final purification of the recombinant human procathepsin S mutant to electrophoretic homogeneity was achieved by gel chromatography on a HiLoad Superdex-75 16/60 column (Amersham Pharmacia Biotech). Fractions containing procathepsin S were pooled and concentrated to 7.0 mg/mL by ultrafiltration (Amicon Centricon Plus-20, 5000 NMWL Millipore-Biomax 5).
Crystallization
The human procathepsin S Cys25 → Ala mutant protein was crystallized using the hanging drop vapor diffusion method. Crystals were obtained by equilibrating 2 μL of a 7.0 mg/mL protein solution with the same amount of reservoir solution (100 mM Tris-HCl, 200 mM magnesium acetate, 20% polyethylene glycol at final pH 7.5–7.8). Within 7 d at 20°C, platelet-shaped crystals appeared which grew to a maximum size of 500 × 250 × 30 μm within 4 wk.
Collection of diffraction data
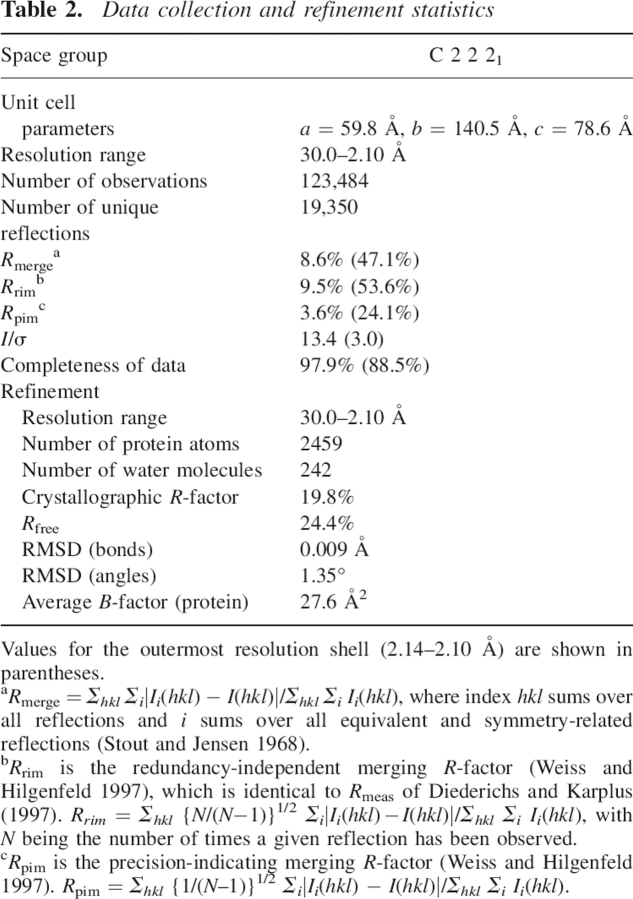
The crystals were flash-cooled at 100 K. No further cryoprotectant was needed in addition to the polyethylene glycol in the mother liquor. Initial data extending to 3 Å were collected using CuKα radiation produced by an FR591 rotating anode generator (Nonius) and used for structure solution. The final data set at 2.1 Å resolution was subsequently collected using the Joint IMB Jena–University of Hamburg–EMBL synchrotron beamline X13 at Deutsches Elektronen-Synchrotron, Hamburg, at a wavelength of 0.803 Å. Diffraction intensities were recorded using a Mar Research CCD detector (X-ray Research). Data collection parameters are given in ▶.
Table 2.
Data collection and refinement statistics
Data processing, structure solution, and refinement
Diffraction data were indexed, integrated, and scaled using the HKL package (Otwinowski and Minor 1997). Structure amplitudes were derived from the measured intensities by TRUNCATE (CCP4 1994). The asymmetric unit contains one molecule, corresponding to a packing density of 2.31 Å3/Da and a solvent content of ∼47% (Matthews 1968). Alignment of the procathepsin S amino acid sequence revealed the highest identity (∼55%) with procathepsin K (LaLonde et al. 1999; Sivaraman et al. 1999). Thus, procathepsin K (PDB entry code 1BY8) was used as the search model in molecular replacement calculations using the program AMoRe (Navaza 1994). The rotation and translation functions, calculated with reflections in the 15–4 Å resolution range, gave a solution with R = 0.461 and a correlation coefficient of 0.418. The model was refined using CNS (Brunger et al. 1998). After rigid body refinement, simulated annealing, and energy minimization in torsion angle space, Rcryst at 3.0 Å could be reduced to 0.35. Synchrotron data were used for further refinement. Five percent of the unique reflections were set aside for the calculations of Rfree (Brunger 1992). Several cycles of refinement, followed by refitting on a graphical workstation using the program O (Jones et al. 1991), resulted in a model that contained nearly all residues of procathepsin S (▶). Only residues 1p–2p and 93p–98p are missing due to poorly defined electron density. After introduction of 242 water molecules, Rcryst and Rfree could be reduced to 0.198 and 0.244, respectively. Refinement statistics are given in ▶. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under entry 2C0Y.
Chemical geometry and interactions
The chemical geometry of the structure was assessed by Ramachandran plots (Ramachandran and Sasisekharan 1968), calculated with the program PROCHECK (Laskowski et al. 1993). The secondary structure was defined using the program DSSP (Kabsch and Sander 1983). Hydrogen bonds, salt bridges, and van der Waals contacts were quantified with the program CONTACTS (CCP4 1994) and inspected with the program O (Jones et al. 1991). C–H…π interactions were identified according to Brandl et al. (2001).
Acknowledgments
We thank Stefan Kreusch for providing the plasmid encoding the human procathepsin S Cys25 → Ala mutant, and Georg Schmid (Hoffmann LaRoche) for the large-scale expression of the recombinant human procathepsin S Cys25 → Ala mutant. We also thank Depnath Pal and Maria Brandl for providing their programs for the identification of C–H…π interactions. This work was supported in part by the Deutsche Forschungsgemeinschaft (Schi 593/1-1). R.H. thanks the Fonds der Chemischen Industrie.
Footnotes
Reprint requests to: Bernd Wiederanders, Institut für Biochemie I, Klinikum der Friedrich-Schiller-Universität Jena, Am Nonnenplan 2, D-07740 Jena, Germany; e-mail: bwie@mti.uni-jena.de; fax 49-3641-938612.
Abbreviations: RMSD, root-mean-square deviation; PBL, prosegment binding loop; Residues and atoms have been named as in the Protein Data Bank entry (e.g., Met68pCE, Cɛ of residue methionine 68p).
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062401806.
References
- Brandl, M., Weiss, M.S., Jabs, A., Suhnel, J., and Hilgenfeld, R. 2001. C-H…π-interactions in proteins. J. Mol. Biol. 307 357–377. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. 1992. Free R-value—A novel statistical quantity for assessing the accuracy of crystal-structures. Nature 355 472–475. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., and Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Carmona, E., Dufour, E., Plouffe, C., Takebe, S., Mason, P., Mort, J.S., and Menard, R. 1996. Potency and selectivity of the cathepsin L propeptide as an inhibitor of cysteine proteases. Biochemistry 35 8149–8157. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S.F., Sivaraman, J., Wang, J., Devanathan, G., Lachance, P., Qi, H., Menard, R., Lefebvre, J., Konishi, Y., and Cygler, M., et al. 2002. Design of noncovalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides. J. Med. Chem. 45 5321–5329. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (CCP4) 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Coulombe, R., Grochulski, P., Sivaraman, J., Menard, R., Mort, J.S., and Cygler, M. 1996. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 15 5492–5503. [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- Diederichs, K. and Karplus, P.A. 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Biol. 4 269–275. [DOI] [PubMed] [Google Scholar]
- Fox, T., de Miguel, E., Mort, J.S., and Storer, A.C. 1992. Potent slow-binding inhibition of cathepsin B by its propeptide. Biochemistry 31 12571–12576. [DOI] [PubMed] [Google Scholar]
- Groves, M.R., Taylor, M.A., Scott, M., Cummings, N.J., Pickersgill, R.W., and Jenkins, J.A. 1996. The prosequence of procaricain forms an α-helical domain that prevents access to the substrate-binding cleft. Structure 4 1193–1203. [DOI] [PubMed] [Google Scholar]
- Groves, M.R., Coulombe, R., Jenkins, J., and Cygler, M. 1998. Structural basis for specificity of papain-like cysteine protease proregions toward their cognate enzymes. Proteins 32 504–514. [PubMed] [Google Scholar]
- Guo, Y.L., Kurz, U., Schultz, J.E., Lim, C.C., Wiederanders, B., and Schilling, K. 2000. The α1/2 helical backbone of the prodomains defines the intrinsic inhibitory specificity in the cathepsin L-like cysteine protease subfamily. FEBS Lett. 469 203–207. [DOI] [PubMed] [Google Scholar]
- Hou, W.S., Bromme, D., Zhao, Y., Mehler, E., Dushey, C., Weinstein, H., Miranda, C.S., Fraga, C., Greig, F., and Carey, J., et al. 1999. Characterization of novel cathepsin K mutations in the pro and mature polypeptide regions causing pycnodysostosis. J. Clin. Invest. 103 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerala, R., Zerovnik, E., Kidric, J., and Turk, V. 1998. pH-induced conformational transitions of the propeptide of human cathepsin LA role for a molten globule state in zymogen activation. J. Biol. Chem. 273 11498–11504. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Karrer, K.M., Peiffer, S.L., and DiTomas, M.E. 1993. Two distinct gene subfamilies within the family of cysteine protease genes. Proc. Natl. Acad. Sci. 90 3063–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulmann, G., Palm, G.J., Schilling, K., Hilgenfeld, R., and Wiederanders, B. 2003. An unfolding/refolding step helps in the crystallization of a poorly soluble protein. Acta Crystallogr. D Biol. Crystallogr. 59 1243–1245. [DOI] [PubMed] [Google Scholar]
- Kirschke, H., Schmidt, I., and Wiederanders, B. 1986. Cathepsin S. The cysteine proteinase from bovine lymphoid tissue is distinct from cathepsin L (EC 3.4.22.15). Biochem. J. 240 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke, H., Barrett, A.J., and Rawlings, N.D. 1995. Proteinases 1: Lysosomal cysteine proteinases. Protein Profile 2 1581–1643. [PubMed] [Google Scholar]
- Kreusch, S., Fehn, M., Maubach, G., Nissler, K., Rommerskirch, W., Schilling, K., Weber, E., Wenz, I., and Wiederanders, B. 2000. An evolutionarily conserved tripartite tryptophan motif stabilizes the prodomains of cathepsin L-like cysteine proteases. Eur. J. Biochem. 267 2965–2972. [DOI] [PubMed] [Google Scholar]
- Lai, C.M., Shen, W.Y., Constable, I., and Rakoczy, P.E. 2000. The use of adenovirus-mediated gene transfer to develop a rat model for photoreceptor degeneration. Invest. Ophthalmol. Vis. Sci. 41 580–584. [PubMed] [Google Scholar]
- LaLonde, J.M., Zhao, B., Janson, C.A., D'Alessio, K.J., McQueney, M.S., Orsini, M.J., Debouck, C.M., and Smith, W.W. 1999. The crystal structure of human procathepsin K. Biochemistry 38 862–869. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Macarthur, M.W., Moss, D.S., and Thornton, J.M. 1993. Procheck—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Matthews, B.W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33 491–497. [DOI] [PubMed] [Google Scholar]
- Maubach, G., Schilling, K., Rommerskirch, W., Wenz, I., Schultz, J.E., Weber, E., and Wiederanders, B. 1997. The inhibition of cathepsin S by its propeptide—Specificity and mechanism of action. Eur. J. Biochem. 250 745–750. [DOI] [PubMed] [Google Scholar]
- McGrath, M.E., Palmer, J.T., Bromme, D., and Somoza, J.R. 1998. Crystal structure of human cathepsin S. Protein Sci. 7 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza, J. 1994. AMoRe—An automated package for molecular replacement. Acta Crystallogr. A 50 157–163. [Google Scholar]
- Ogino, T., Kaji, T., Kawabata, M., Satoh, K., Tomoo, K., Ishida, T., Yamazaki, H., Ishidoh, K., and Kominami, E. 1999. Function of the propeptide region in recombinant expression of active procathepsin L in Escherichia coli. J. Biochem. 126 78–83. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Pauly, T.A., Sulea, T., Ammirati, M., Sivaraman, J., Danley, D.E., Griffor, M.C., Kamath, A.V., Wang, I.K., Laird, E.R., and Seddon, A.P., et al. 2003. Specificity determinants of human cathepsin S revealed by crystal structures of complexes. Biochemistry 42 3203–3213. [DOI] [PubMed] [Google Scholar]
- Pietschmann, S., Fehn, M., Kaulmann, G., Wenz, I., Wiederanders, B., and Schilling, K. 2002. Foldase function of the cathepsin S proregion is strictly based upon its domain structure. Biol. Chem. 383 1453–1458. [DOI] [PubMed] [Google Scholar]
- Quraishi, O. and Storer, A.C. 2001. Identification of internal autoproteolytic cleavage sites within the prosegments of recombinant procathepsin B and procathepsin S. Contribution of a plausible unimolecular autoproteolytic event for the processing of zymogens belonging to the papain family. J. Biol. Chem. 276 8118–8124. [DOI] [PubMed] [Google Scholar]
- Ramachandran, G.N. and Sasisekharan, V. 1968. Conformation of polypeptides and proteins. Adv. Protein Chem. 23 283–438. [DOI] [PubMed] [Google Scholar]
- Riese, R.J., Wolf, P.R., Bromme, D., Natkin, L.R., Villadangos, J.A., Ploegh, H.L., and Chapman, H.A. 1996. Essential role for cathepsin S in MHC class II-associated invariant chain processing and peptide loading. Immunity 4 357–366. [DOI] [PubMed] [Google Scholar]
- Riese, R.J., Mitchell, R.N., Villadangos, J.A., Shi, G.P., Palmer, J.T., Karp, E.R., De Sanctis, G.T., Ploegh, H.L., and Chapman, H.A. 1998. Cathepsin S activity regulates antigen presentation and immunity. J. Clin. Invest. 101 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter, I. and Berger, A. 1967. On the size of the active site in proteases. I Papain. Biochem. Biophys. Res. Commun. 27 157–162. [DOI] [PubMed] [Google Scholar]
- Schilling, K., Pietschmann, S., Fehn, M., Wenz, I., and Wiederanders, B. 2001. Folding incompetence of cathepsin L-like cysteine proteases may be compensated by the highly conserved, domain-building N-terminal extension of the proregion. Biol. Chem. 382 859–865. [DOI] [PubMed] [Google Scholar]
- Shi, G.P., Munger, J.S., Meara, J.P., Rich, D.H., and Chapman, H.A. 1992. Molecular cloning and expression of human alveolar macrophage cathepsin S, an elastinolytic cysteine protease. J. Biol. Chem. 267 7258–7262. [PubMed] [Google Scholar]
- Sivaraman, J., Lalumiere, M., Menard, R., and Cygler, M. 1999. Crystal structure of wild-type human procathepsin K. Protein Sci. 8 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraman, J., Nagler, D.K., Zhang, R., Menard, R., and Cygler, M. 2000. Crystal structure of human procathepsin X: A cysteine protease with the proregion covalently linked to the active site cysteine. J. Mol. Biol. 295 939–951. [DOI] [PubMed] [Google Scholar]
- Smith, S.M. and Gottesman, M.M. 1989. Activity and deletion analysis of recombinant human cathepsin L expressed in Escherichia coli. J. Biol. Chem. 264 20487–20495. [PubMed] [Google Scholar]
- Stout, G.H. and Jensen, L.H. 1968. X-ray structure determination: A practical guide. Macmillan, New York.
- Taylor, M.A., Baker, K.C., Briggs, G.S., Connerton, I.F., Cummings, N.J., Pratt, K.A., Revell, D.F., Freedman, R.B., and Goodenough, P.W. 1995. Recombinant pro-regions from papain and papaya proteinase IV are selective high affinity inhibitors of the mature papaya enzymes. Protein Eng. 8 59–62. [DOI] [PubMed] [Google Scholar]
- Turkenburg, J.P., Lamers, M.B., Brzozowski, A.M., Wright, L.M., Hubbard, R.E., Sturt, S.L., and Williams, D.H. 2002. Structure of a Cys25 → Ser mutant of human cathepsin S. Acta Crystallogr. D Biol. Crystallogr. 58 451–455. [DOI] [PubMed] [Google Scholar]
- Turnsek, T., Kregar, I., and Lebez, D. 1975. Acid sulphydryl protease from calf lymph nodes. Biochim. Biophys. Acta 403 514–520. [DOI] [PubMed] [Google Scholar]
- Vernet, T., Berti, P.J., de Montigny, C., Musil, R., Tessier, D.C., Menard, R., Magny, M.C., Storer, A.C., and Thomas, D.Y. 1995. Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J. Biol. Chem. 270 10838–10846. [DOI] [PubMed] [Google Scholar]
- Volkel, H., Kurz, U., Linder, J., Klumpp, S., Gnau, V., Jung, G., and Schultz, J.E. 1996. Cathepsin L is an intracellular and extracellular protease in Paramecium tetraurelia. Purification, cloning, sequencing and specific inhibition by its expressed propeptide. Eur. J. Biochem. 238 198–206. [DOI] [PubMed] [Google Scholar]
- Ward, Y.D., Thomson, D.S., Frye, L.L., Cywin, C.L., Morwick, T., Emmanuel, M.J., Zindell, R., McNeil, D., Bekkali, Y., and Girardot, M., et al. 2002. Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors. J. Med. Chem. 45 5471–5482. [DOI] [PubMed] [Google Scholar]
- Weiss, M.S. and Hilgenfeld, R. 1997. On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 30 203–205. [Google Scholar]
- Wex, T., Levy, B., Wex, H., and Bromme, D. 2000. Human cathepsins W and F form a new subgroup of cathepsins that is evolutionary separated from the cathepsin B- and L-like cysteine proteases. Adv. Exp. Med. Biol. 477 271–280. [DOI] [PubMed] [Google Scholar]
- Wiederanders, B. 2000. The function of propeptide domains of cysteine proteinases. Adv. Exp. Med. Biol. 477 261–270. [DOI] [PubMed] [Google Scholar]
- Wiederanders, B., Bromme, D., Kirschke, H., von Figura, K., Schmidt, B., and Peters, C. 1992. Phylogenetic conservation of cysteine proteinases. Cloning and expression of a cDNA coding for human cathepsin S. J. Biol. Chem. 267 13708–13713. [PubMed] [Google Scholar]
- Yamamoto, Y., Watabe, S., Kageyama, T., and Takahashi, S.Y. 1999. Proregion of Bombyx mori cysteine proteinase functions as an intramolecular chaperone to promote proper folding of the mature enzyme. Arch. Insect Biochem. Physiol. 42 167–178. [DOI] [PubMed] [Google Scholar]
- Zhu, X.L., Ohta, Y., Jordan, F., and Inouye, M. 1989. Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature 339 483–484. [DOI] [PubMed] [Google Scholar]