Abstract
Understanding substrate specificity and identification of natural targets of transglutaminase 2 (TG2), the ubiquitous multifunctional cross-linking enzyme, which forms isopeptide bonds between protein-linked glutamine and lysine residues, is crucial in the elucidation of its physiological role. As a novel means of specificity analysis, we adapted the phage display technique to select glutamine-donor substrates from a random heptapeptide library via binding to recombinant TG2 and elution with a synthetic amine-donor substrate. Twenty-six Gln-containing sequences from the second and third biopanning rounds were susceptible for TG2-mediated incorporation of 5-(biotinamido)penthylamine, and the peptides GQQQTPY, GLQQASV, and WQTPMNS were modified most efficiently. A consensus around glutamines was established as pQX(P,T,S)l, which is consistent with identified substrates listed in the TRANSDAB database. Database searches showed that several proteins contain peptides similar to the phage-selected sequences, and the N-terminal glutamine-rich domain of SWI1/SNF1-related chromatin remodeling proteins was chosen for detailed analysis. MALDI/TOF and tandem mass spectrometry-based studies of a representative part of the domain, SGYGQQGQTPYYNQQSPHPQQQQPPYS (SnQ1), revealed that Q6, Q8, and Q22 are modified by TG2. Kinetic parameters of SnQ1 transamidation (KMapp = 250 μM, kcat = 18.3 sec−1, and kcat/KMapp = 73,200) classify it as an efficient TG2 substrate. Circular dichroism spectra indicated that SnQ1 has a random coil conformation, supporting its accessibility in the full-length parental protein. Added together, here we report a novel use of the phage display technology with great potential in transglutaminase research.
Keywords: transglutaminase 2, phage display, glutamine-donor substrate, chromatin remodeling proteins, glutamine-rich
Transglutaminases (TGs; EC 2.3.2.13) (Aeschlimann and Paulsson 1994; Lorand and Graham 2003) catalyze the post-translational modification of proteins via a Ca2+-dependent acyl-transfer reaction between the γ-carboxamide group of protein-bound glutamine (Q-donor) and the ɛ-amino group of protein-bound lysine (K-donor) residues, which leads to the formation of a protease-resistant interchain ɛ(γ-glutamyl)lysine isopeptide bond, cross-linking the two proteins. TGs also incorporate naturally occurring polyamines into proteins, and can deamidate glutamines (Folk and Finlayson 1977).
Transglutaminase 2 (or tissue-type transglutaminase, TG2; for review, see Fesus and Piacentini 2002; Griffin et al. 2002) is a ubiquitous member of the transglutaminase family, acting at various locations as a multifunctional protein. Apart from a few cell types that sustain high levels of TG2 (Thomázy and Fesus 1989), expression of the protein is regulated by a range of transcription factors (retinoic acid, TGFβ, NF-κB, and AP) that control cell defense and maturation (Szegezdi et al. 2000). When activated by Ca2+ in the cytosol, TG2 modifies the cytoskeleton to induce stress fiber assembly and cell adhesion (Shing et al. 2001). In apoptotic cells, Ca2+ influx triggers extensive polymerization of intracellular proteins by TG2, resulting in the formation of detergent-insoluble structures (Pirreda et al. 1997). At membrane locations, TG2 moonlights as a large G-protein (Gh) participating in adrenergic receptor signaling via phospholipase Cδ1 activation (Nakaoka et al. 1994). Nuclear translocation of the enzyme leads to modification of histones and pRB, reflecting its possible roles in chromatin remodeling and gene regulation (Lesort et al. 1998; Kim et al. 2002a). Once exported to the cell surface, TG2 interacts with β-integrins and serves as a coreceptor for fibronectin, facilitating cell adhesion, spreading, and motility (Gaudry et al. 1999; Akimov and Belkin 2001). Also, the enzyme mediates remodeling and stabilization of the ECM via cross-linking (Aeschlimann and Thomazy 2000). TG2 is thereby implicated in neuronal growth and regeneration, bone development, angiogenesis, wound healing, cell proliferation, differentiation, and apoptosis (Aeschlimann and Thomazy 2000; Mahoney et al. 2000; Fesus and Piacentini 2002; Griffin et al. 2002). Recently, the pathophysiology of Celiac Sprue, autoimmune diseases, inflammation, cancer metastasis, fibrosis, and neurodegenerative conditions such as HD, AD, and PD was found to be associated with dysregulation of TG2 (Anderson et al. 2000; Nanda et al. 2001; Fesus and Piacentini 2002; Kim et al. 2002b; Shan et al. 2002; Andringa et al. 2004; Nemes et al. 2004).
A crucial step in understanding the biological role of TG2 is identification of its physiological substrates (for review, see Esposito and Caputo 2005). The majority of the so far known TG2-modified proteins are involved in cell motility (e.g., actin myosin, RhoA GTPase), cell–extracellular matrix interactions (e.g., fibronectin, collagen, laminin), and energetic intermediate metabolism of cells (e.g., glycolytic enzymes), while only a few organelle proteins have been identified as its substrates (Fesus and Piacentini 2002; Griffin et al. 2002; Esposito and Caputo 2005). An intriguing question, for example, is whether TG2 can act on poly(Q) and Q-rich domains found in a group of eukaryotic transcription factors (Freiman and Tjian 2002), or on extended poly(Q) stretches characteristic of (CAG)-repeat expansion diseases (Cooper et al. 2002).
The hunt for TG2 substrates is intense and is typically achieved by penetrating intact cells with labeled synthetic amine-donors [e.g., 5-(biotinamido)penthylamine, 5BP] and Q-donors (e.g., biotinyl-TVQQEL peptide) to isolate and identify in situ tagged proteins using proteomics (Orru et al. 2003; Ruopollo et al. 2003). However, the mechanism by which the enzyme selects substrate glutamines and lysines is still an enigma. A regularly updated list of identified TG substrate proteins (currently 150 entries) can be found in the Transglutaminase Substrate Database (TRANSDAB, http://www.biochem.dote.hu/TRANSDAB), and TG sites (currently 115 entries) are available in a searchable format at the Transglutamination Sites Database (TRANSIT, http://bioinformatica.isa.cnr.it/TRANSIT) (Facchiano et al. 2003). Comparison of the primary sequence of these modification sites or studies using synthetic peptides to address specificity at the level of individual amino acid positions around reactive residues have not concluded in any consensus recognition pattern (Aeschlimann et al. 1992; Coussons et al. 1992; Grootjans et al. 1995; Pastor et al. 1999). Our current knowledge is still limited to the fact that TG2 has higher specificity for Q-donor than K-donor substrates, that reactive Q and K residues must be accessible, and that some residue types are preferred or discouraging around them (Coussons et al. 1992). Interestingly, continuous stretches of glutamines (Qn) are readily modified in vitro (Cooper et al. 1999), and Qs found in the contexts of proline-rich plant storage proteins, glutens and gliadins, are also TG2-reactive (Piper et al. 2002; Vader et al. 2002). Unfortunately, the recently solved X-ray structure of TG2 (Liu et al. 2002) did not confer an ultimate explanation for the mechanism of substrate binding in the active site, although structural information on other TG family members is rapidly accumulating (Avhazi and Steinert 2003).
This study reports on the first use of a fully combinatorial approach to address TG2 specificity at the peptide sequence level via adaptation of the phage display technique, which is an effective and simple tool to find peptides showing specific binding affinities or activities toward target proteins (Burritt et al. 1996). We demonstrate that by using recombinant human TG2 as the target and specific elution to recover sequences binding to the active site, preferential selection of Q-donor substrates from a random heptapeptide library can be achieved, and that this procedure (1) can provide insight into the general criteria of TG2 substrate recognition, (2) has the potential to predict its natural substrates, and ultimately (3) may be suitable for obtaining highly efficient “super” substrates for the enzyme.
Results
Phage display selection of TG2-specific peptides
For selection of peptides, we used a commercially available 7-mer random library displayed on the minor coat protein (g3p) of phage M13 with a diversity of 2.8 × 109. Recombinant human TG2 was immobilized on glutathione-Sepharose beads via an N-terminal GST-fusion tag (Ambrus et al. 2001) and was incubated with an aliquot of the library (2 × 1012 phage particles). After unbound phages were washed off the matrix, the population remaining attached was nonspecifically eluted by low pH. The eluate was then amplified, and the above procedure was repeated twice except that a synthetic amine-donor TG2 substrate, 5-(biotinamido)penthylamine (5BP), was used in the elution steps as an attempt to specifically release phage clones displaying glutamine-donor substrate peptides. During the course of selection, the relative yield of phage recovery improved from 0.12% (first round) to 6.5% (third round), which is consistent with enrichment in binding clones. Therefore, individual phage clones from the second and third rounds (50 total) were amplified for further analysis. First, the affinity of each clone for TG2 was tested by ELISA, and those that showed concentration-dependent binding (44) (data not shown) were then subjected to DNA sequence analysis to identify the displayed peptide. The results revealed that while out of the 24 sequences selected in the second round only 11 contained one or more glutamines, this ratio increased to 15 in 20 after the third cycle. The enrichment of Q-containing sequences during the consecutive rounds is shown in the Supplemental Material.
The 26 Q-containing peptides identified are listed in ▶. The representation of glutamine within the selected pools increased from 5.1% (initial library) to 8.9% in the second round and to 16.5% in the third. Also, the proportion of glutamine-containing sequences was higher after rounds two (46%) and three (75%) than the calculated initial value (35.7%). In addition, glutamine occurs more than once in 10 of the peptides, and remarkably these residues are adjacent in seven of them. The above findings support the contention that our selection design facilitates enrichment of phage particles displaying glutamine-containing peptides.
Table 1.
Deduced amino acid sequence and glutamine-donor substrate character of peptides selected from a phage display library using transglutaminase 2

Transamidation of phage-displayed peptides by TG2
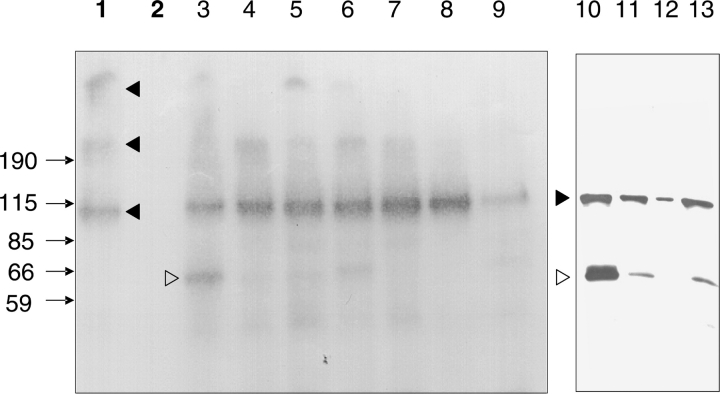
To test whether the above sequences (▶) are recognized by TG2 as glutamine-donor substrates, the amplified phage clones were subjected to in vitro transamidation using 5BP as the amine-donor. The incorporation of 5BP into phage particles was monitored by Western blotting, and, as shown in ▶ for representative examples, it gave rise to an avidin-reactive band at 62 kDa, which corresponds to the apparent molecular weight of the phage g3p protein. Two phage clones displaying the MPPPMRS and LMAKPTR peptides were used as negative controls and were found to be unsusceptible to transamidation (▶). This indicates that the phage does not expose efficient TG2-reactive glutamine residues, and therefore transamidation is restricted to the displayed sequence. Based on the intensity of luminescence by the 62-kDa band, peptides were classified as efficient or inefficient TG2 substrates (▶). Strong TG2-mediated 5BP incorporation was demonstrated for half of the peptides, including those containing two or more adjacent glutamine residues, and the strongest signals were obtained for PhQ2.7 (GQQQTPY), PhQ2.2 (GLQQASV), and PhQ3.11 (WQTPMNS).
Figure 1.
TG2-catalyzed modification of phage-displayed peptides from selection rounds 2 and 3. Phage particles were incubated at 37°C for 30 min in the presence of 5 mM CaCl2, 2 mM 5-(biotinamido)penthylamine (5BP), and 2 μg of GST-transglutaminase 2 (GST-TG2). Phage proteins were resolved on 10% SDS-PAGE and analyzed by Western blotting. Biotinylated protein bands were detected by streptavidin-biotinylated horseradish peroxidase conjugate and visualized by a chemiluminescent technique. (Lane 1) GST-TG2 incubated with 5BP; (lane 2) GST-TG2 incubated with 5BP and 5 mM EDTA as a negative control; (lanes 3–13) phage clones incubated with GST-TG2 and 5BP; (lane 3) PhQ2.7 (GQQQTPY); (lane 4) PhQ2.8 (GLQATPA); (lane 5) PhQ2.10 (VQETQRP); (lane 6) PhQ2.2 (GLQQASV); (lane 7) PhQ2.5 (QGRIPTS); (lane 8) phage clone displaying the peptide MPPPMRS as non-Q control; (lane 9) PhQ2.4 (VTQRLPL); (lane 10) PhQ3.11 (WQTPMNS); (lane 11) PhQ3.13 (QQLHLEA); (lane 12) phage clone displaying the peptide LMAKPTR as non-Q control; (lane 13) PhQ3.19 (SQLTLLP). Closed arrowheads mark bands corresponding to GST-TG2 species with incorporated 5BP. The open arrowhead marks the position of the gene III protein of M13 phage, which runs at an apparent molecular mass of 62 kDa. Lanes 1–9 and lanes 10–13 are from separate experiments.
Sequence analysis and comparison of the glutamine-donor peptides with known TG2 substrates
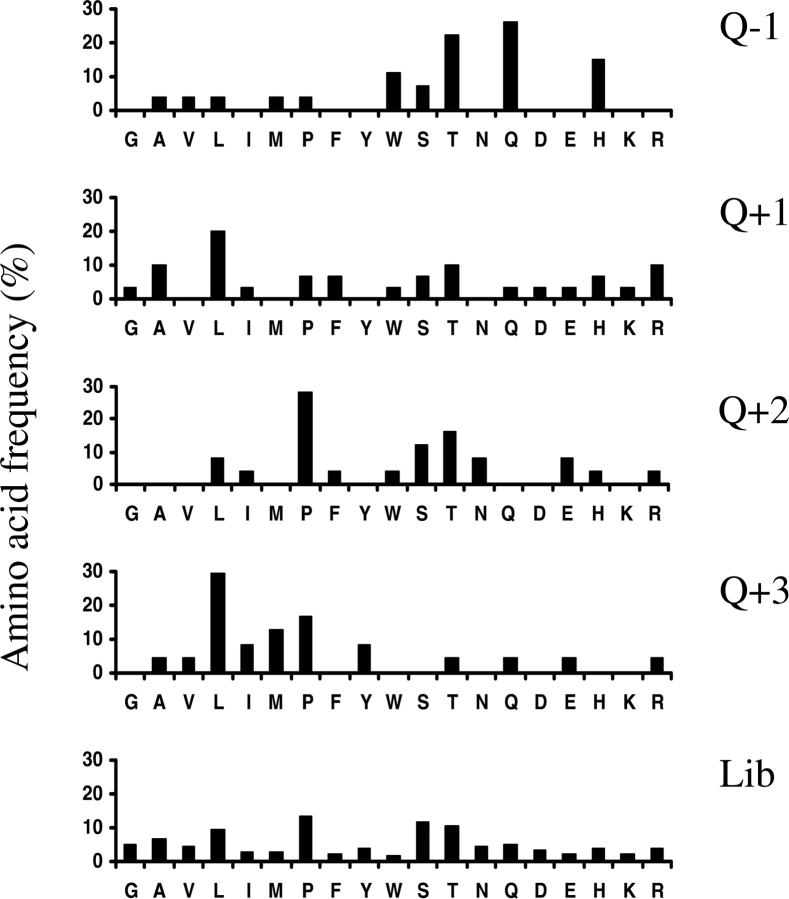
When binding peptides are selected from phage libraries, a consensus sequence generally appears after several consecutive cycles (Burritt et al. 1996). We aimed to identify such features in our peptides and, more importantly, to investigate whether they are functionally linked to TG2 substrate properties. At first inspection, no obvious convergence was observable apart from the reoccurrence of two motifs, WQXP and SQLXLLP (see phage clones PhQ3.3, PhQ3.11, PhQ3.18, and PhQ3.5, PhQ3.19, respectively, in ▶). As a more thorough approach, we chose to analyze whether the distribution of amino acids is different from that of the initial random library. The occurrence of amino acids was determined at individual positions around glutamines, and where adjacent Q residues were present, the most C-terminally located one was chosen arbitrarily as the point of reference (see ▶). As shown by the bar diagrams in ▶, significant deviations from random distribution were observed at relative positions Q − 1, where most of the residues are polar (70%); at Q + 2, where serine, threonine, and proline are over-represented (56%); and at Q + 3, where residues are predominantly aliphatic (75%). This is consistent with a consensus sequence of pQX(P,T,S)l, which is indicated in ▶.
Figure 2.
Relative occurrence of amino acids in the environment of glutamines within the selected glutamine-containing peptides compared to those of the initial library. Amino acid frequencies are shown for relative positions Q − 1, Q + 1, Q + 2, and Q + 3 (top diagrams), and the values for the initial library are presented in the bottom diagram.
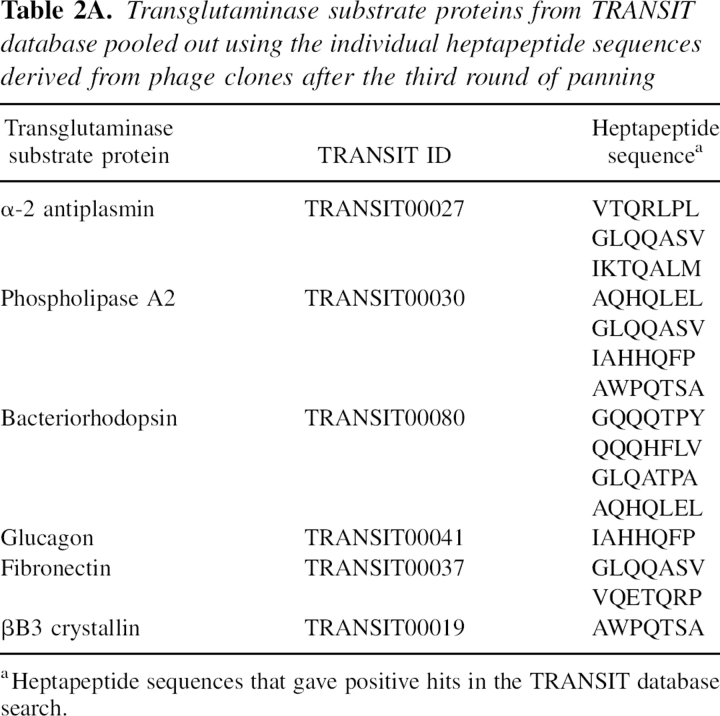
Next, the TRANSIT database (Facchiano et al. 2003), which contains 115 known transamidating sites for six different transglutaminases, was threaded using each of the phage-selected Q-peptides. As shown in Table ▶, 10 peptides gave positive hits matching six transglutaminase substrate proteins: phospholipase A2, glucagon, fibronectin, βA3 crystallin, α2-plasmin inhibitor, and bacteriorhodopsin. It has to be mentioned that the database is incomplete since modification sites in a large number of known TG2 substrates remain to be identified (Esposito and Caputo 2005).
Table 2A.
Transglutaminase substrate proteins from TRANSIT database pooled out using the individual heptapeptide sequences derived from phage clones after the third round of panning
As a broader approach, the phage-selected peptides were screened against the general features found in the primary sequence environment of TG2-modifiable glutamines as derived from in vitro experiments using natural and synthetic peptides (Pastor et al. 1999; Vader et al. 2002). Such characteristics include the transamidation-enhancing effect of charged and polar residues near reactive glutamines, adjacent glutamines, glycine at positions Q − 1, Q + 1, or Q + 2, proline at Q + 2, or hydrophobic residues at Q + 3 (Aeschlimann et al. 1992; Coussons et al. 1992; Grootjans et al. 1995; Pastor et al. 1999; Vader et al. 2002). In contrast, positively charged residues flanking Q, or proline at positions Q + 1 or Q + 3, can be discouraging for TG2 (Ballestar et al. 1996; Pastor et al. 1999; Vader et al. 2002). The dominating sequence pattern pQX(P,T,S,)l identified in the selected peptides perfectly matches the abovementioned transamidation-enhancing characteristics, and there is a good correlation between the presence of discouraging features (underlined in ▶) and low efficiency of TG2-catalyzed modification of the corresponding sequences.
Additionally, the so far identified transglutaminase substrate sequences listed in the TRANSDAB database were examined if they contain the pQX(P,T,S,)l pattern. The results clearly demonstrate that the consensus pattern deduced from the heptapeptides obtained in the third round of biopanning appears around known substrate glutamines (Table ▶).
Table 2B.
Protein sequence of known transglutaminase proteins listed in TRANSDAB (http://www.biochem.dote.hu/TRANSDAB)
The reactive glutamines are in italic, and the amino acid residues corresponding to the consensus pattern pQX(P,T,S,)l deduced using the selected phage clones are indicated with boldface.
Transamidation of GQQQTPY-like motifs within a native peptide
Using the peptide PhQ2.7 as a representative example of the efficient TG2 substrates selected from the library, we chose to investigate whether such sequences can be considered as TG2-modified motifs within native proteins. To address this, first we looked for human proteins that contain regions similar to PhQ2.7 by a BLAST search (NCBI, “short nearly exact sequence match” option) in the PIR (Protein Information Resource) database. Among the significant hits listed in ▶, the best was a group of SWI1/SNF1-related chromatin remodeling factors (Dallas et al. 2000), which contain two repeats of PhQ2.7-like sequences (80GQQGQTPY87 and 97QQQPPY102 in p270) within one of their two conserved glutamine/proline-rich domains (residues 1–220 and 944–1021 in p270). Therefore, for the purpose of in vitro cross-linking studies, we synthesized a 27-mer peptide (SnQ1) with the sequence 1SGYGQQGQTPYYNQQSPHPQQQQPPYS27 spanning the S77–S103 segment of p270 to incorporate both of the above repeats, and in total, nine glutamine residues.
Table 3.
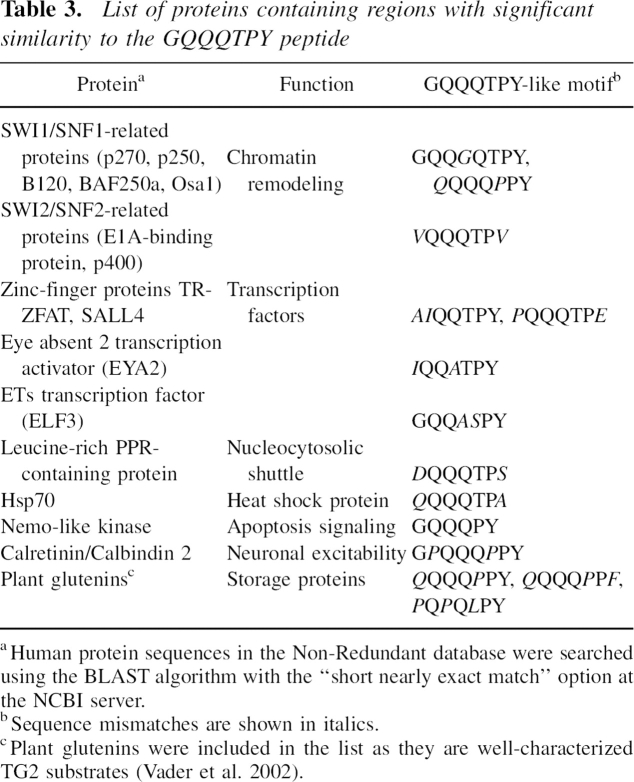
List of proteins containing regions with significant similarity to the GQQQTPY peptide
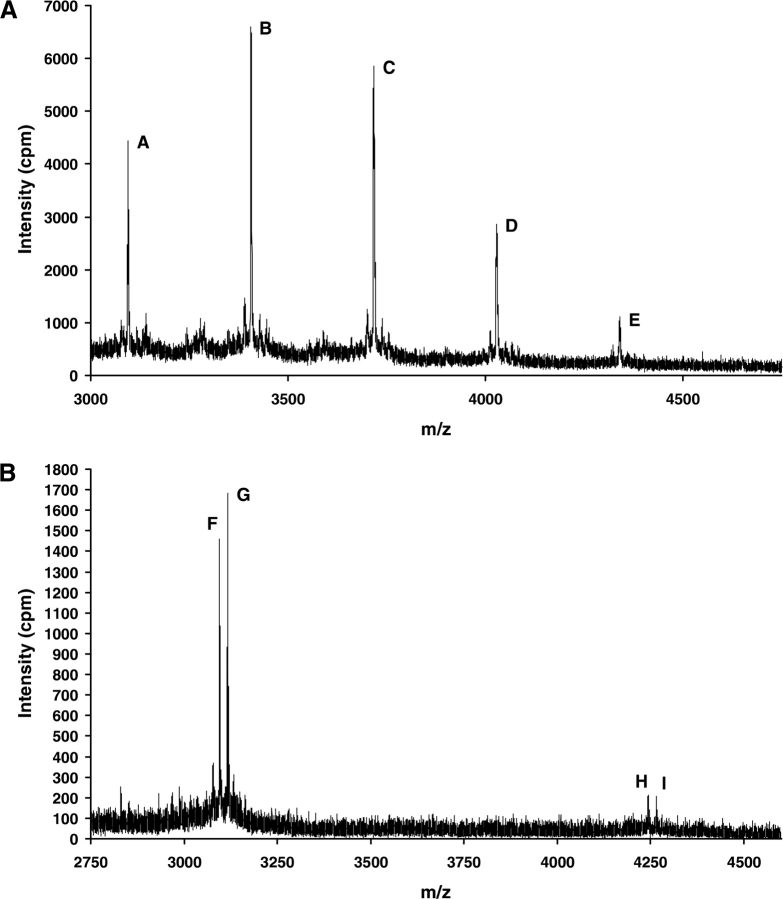
SnQ1 was treated with recombinant TG2 in the presence of 5BP or a biotinylated lysine-donor decapeptide, biotin-GPAVTAAPKK (BKP). Following incubation with added calcium for transglutaminase activation, aliquots were analyzed by MALDI/MS. ▶ shows the mass spectrometric traces obtained for the modification reactions, and peak listing and assignments for parental peptides and cross-linked products are presented in ▶. When SnQ1 is cross-linked to the amine donor, it is expected to show a mass increase of either ∼1148.7 Da or 311.5 Da, which equals the mass of either BKP (1165.6 Da, [M + H]+) or 5BP (329.5 Da, [M + H]+) minus the mass of NH3 (17.03 Da), which is released during the transamidation reaction. Accordingly, the MALDI/MS signals observed for the SnQ1–5BP (▶) and the SnQ1–BKP reactions (▶) could be assigned to cross-linked products (▶), resulting from the attachment of up to four molecules of 5BP (peaks B–E in ▶) or one molecule of BKP to SnQ1 (peaks H and I in ▶). The fraction of peptide converted to products during 1 h of incubation at 37°C was semiquantitatively estimated on the basis of the corresponding MS peak areas as 75% and 10% when using 5BP and BKP as amine-donor substrates, respectively.
Figure 3.
Mass spectrometric product analysis of TG2-catalyzed cross-linking reactions between the SnQ1 peptide and amine-donor compounds. MALDI/MS spectrum of aliquots from reactions between SnQ1 and (A) 5-(biotinamido)penthylamine (5BP) or (B) biotinyl-GPAVTAAPKK (BKP) after incubation in the presence of TG2. Signal assignments are given in ▶.
Table 4.
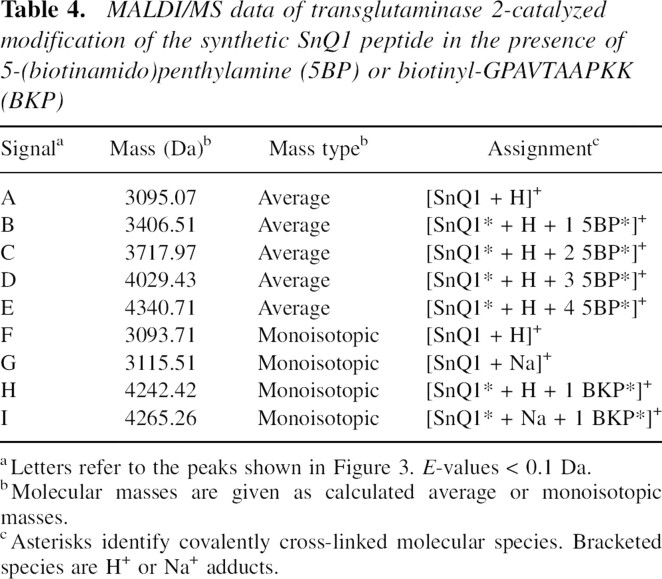
MALDI/MS data of transglutaminase 2-catalyzed modification of the synthetic SnQ1 peptide in the presence of 5-(biotinamido)penthylamine (5BP) or biotinyl-GPAVTAAPKK (BKP)
Identification of reactive glutamine residues in SnQ1
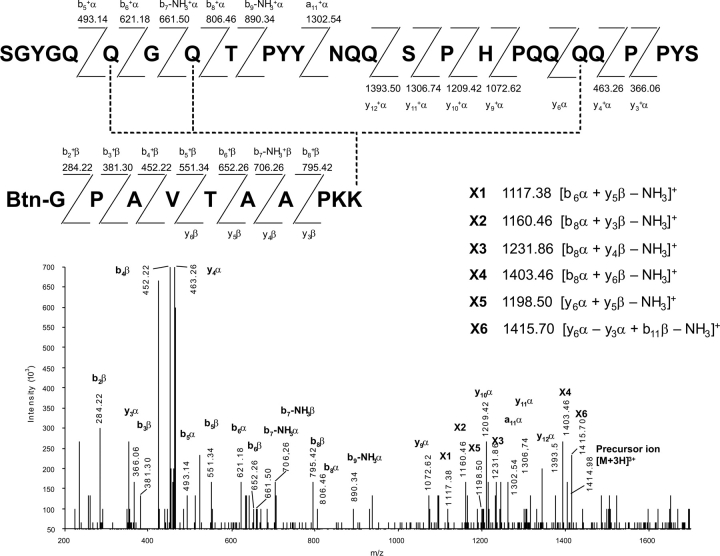
A more detailed mass spectrometric study was carried out on the SnQ1–BKP cross-linked product to identify modified glutamine residues. Reaction components were separated (RP-HPLC, C18 microbore column), and the peak corresponding to SnQ1 conjugated with amine donors was selected for fragmentation in a μLC/MS/MS system (Q-TRAP, Applied Biosystems, Inc.). An EPI spectrum of SnQ1–BKP (molecular mass 4244.1 Da, [M + 3H]3+) and signal assignments are presented in ▶. When interpreting this data, it has to be considered that the parental cross-linked species (SnQ1–BKP) probably consists of a population of isomeric compounds, since BKP can be attached to any of the four Q residues proven to be reactive using 5BP as the amine-donor substrate (▶). Although inherent properties of the parental SnQ1 peptide (trace not shown) hindered efficient fragmentation of SnQ1–BKP, the analysis software successfully assigned several signals to parental fragments (▶), and in addition, six molecular species with masses not assignable to fragments of either SnQ1 or BKP alone were also detectable (peaks X1–X6 in ▶). The latter species could be structurally explained as SGYGQQ* − AAPKK* (X1), SGYGQQGQ* + PKK* (X2), SGYGQQGQ* + APKK* (X3), SGYGQQGQ* + TAAPKK* (X4), Q*QPPYS + AAPKK* (X5), and Q*QP + TAAPKK* (X6), where fragments from SnQ1 and BKP are cross-linked by single isopeptide bonds between the Q and K residues marked with asterisks.
Figure 4.
Tandem mass spectrometric identification of TG2-modified glutamine residues in SnQ1. An aliquot of the TG2 reaction mixture of SnQ1 and BKP was resolved using μLC-ESI/MS, and the triply charged ion at m/z 1414.98, originating from the SnQ1 peptide cross-linked to biotinyl-GPAVTAAPKK (BKP), was selected for fragmentation. The MS/MS spectrum obtained from the fragmentation of the above precursor ion is shown below. Peptide sequences and interpretation are given above. Signals X1–X6 are assigned to cross-linked fragments, which are structurally explained on the right. Dashed lines depict the three alternative ɛ(γ-glutamyl)lysine isopeptide bonds between the two peptides as deduced from fragments X1–X6.
The data presented in ▶ and their analysis unequivocally demonstrate that Q6, Q8, and Q22 located within the PhQ2.7-like motifs of SnQ1 are TG2-reactive glutamines, while the fourth modification site could not be identified. It is noteworthy that SnQ1 has several glutamines that satisfy previously established transamidation criteria (Piper et al. 2002; Vader et al. 2002): (1) there are Q2 and Q4 stretches in its sequence; (2) residues Q5, Q6, and Q8 have G in their vicinity; and (3) Q15 and Q22 satisfy the QXP, while Q8 satisfies the QXP(Y) condition for efficient deamidation. On the same basis, Q14 and Q23 are less preferred targets for TG2.
Analysis of the structure of SnQ1
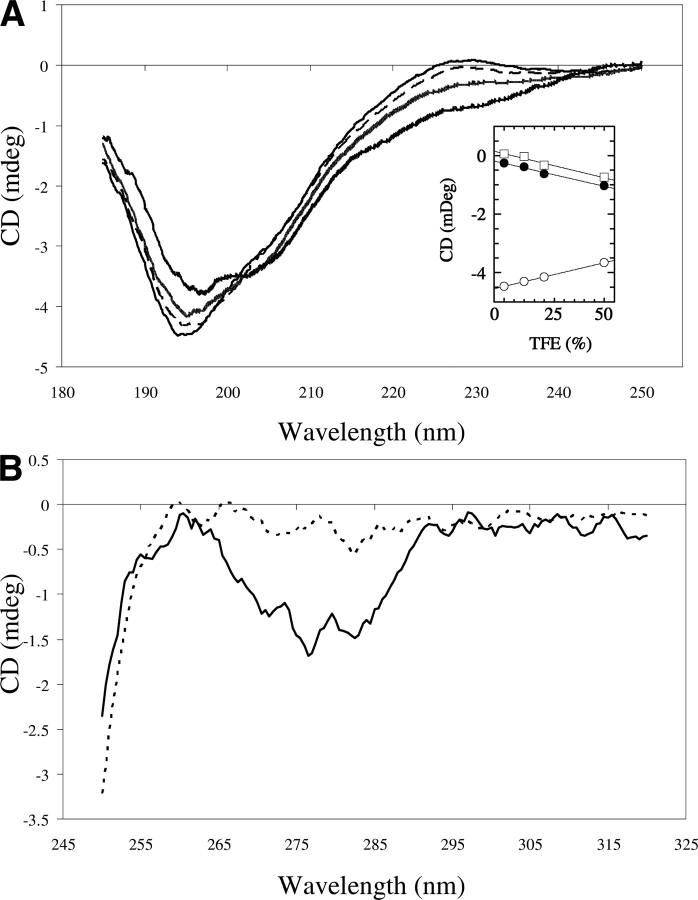
Structural characterization of the SnQ1 peptide was conducted in order to understand accessibility of the TG2-reactive Q residues in this sequence and to extrapolate to the parental domain in SWI1/SNF1-related proteins. The secondary structure of the N-terminal Q-P-rich domain was calculated using the PredictProtein tool. As expected for proline-rich regions, the domain has no ordered secondary structural elements detectable by the algorithm. To confirm this, SnQ1 was subjected to circular dichroism spectroscopic analysis. ▶ shows the far- and near-UV spectra of the peptide in the absence and presence of trifluoroethanol (TFE), which is a commonly used secondary structure inducer.
Figure 5.
Circular dichroism–based analysis of the structure of the SnQ1 peptide. (A) Far-UV CD spectra of SnQ1 (1 mg/mL) recorded in a 0.1-mm cuvette (Hellma), with 8-sec response time, in 10 mM phosphate buffer (pH 7.0) in the absence (black line) and the presence of 10% (black dashed line), 20% (light gray), and 50% (dark gray) trifluoroethanol. The inset shows a plot of ellipticity at 195 nm (open circle), 222 nm (closed circle), and 228 nm (open square) against the TFE concentration. (B) Near-UV CD spectra of the peptide (0.1 mg/mL, 1-mm cuvette) in the absence (continuous line) and the presence of 50% TFE (dashed line).
The far-UV spectrum (▶) recorded in phosphate buffer at pH 7.0 shows a minimum at 195 nm, which reports a predominant random coil conformation. An additional small positive peak at 228 nm indicates some Polyproline II helix character in the peptide. Increasing concentration of TFE (from 0% to 50%) caused linearly proportional elevation in CD signals at 208 nm and 222 nm (▶, inset), which represents α-helix formation. Such behavior is generally consistent with a natively unfolded peptide with inducible secondary structure. The near-UV spectrum (▶) shows distinct negative peaks at 282 nm and 278 nm, which may be attributed to either tertiary structure content or local stacking of Y residues (e.g., Y12 and Y13 are adjacent) in the peptide. In the presence of 50% TFE, these near-UV signals essentially disappear, showing that secondary structure formation disrupts rather than stabilizes the above interactions.
Finally, the peptide was subjected to a temperature unfolding study to clarify whether it contains any defined structure. Far- and near-UV CD spectra of SnQ1 were recorded at temperatures between 5°C and 95°C (5°C increments). The changes observed in CD signals were linearly proportional to the incubation temperature, which excludes cooperative unfolding (data not shown). All the above results correlate with a random-coil conformation for SnQ1 and supposedly for the Q-P-rich domain of SWI1/SNF1-related proteins, which may maximize accessibility of the reactive glutamines identified above.
Kinetics of SnQ1 binding and transamidation
Binding of SnQ1 to TG2 was studied by surface plasmon resonance spectroscopy using a streptavidin-coated gold chip (Biacore SA-Chip) charged with an N-terminally biotinylated version of the peptide (biotinyl-SnQ1). GST-TG2 was passed over this surface under conditions described in the Materials and Methods section, and the profiles obtained for the three channels charged with an increasing amount of the peptide (190, 630, and 900 RU) are shown in ▶. The results reflect that TG2 binds to SnQ1 in a concentration-dependent manner with complex kinetics. Dissociation constant and association/dissociation rate constants for channel 3 were estimated as ka = 9.7 × 10−4 M−1 sec−1, kd = 1.7 × 10−6 sec−1, and KD = 1.75 mM. Detailed kinetic analysis of the interaction was hindered by the fact that a large proportion of TG2 remained strongly attached to the chip surface, supposedly because of either covalent attachment (Fleckenstein et al. 2004) or a change in the affinity for SnQ1 as a result of modification arising from residual TGase activity (Piper et al. 2002).
Figure 6.
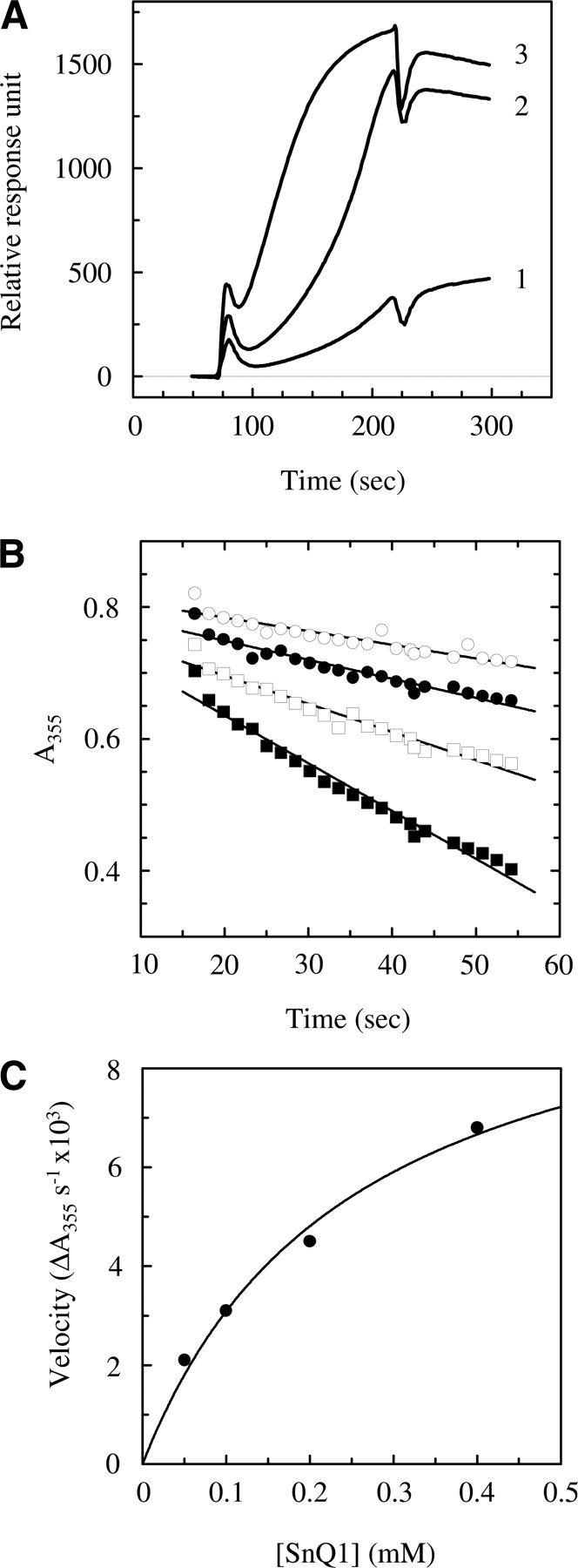
Kinetics of binding and transamidation of the SnQ1 peptide by TG2. (A) For surface plasmon resonance spectroscopic analysis of binding, a streptavidin-coated (SA) chip was charged with biotinyl-SnQ1 (2 μg/mL) to reach a 190, 630, and 900 response unit increase in channels 1, 2, and 3, respectively. Five microliters of GST-TG2 (2 μg/mL) was passed over the chip surface in running buffer (10 mM Tris-HCl at pH 7.4, 150 mM NaCl, 3.4 mM EDTA, 0.01% P20). The profiles for channels 1, 2, and 3 are shown after subtraction of the control. (B) Analysis of the progress of ammonia release when TG2 is incubated with 50 μM (open circles), 100 μM (closed circles), 200 μM (open squares), and 400 μM (closed squares) SnQ1 in the presence of 80 mM ethylamine as the amine-donor substrate and 5 mM CaCl2, using a continuous spectrophotometric linked enzyme assay as described in Materials and Methods. (C) Determination of kinetic parameters (Vmax and KM) of SnQ1 transamidation by nonlinear fitting of the Michaelis-Menten equation onto the initial velocities calculated by linear fitting of the corresponding progress curves and plotted against SnQ1 concentration.
The kinetic parameters of SnQ1 transamidation were determined by a continuous spectrophotometric linked enzyme assay (Day and Keillor 1999) using ethylamine as the amine-donor substrate and varying concentrations of the peptide (▶). Parameters were calculated by nonlinear fitting of the data (▶) as KMapp = 250 ± 66 μM, kcat = 18.3 ± 1.9 sec−1, and kcat/KMapp = 73,200, which are comparable to those of characterized peptide substrates (Day and Keillor 1999).
Database searches to identify potential glutamine-donor substrate proteins
To further assess whether the technique can provide predictions toward yet unidentified TG2 substrates, we carried out database searches by feeding in each of the efficient Q-donor sequences from ▶. Protein hits gathered by the “short nearly exact matches” algorithm of the NCBI-BLAST (National Center for Biotechnology Information Basic Local Sequence Alignment Tool) were kept if (1) the significance of similarity exceeded a set level (E < 2500, the high value is a consequence of using short sequences in the search tool), (2) at least one Q residue was retained from the parental sequence, (3) the calculated hydrophobicity of the respective protein region supported solvent exposition (Chothia 1976), and (4) were of human or mammalian origin. In addition, significant hits from other source organisms with anticipated biological relevance were also considered.
Out of the 61 proteins that satisfied the above conditions, 14 are already known TG2 targets (e.g., pRB1, huntingtin, L-TBP, hsp70, spectrins, importin β, ribosomal subunits, viral envelope proteins, and plant glutens) (Esposito and Caputo 2005). It was also apparent that the majority of the resulting proteins (45 out of 61) fall into a few functional groups, transcription factors and chromatin remodeling complex proteins (13), intracellular signaling and receptor proteins (14), cytoskeletal and extracellular matrix proteins (12), and intracellular traffic-related proteins (6). The rest of the proteins are related to protein folding and synthesis, metabolism, and in addition, several are of viral or pathogenic bacterial origin (see Supplemental Table S2).
Five native peptides (SnQ2–6) randomly chosen from database hits were synthesized and analyzed for TG2 reactivity using the same MALDI-TOF approach as described above for SnQ1 and BKP as the amine-donor partner. The results of cross-linking experiments (data not shown) revealed that SnQ1 is the most efficient TG2 substrate, followed by RRGLQQSSVN (SnQ3) > GALQQASVII (SnQ2) > AEVQETQRTPSA (SnQ5), while peptides SGGMQATPAT (SnQ4) and GVEETQRPPTL (SnQ6) were not modified by the enzyme (the origins of sequences are described in Materials and Methods). The extent of modification in each case was in agreement with the reactivity of the parental phage-selected sequences, namely, SnQ1, SnQ2, SnQ3, and SnQ5 show similarity to efficient heptapeptides (PhQ2.7, PhQ2.2, and PhQ2.10), while SnQ6 lacks one of the glutamines from PhQ2.10, and SnQ4 is derived from the inefficient PhQ2.8 clone.
Discussion
Transglutaminase 2 plays versatile roles in several physiological processes by catalyzing post-translational modification of a wide range of protein substrates at distinct subcellular and extracellular locations (Fesus and Piacentini 2002; Griffin et al. 2002; Esposito and Caputo 2005). Although the broad specificity of the enzyme for its targets may provide the flexibility needed to achieve the variety of functions, it also necessitates that the selection of a specific subset of proteins related to a particular biological event must be controlled by additional factors like cell type- and tissue-dependent abundance and localization of the enzyme and its substrates, local availability of Ca2+ and absence of inhibitors, and finally, the physical accessibility of modification sites on the individual molecules. While the discrimination between target and nontarget residues is primarily governed by secondary and/or tertiary structural features of the substrates, the enzyme also exerts preference at the level of primary sequence, especially around reactive glutamines (Aeschlimann et al. 1992; Coussons et al. 1992). Although some general rules have been established for the recognition of modification sites by TG2, its mechanism has not been explained at the molecular level. Better understanding might emerge from current structural studies on transglutaminases (Liu et al. 2002; Avhazi and Steinert 2003), but until enzyme–ligand complex structures become available, studies using peptide substrates are crucial in dissecting binding mechanisms (Pastor et al. 1999; Vader et al. 2002).
To address TG2 substrate specificity, we developed a phage display/biopanning technology based on a commercially available random library to select a range of glutamine-donor heptapeptides via biopanning on recombinant human TG2 and eluting bound phage particles with an amine-donor TG2 substrate. Phage clones recovered after the second and third biopanning rounds were then subjected to screening for TG2 reactivity, which is based on the detection of the incorporation of a labeled amine-donor into phage particles by Western blotting. By this means, we could categorize the phage-displayed heptapeptides as efficient or inefficient glutamine-donor substrates (▶; ▶) and also identified several highly TG2-reactive sequences, e.g., GQQQTPY (PhQ2.7), GLQQASV (PhQ2.2), and WQTPMNS (PhQ3.11).
The analysis of the sequence environment of glutamines in the selected peptides revealed a rough consensus pattern, pQX(P,T,S)l (▶; ▶), which is in agreement with the characteristics of previously identified TG2 modification sites. For example, such general rules as that the vicinity of polar residues, proline at Q + 2, or hydrophobic amino acids at Q + 3 ensure enhanced modification of substrate glutamines are met, and the pattern is similar to the highly TG2-reactive motif QXP(F,Y,W,M,L,I,V) found in gluten peptides (Pastor et al. 1999). In addition, searches in the TRANSIT database using the phage-selected peptides gave positive hits for six known transglutaminase sites (Table ▶), and the above consensus appears in several glutamine-donor substrate proteins listed in the TRANSDAB database (Table ▶).
While the final version of our manuscript was prepared, similar observations were reported by Sugimura et al. (2006), who screened a dodecapeptide phage display library for preferred TG2 substrate sequences and found marked tendencies in glutamine-donors as Q-X-P-ϕ-D-(P), Q-X-P-ϕ, and Q-X-X-ϕ-D-P (where X and ϕ represent nonconserved and hydrophobic amino acids, respectively). Although they used a different affinity approach, namely, their selection was based on (1) the incorporation of a biotinylated primary amine by the enzyme and (2) mono-avidin affinity chromatographic purification of the biotinylated phage particles, their results are in accordance with our findings [p-Q-X-(P,T,S)-l, where p and l stand for polar and aliphatic amino acids, respectively].
Some features of the consensus identified here may improve our present knowledge about the substrate recognition mechanism of TG2. For example, the over-representation of polar residues at position Q − 1 can emphasize H-bond formation with active-site residues at the corresponding site. Also, alternative binding conformations or mechanisms may exist for substrates with proline as opposed to serine or threonine at Q + 2. Although aliphatic residues at Q + 3 suggest a hydrophobic pocket, some of our peptides have a proline at this position.
To put these theories into structural context, we used molecular modeling to investigate the TG2 transamidating site. Since X-ray structure of the catalytically active form is not available for TG2, we investigated the calcium-bound TG3 structure combined with the results of a modeling study on glutamine substrate binding by TG3 (Avhazi and Steinert 2003) to identify active site residues in the vicinity of distinct amino acid positions around the reactive glutamine in a bound substrate. In TG3, Gln271 is close to Q − 1, while Ser325 and Val326 are in proximity to Q + 3. The corresponding TG2 residues are Gln276, Met330, and Ile331, respectively. In a TG3-based homology model of TG2, the side chains of Met330 and Ile331 form a flat hydrophobic surface rather than a pocket, which can interact with various aliphatic amino acids, including a proline ring at the Q + 3 position, while the polar residues at Q − 1 can form H-bonds with Gln276.
To further investigate whether the technique has a potential to predict TG2-modification sites in native protein sequences, we chose one of the most efficient Q-donor peptides, PhQ2.7 (GQQQTPY), as a representative example. A search in the human protein database revealed that several chromatin remodeling complex proteins and transcription factors contain stretches that are similar to PhQ2.7. The best match was found in the N-terminal glutamine-rich domain of SWI1/SNF1-related proteins (p270, BAF250a, p250, hOsa1), which are involved in unique gene regulatory activities in, i.e., myelocyte differentiation, and contain two conserved glutamine/proline-rich domains with yet unclear function (Dallas et al. 2000). Since existing recombinant versions of the above proteins do not contain the Q-P-rich regions, we decided to synthesize a 27-mer model peptide (SnQ1). MALDI/MS-based analysis revealed that at least four of the nine glutamines in SnQ1 are readily transamidated by TG2 in vitro, and by a tandem mass spectrometric study, we identified three modified glutamines as lying within the two GQQQTPY-like motifs (GQQGQTPY and QQQQPPY).
Remarkably, the above domain of SWI1/SNF1-like proteins resembles the sequence of the glutamine-rich plant storage proteins, glutenins and gliadins (see BLAST Link at NCBI), and this similarity is manifested in a common spatial arrangement of Q, P, and aromatic amino acids (e.g., common QXPY motifs in SWI1/SNF1 vs. QXPF/PY in glutenins; ▶). Importantly, positioning of P relative to Q is an underlining condition in determining TG2 reactivity for gluten peptides (Piper et al. 2002; Vader et al. 2002). Considering the efficient TG2-catalyzed deamidation reported for long gluten-derived peptides, the above sequence relationship adds to the hypothesis that the Q-P-rich domains of SWI-SNF-related proteins can be modified by the enzyme, although mass spectrometric analysis failed to detect deamidation products when SnQ1 was treated with TG2 in the absence of amine-donors.
Far- and near-UV CD spectroscopic studies elucidated the fact that SnQ1 has a predominantly random-coil conformation with a slight polyproline II-helical character. In addition, using TFE, some inducible α-helical feature was also detected that might have functional significance in the full-length parental proteins. Coincidently, a CD- and NMR-based study of the structure of immunogenic gluten peptides and their analogs also revealed that they are in an extended conformation with strong polyproline II helical propensity, which correlates with TG2 specificity for such sequences (Parrot et al. 2002).
The kinetics of binding SnQ1 to TG2 was also investigated partly in light of previous research that demonstrated that TG2 can form stable noncovalent adducts with long gluten-derived dietary peptides under steady-state conditions (Piper et al. 2002; Hausch et al. 2003). A surface plasmon resonance spectroscopic analysis was carried out using a biotinylated version of SnQ1 binding to TG2 with complex kinetics and calculated the dissociation constant for the process as KD = 1.75 mM. In addition, we observed permanent attachment of TG2 to the peptide, which may reflect the formation of adducts in the absence of a suitable amine-donor partner. Finally, we used a continuous spectrophotometric transglutaminase assay to determine the apparent kinetic parameters of SnQ1 transamidation. The observed values of KMapp as 250 μM, kcat as 18.3 sec−1, and kcat/KMapp as 73,200 represent the overage of the conversion process, which may involve more than one glutamine residue within a single peptide. The above parameters are comparable to those determined for various peptide substrates (Day and Keillor 1999) and classify SnQ1 as an efficient substrate sequence for TG2.
The similarity searches carried out with the phage-selected peptides revealed several TG2 substrate proteins and other proteins that fall into distinct functional categories. It is remarkable that most of those categories correlate with established roles of TG2 in G-protein signaling and organization of the cytoskeleton and extracellular matrix, as well as receptor-mediated endocytosis in different cellular systems (Fesus and Piacentini 2002; Griffin et al. 2002), while apart from a few known examples of nuclear TG2 targets, e.g., pRB, histones or huntingtin, transcription factors and chromatin remodeling proteins may represent a novel range of substrates, underlining the potential involvement of TG2 in nuclear processes, as already proposed by previous research (Shimizu et al. 1996; Zhang et al. 1998).
In summary, using in vitro cross-linking studies, structural characterization, and binding and kinetic analysis, we demonstrated that a Q-P-rich domain of SWI/SNF1 family chromatin remodeling factors contains a flexible sequence (SnQ1) that makes glutamine residues highly efficient substrates for TG2-catalyzed transamidation. The spatial arrangement of Q and P residues in the above domain promotes an overall extended conformation preferred by TG2 with concomitant accessibility of Q residues. In general, glutamine-proline-rich domains just like homopolymeric glutamine segments are primarily associated with a large family of eukaryotic transcription factors (including SP1, nuclear hormone receptors, TATA binding protein, etc.) and function as activation domains working from proximal promoter positions in conjunction with an enhancer element (Gerber et al. 1994; Freiman and Tjian 2002). Potentially, any of the four types of transglutaminase-catalyzed reactions can occur on these sites, including homopolymerization or cross-linking to lysine-donor substrate proteins, polyaminylation, which results in the introduction of positive charge, or deamidation leading to increased negative charge via Q-to-E conversion. The above post-translational modifications can affect protein structure and function in various ways, exemplified by activation of PLA2, latent TGFβ, RhoA, inhibition of pRB, modulation of SP1 activity, and down-regulation of the androgen receptor via targeting to proteasomes (Fesus and Piacentini 2002; Griffin et al. 2002). TG2-mediated alterations on the abovementioned sites can interfere with protein–protein or protein–DNA interactions, all with possible consequences on chromatin condensation and gene regulation (for histone cross-linking, see also Ballestar et al. 1996; Shimizu et al. 1996; Zhang et al. 1998). Ultimately, they can contribute to overall structural condensation of the nucleus taking place in terminally differentiating or apoptotic cells. Although the question whether these proteins are actually accessed under physiological conditions remains elusive, the above data present a reasonable and testable hypothesis.
This study reports a novel use of the phage display technology in transglutaminase research. We demonstrated that (1) the strategy reported here is successful in preferential selection of glutamine-containing sequences, (2) such peptides can simply be screened for glutamine-donor function via incorporation of a labeled amine-donor by transglutaminase directly on phage particles, (3) the sequence information obtained thereby is relevant to the analysis of substrate recognition, and (4) it has the potential to predict previously unidentified transglutaminase substrate proteins. Nonetheless, this system is suitable for finding highly efficient Q-donor sequences that can then be used in applications like in situ tagging and identification of natural substrate proteins of TG2.
Materials and methods
Phage biopanning
Peptides were selected from a commercially available 7-mer random library that is displayed on phage M13 via N-terminal fusion to the minor coat protein, g3p, with a diversity of 2.8 × 109 (Ph.D.-7 Phage Display Peptide Library Kit, NEB). Recombinant human transglutaminase 2 was produced as a GST-fusion protein (GST-TG2) in Escherichia coli DH5α using a pGEX-2T vector construct, purified as described earlier (Ambrus et al. 2001), and served as the target in the selection process. One microgram of GST-TG2 in 50 μL of TBS [50 mM Tris(hydroxymethyl) aminomethane Tris-HCl at pH 7.5, 150 mM NaCl] was immobilized on glutathione-Sepharose 4B beads (50%, 50 μL) (Amersham). The slurry was washed three times with 1 mL of TBS by centrifugation (3000 rpm, 10 min) and decantation, blocked with 100 μL of blocking buffer (0.5% BSA in TBS) for 1 h at 4°C, and washed as before using TBST (TBS containing 0.1% Tween 20). A 50-μL reaction mix consisting of 5 mM CaCl2, 10 mM dithiothreitol (DTT, Sigma), and 10 μL of the phage library (2 × 1012 plaque-forming units, PFUs) in TBS was added to the beads and incubated for 1 h at 25°C with continuous shaking. To remove unbound phages, the slurry was washed 10 times with 1 mL of TBST, and the phage population remaining attached was eluted with shaking in 100 μL of 0.1 M glycine-HCl (pH 2.0) (10 min at 25°C) and was neutralized with 150 μL of 1 M Tris-HCl (pH 8.0). The eluate was amplified in E. coli ER2537 cells, purified by precipitation with polyethylene glycol PEG/NaCl (PEG # 8000, Sigma), titrated as described in the standard protocol (NEB), and used (1011 PFUs) in the next selection cycle. In the second and third biopanning rounds, bound phages were eluted with 100 μL of 5 mM 5-(biotinamido)penthylamine in TBS (Pierce) by 30 min of shaking at 25°C.
After the second and third rounds, individual phage clones were isolated, and ssDNA was prepared according to the standard protocol (NEB). The insert sequence was determined using the “-96” M13 sequencing primer (NEB) and an ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems).
Enzyme-linked immunosorbent assay (ELISA)
Phage clones from randomly picked plaques were amplified and purified by precipitation with PEG/NaCl. The concentration of phage stocks was estimated from the absorbance at 260 nm (when A260 = 1, [phage] = 1.1 × 10−8 M) and dilution series (10−4–10 nM) were prepared in blocking buffer (0.5% BSA in TBS) containing 10 mM DTT and 5 mM CaCl2. Ninety-six-well microtiter plates were coated with 10 μg/mL GST-transglutaminase-2 in TBS containing 10 mM DTT (2 h, 25°C), and blocked with 200 μL of blocking buffer (2 h, 25°C). After three washes with TBST, 50-μL aliquots of the phage solutions were incubated in the plate for 1 h at 25°C, followed by 10 washes with TBST. Bound phage particles were quantified adding a horseradish peroxidase-conjugated anti-M13 antibody (Amersham) (50 μL of 1:5000 dilution in 0.25% BSA/TBS) for 1 h at 25°C. After washing, the plates were incubated with 1 mg/mL tetramethyl-benzydine (Sigma) (100 μL/well) for 10 min at 25°C, the color was developed by adding 50 μL of 2 M H2SO4, and the absorbance was measured at 450 nm in a Wallac 1420 Victor2 microtiter plate reader.
In vitro transamidation
To test for TG2-catalyzed transamidation of phage-displayed peptides, aliquots of amplified phage clones (5 × 1010 PFUs) were dissolved in XL1 reaction buffer (150 mM Tris-HCl at pH 8.5, 15% [v/v] glycerol, 10 mM DTT) containing 1 mM 5BP as the amine-donor substrate, and 2 μg of GST-TG2. Reactions were initiated by the addition of 5 mM CaCl2, and were allowed to proceed for 30 min at 37°C. In control samples, CaCl2 was replaced by 5 mM EDTA. Reaction products were analyzed by Western blotting. Synthetic peptides were dissolved to a final concentration of 200 μM in XL2 reaction buffer (10 mM Tris-HCl at pH 8.5, 5 mM DTT) containing either 1 mM biotinyl-GPAVTAAPKK (lysine-donor peptide) or 1 mM 5BP (amine-donor substrate), and 5 mM CaCl2, in a final volume of 250 μL. Reactions were initiated by the addition of 5–10 μg of GST-TG2 and incubated for 1 h at 37°C. High molecular mass (>10 kDa) components of the reaction mixtures were partially removed by ultrafiltration (Centricon YM-10), and reaction products were analyzed by mass spectrometry.
Immunoblotting
Aliquots of in vitro TG2-labeling reactions containing 5 × 1010 phage particles were run on 10% SDS-PAGE (Mini-Protean III, Bio-Rad). Protein bands were stained by Coomassie Brilliant Blue R (Sigma) or transferred to an Immobilon-P PVDF membrane (Millipore) in a Hoefer Semi-Dry Blotting apparatus (Bio-Rad). Visualization of biotinylated protein species was carried out using horseradish peroxidase (HRP)-conjugated streptavidin (Vectastain Western Blotting Kit, Vector Laboratories, Inc.), followed by chemiluminescent staining (ECL Western Blot Detection Kit, Amersham) and detection in an AlphaImager Gel Documentation System (Alpha Innotech).
Database search
The heptapeptide sequences obtained from the selected phage clones after the third round of biopanning were examined for a consensus sequence for TG2 modification. Because no consensus sequence was observed, we have analyzed whether the distribution of amino acids is different from that expected in the initial random library. The occurrence of amino acids was determined at individual positions around glutamines, and where adjacent Q residues were present, the most C-terminally located one was chosen arbitrarily as the point of reference. The deduced consensus pattern was used to further examine if it is also present in sequences around glutamine residues modified by transglutaminase in known substrate proteins listed in TRANSIT and TRANSDAB transglutaminase substrate databases. It was also examined whether the phage-derived sequences match any transglutaminase substrate protein listed in the TRANSIT database using the Sitematcher searching tool and considering only those results that match with the reactive glutamine residue(s).
As a broader approach, each heptapeptide was searched against the Non-Redundant protein database using the BLAST algorithm with the “short nearly exact match” option at the NCBI server to see whether this technique is suitable to predict novel, yet unidentified potential transglutaminase substrates. To address this question, peptide Ph.Q2.7 (GQQQTPY) was chosen from modified heptapeptides as a representative substrate sequence, and proteins that contain this sequence were searched from the PIR database using the abovementioned BLAST algorithm with the “short nearly exact match” option at the NCBI server. The group of SWI1/SNF1-related chromatin-remodeling factors was chosen for further investigation.
Besides the GQQQTPY sequence, randomly chosen native peptides from the above-mentioned database searches were also examined to see whether they are modified by TG2.
Peptide synthesis
The peptides––SnQ1, SGYGQQGQTPYYNQQSPHPQQQQPPYS from human SWI/SNF chromatin remodeling complex protein p270 (Accession no. O14497, S77-S103); SnQ2, GALQQASVII from Caenorhabditis elegans Dynamin-like protein (Accession no. NP_500826, G687-I696); SnQ3, RRGLQQSSVN from Drosophila G-protein homolog waclaw protein (Accession no. NP_523431, R526-N535); SnQ4, SGGMQATPAT from human Zn-finger-type transcription factor (Accession no. NP_057507, S140-T149); SnQ5, AEVQETQRTPSA from human SH3 binding protein SNP70 (Accession no. XP_225186, A-A); SnQ6, GVEETQRPPTL from human NORE-1 protein (Accession no. NP_113625, G196-L206); biotinyl-SnQ1; and biotinyl-GPAVTAAPKK (BKP, sequence is derived from the C-terminal 10 amino acids of the lysine-donor TG2 substrate protein, bovine αB-crystallin; Grootjans et al. 1995)––were synthesized by Joe Gray at the Molecular Biology Unit, University of Newcastle upon Tyne. The peptides were purified using RP-HPLC, and >90% purity was confirmed by MALDI/MS.
Mass spectrometry
MALDI-TOF mass spectra were recorded on a Voyager DE STR mass spectrometer (Applied Biosystems). A mixture of 1 μL of sample solution and 1 μL of α-cyano-4-hydroxycinnamic acid (10 mg/mL in acetonitrile/0.2%TFA = 7/3 [v/v]) was applied to the sample plate and dried. Mass calibration was performed using the Sequazyme Peptide Mass Standard Kit Calmix 2 (Applied Biosystems) and a matrix peak (379.1 Da) as internal standard. Raw data were analyzed using computer software provided by the manufacturers and reported as average or monoisotopic masses. Liquid chromatography/mass spectrometry (μLCMS) analyses were performed using a Q-TRAP μLCMS/MS ion trap mass spectrometer (Applied Biosystem MDS Sciex), equipped with a turbo electrospray ion source. The eluting system consisted of 0.25% formic acid, water (eluent A), and 0.25% formic acid, in 70% acetonitrile (eluent B). The 100-μL aliquots of each peptide mixture were injected onto an RP-HPLC C18 column Tagra (100 × 0.3 mm, 3 μm; Higgins Analytical, Inc.) and fractionated by performing a linear gradient of eluent B in eluent A from 0% to 100% B in 10 min, at a flow rate of 6 μL/min. Spectra were acquired from 200 to 1700 Da. The resulting mass data were elaborated using the Analyst software provided by the manufacturer. The mass range was calibrated using an Ultramark solution provided by the manufacturer.
Circular dichroism
Far- and near-UV circular dichroism (CD) spectra were measured on a Jasco J-810 spectropolarimeter. The temperature was controlled using a Peltier unit set to 25°C. CD spectra were recorded in 0.1-mm or 10-mm pathlength circular quartz cuvettes (Hellma) at a data acquisition rate of 8 sec/nm. SnQ1 peptide concentrations (in 10 mM phosphate buffer at pH 7.0) were between 0.1 and 1 mg/mL. For temperature unfolding experiments, the temperature of the cuvette was raised from 5°C to 95°C in 5°C increments. The effect of trifluoroethanol (TFE) on secondary structure of the peptide was investigated using TFE concentrations of 10, 20, and 50% (v/v).
Surface plasmon resonance spectroscopy
Binding of GST-TG2 to SnQ1 peptide was analyzed on a Biacore 2000 instrument. Three flow channels of a Biacore (Bia Technologies, Inc.) Streptavidin (SA)-coated chip were charged with ligand by passing through the biotinyl-SnQ1 peptide at a concentration of 2 μg/mL in running buffer (10 mM Tris, 150 mM NaCl, 3.4 mM EDTA at pH 7.4) containing 0.01% P20 (Bia Technologies, Inc.), at a flow rate of 5 μL/min, until a response unit (RU) increase of 190, 630, and 900 was achieved in channels 1, 2, and 3, respectively. The reference channel was not charged with the ligand. As the analyte, 5 μL of purified GST-TG2 was injected into all channels, at a concentration of 2 μg/mL in running buffer with a flow rate of 2 μL/min.
Continuous spectrophotometric linked transamidation assay
Transglutaminase activity was measured by quantifying the ammonia released in the reaction by coupling to glutamate dehydrogenase-catalyzed NADPH depletion with a continuous UV test as described earlier (Day and Keillor 1999) with modifications. Briefly, a reaction mix containing 60 mM HEPES (pH 7.0), 10 mM DTT, 7.5 mM α-ketoglutarate, 0.7 mM NADPH, 0.75 mM ADP, 30 U/mL glutamate dehydrogenase, 80 mM ethylamine, 50–400 μM SnQ1 peptide, and 5 mM CaCl2 (or 5 mM EDTA in the control samples) at a final volume of 200 μL was pre-incubated for 10 min at 25°C. Reactions were initiated by the addition of 2 μg of GST-TG2. NADPH depletion was monitored at 355 nm. The initial rates and kinetic parameters of transamidation were determined by linear and nonlinear fitting of the data, respectively, using the software Grafit (Leatherbarrow 1990).
Electronic supplemental material
Supplemental Table S1 shows the enrichment of the glutamine-containing heptapeptides in the consecutive rounds of the biopanning. Supplemental Table S2 lists the functional classification of human or mammalian proteins identified in database searches to contain regions similar to phage-selected glutamine-containing peptides. Supplemental Table S3 summarizes the source and TG2 reactivity of the synthetic peptides used in this study (SnQ1–6) as determined by MALDI/MS using BKP as the amine-donor substrate.
Abbreviations
TG, transglutaminase; TG2, tissue-type transglutaminase; RA, retinoic acid; HD, Huntington's disease; AD, Alzheimer's disease; PD, Parkinson's disease; TRANSDAB, Transglutaminase Substrate Database; TRANSIT, Transglutamination Sites Database; SnQ, synthetic glutamine-containing peptide derived from native protein sequence; MALDI, matrix-assisted laser desorption ionization; TOF, time of flight; RP-HPLC, reverse-phase high pressure liquid chromatography; GST, glutathione sulfotransferase; DTT, dithiothreitol; 5BP, 5-(biotinamido)penthylamine; HRP, horseradish peroxidase; TBS, Tris buffer saline; ELISA, enzyme linked immunoassay; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; BLAST, basic local alignment sequence tool; BKP, biotinylated lysine-donor decapeptide (biotinyl-GPAVTAAPKK); TFA, trifluoroacetic acid; μLCMS/MS, capillary liquid chromatography/tandem mass spectrometry; ESI, electrospray ionization; CD, circular dichroism; TFE, trifluoro-ethanol; SA, streptavidin; RU, response unit; g3p, minor phage coat protein; PhQ, phage-displayed glutamine-containing peptide; L-TBP, latent TGFβ-binding protein; pRB, retinoblastoma gene product.
Acknowledgments
This research was financially supported by the National Scientific Research Fund of Hungary (OTKA F 32994, TS44798, T043083), ETT, and a grant from EU (QLK3-CT-2002-02017). Z.K. was funded by a Bolyai Janos Research Fellowship, Hungary, and a European Science Foundation Protein Cross-Linking Travel Grant.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: László Fésüs, Department of Biochemistry and Molecular Biology, Medical and Health Sciences Centre, University of Debrecen, Life Science Building, Egyetem ter 1, Debrecen H-4010, Hungary; e-mail: fesus@indi.biochem.dote.hu; fax: 36-52-314989.
Abbreviations: See “Abbreviations” section at the end of Materials and Methods.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051818406.
References
- Aeschlimann, D. and Paulsson, M. 1994. Transglutaminases: Protein cross-linking enzymes in tissues and body fluids. Thromb. Haemost. 71 402–415. [PubMed] [Google Scholar]
- Aeschlimann, D. and Thomazy, V. 2000. Protein crosslinking in assembly and remodelling of extracellular matrices: The role of transglutaminases. Connect. Tissue Res. 41 1–27. [DOI] [PubMed] [Google Scholar]
- Aeschlimann, D., Paulsson, M., and Mann, K. 1992. Identification of Gln726 in nidogen as the amine acceptor in transglutaminase-catalyzed cross-linking of laminin–nidogen complexes. J. Biol. Chem. 267 11316–11321. [PubMed] [Google Scholar]
- Akimov, S.S. and Belkin, A.M. 2001. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood 99 1567–1576. [DOI] [PubMed] [Google Scholar]
- Ambrus, A., Bányai, I., Weiss, M.S., Hilgenfeld, R., Keresztessy, Zs, Muszbek, L., and Fésüs, L. 2001. Calcium binding of transglutaminases: A 43Ca NMR study combined with surface polarity analysis. J. Biomol. Struct. Dyn. 19 59–74. [DOI] [PubMed] [Google Scholar]
- Anderson, R.P., Degano, P., Godkin, A.J., Jewell, D.P., and Hill, A.V.S. 2000. In vivo antigen challenge in coeliac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat. Med. 6 337–342. [DOI] [PubMed] [Google Scholar]
- Andringa, G., Lam, K.Y., Chegary, M., Wang, X., Chase, T.N., and Bennett, M.C. 2004. Tissue transglutaminase catalyzes the formation of α-synuclein crosslinks in Parkinson's disease. FASEB J. 18 932–934. [DOI] [PubMed] [Google Scholar]
- Avhazi, B. and Steinert, P.M. 2003. A model for the reaction mechanism of transglutaminase 3. Exp. Mol. Med. 35 228–242. [DOI] [PubMed] [Google Scholar]
- Ballestar, E., Abad, C., and Franco, L. 1996. Core histones are glutaminyl substrates for tissue transglutaminase. J. Biol. Chem. 271 18817–18824. [DOI] [PubMed] [Google Scholar]
- Burritt, J.B., Bond, C.W., Doss, K.W., and Jesaitis, A.J. 1996. Filamentous phage display of oligopeptide libraries. Anal. Biochem. 238 1–13. [DOI] [PubMed] [Google Scholar]
- Chothia, C. 1976. The nature of accessible and buried surfaces in proteins. J. Mol. Biol. 105 1–12. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J., Sheu, K.F., Burke, J.R., Strittmatter, W.J., Gentile, V., Peluso, G., and Blass, J.P. 1999. Pathogenesis of inclusion bodies in (CAG)n/Qn-expansion diseases with special reference to the role of tissue transglutaminase and to selective vulnerability. J. Neurochem. 72 889–899. [DOI] [PubMed] [Google Scholar]
- Cooper, A.J., Jeitner, T.M., Gentile, V., and Blass, J.P. 2002. Cross linking of polyglutamine domains catalyzed by tissue transglutaminase is greatly favored with pathological-length repeats: Does transglutaminase activity play a role in (CAG)n/Qn-expansion diseases? Neurochem. Int. 40 53–67. [DOI] [PubMed] [Google Scholar]
- Coussons, P.J., Price, N.C., Kelly, S.M., Smith, B., and Sawyer, L. 1992. Factors that govern the specificity of transglutaminase-catalysed modification of proteins and peptides. Biochem. J. 282 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas, P.B., Pacchione, S., Wilsker, D., Bowrin, V., Kobayashi, R., and Moran, E. 2000. The human SWI–SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol. Cell. Biol. 20 3137–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, N. and Keillor, J.W. 1999. A continuous spectrophotometric linked enzyme assay for transglutaminase activity. Anal. Biochem. 274 141–144. [DOI] [PubMed] [Google Scholar]
- Esposito, C. and Caputo, I. 2005. Mammalian transglutaminases: Identification of substrates as a key to physiological function and physiopathological relevance. FEBS J. 272 615–631. [DOI] [PubMed] [Google Scholar]
- Facchiano, A.M., Facchiano, A., and Facchiano, F. 2003. Active Sequences Collection (ASC) database: A new tool to assign function to protein sequences. Nucleic Acids Res. 31 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesus, L. and Piacentini, M. 2002. Transglutaminase 2: An enigmatic enzyme with diverse functions. Trends Biochem. Sci. 27 534–538. [DOI] [PubMed] [Google Scholar]
- Fleckenstein, B., Qiao, S.-W., Larsen, M.R., Jung, G., Roepstoff, P., and Sollid, L.M. 2004. Molecular characterisation of covalent complexes between tissue transglutaminase and gliadin peptides. J. Biol. Chem. 279 17607–17616. [DOI] [PubMed] [Google Scholar]
- Folk, J.E. and Finlayson, J.S. 1977. The ɛ(γ-glutamyl)lysine crosslink and the catalytic role of transglutaminases. Adv. Protein Chem. 31 1–133. [DOI] [PubMed] [Google Scholar]
- Freiman, R.N. and Tjian, R. 2002. Neurodegeneration: A glutamine-rich trail leads to transcription factors. Science 296 2149–2150. [DOI] [PubMed] [Google Scholar]
- Gaudry, C.A., Verderio, E., Aeschlimann, D., Cox, A., Smith, C., and Griffin, M. 1999. Cell surface localization of tissue transglutaminase is dependent on a fibronectin-binding site in its N-terminal β-sandwich domain. J. Biol. Chem. 274 30707–30714. [DOI] [PubMed] [Google Scholar]
- Gerber, H.-P., Seipel, K., Georgiev, O., Hofferer, M., Hug, M., Rusconi, S., and Schaffner, W. 1994. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science 263 808–811. [DOI] [PubMed] [Google Scholar]
- Griffin, M., Casadio, R., and Bergamini, C.M. 2002. Transglutaminases: Nature's biological glues. Biochem. J. 368 377–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans, J.J., Groenen, P.J.T.A., and de Jong, W.W. 1995. Substrate requirements for transglutaminases: Influence of the amino acid preceding the amine donor lysine in a native protein. J. Biol. Chem. 270 22855–22858. [DOI] [PubMed] [Google Scholar]
- Hausch, F., Halttunen, T., Maki, M., and Khosla, C. 2003. Design, synthesis, and evaluation of gluten peptide analogs as selective inhibitors of human tissue transglutaminase. Chem. Biol. 10 225–231. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Nam, K.H., Kwon, O.-S., Kim, I.G., Bustin, M., Choy, H.E., and Park, S.C. 2002a. Histone cross-linking by transglutaminase. Biochem. Biophys. Res. Commun. 293 1453–1457. [DOI] [PubMed] [Google Scholar]
- Kim, S.-Y., Jeitner, T.M., and Steinert, P.M. 2002b. Transglutaminases in disease. Neurochem. Int. 40 85–103. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow, R.J. 1990. Grafit version 2.0. Erithacus Software Ltd, Staines, UK.
- Lesort, M., Attanavanich, K., Zhang, J., and Johnson, G.V. 1998. Distinct nuclear localization and activity of tissue transglutaminase. J. Biol. Chem. 273 11991–11994. [DOI] [PubMed] [Google Scholar]
- Liu, S., Cerione, R.A., and Clardy, J. 2002. Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity. Proc. Natl. Acad. Sci. 99 2743–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand, L. and Graham, R.M. 2003. Transglutaminases: Crosslinking enzymes with pleiotropic functions. Nat. Rev. Mol. Cell Biol. 4 140–156. [DOI] [PubMed] [Google Scholar]
- Mahoney, S., Wilkinson, M., Smith, S., and Haynes, L.W. 2000. Stabilization of neurites in cerebellar granule cells by transglutaminase activity: Identification of midkine and galectin-3 as substrates. Neuroscience 101 141–155. [DOI] [PubMed] [Google Scholar]
- Nakaoka, H., Perez, D.M., Baek, K.J., Das, T., Husain, A., Misono, K., Im, M.-J., and Graham, R.M. 1994. Gh: A GTP-binding protein with transglutaminase activity and receptor signaling function. Science 264 1593–1596. [DOI] [PubMed] [Google Scholar]
- Nanda, N., Iismaa, S.E., Owens, W.A., Husain, A., Mackay, F., and Graham, R.M. 2001. Targeted inactivation of Gh/tissue transglutaminase II. J. Biol. Chem. 276 20673–20678. [DOI] [PubMed] [Google Scholar]
- Nemes, Z., Devreese, B., Steinert, P.M., Van Beeumen, J., and Fesus, L. 2004. Cross-linking of ubiquitin, HSP27, parkin and α-synuclein by γ-glutamyl-ɛ-lysine bonds in Alzheimer's neurofibrillary tangles. FASEB J. 18 1135–1137. [DOI] [PubMed] [Google Scholar]
- Orru, S., Caputo, I., D'Amato, A., Ruoppolo, M., and Esposito, C. 2003. Proteomics identification of acyl-acceptor and acyl-donor substrates for transglutaminase in a human intestinal epithelial cell line: Implications for Celiac disease. J. Biol. Chem. 278 31766–31773. [DOI] [PubMed] [Google Scholar]
- Parrot, I., Huang, P.C., and Khosla, C. 2002. Circular dichroism and nuclear magnetic resonance spectroscopic analysis of immunogenic glutene peptides and their analogs. J. Biol. Chem. 277 45572–45578. [DOI] [PubMed] [Google Scholar]
- Pastor, M.T., Diez, A., Perez-Paya, E., and Abad, C. 1999. Addressing substrate glutamine requirements for tissue transglutaminase using substance P analogues. FEBS Lett. 451 231–234. [DOI] [PubMed] [Google Scholar]
- Piper, J.L., Gray, G.M., and Khosla, C. 2002. High selectivity of human tissue transglutaminase for immunoactive gliadin peptides: Implications for Celiac Sprue. Biochemistry 41 386–393. [DOI] [PubMed] [Google Scholar]
- Pirreda, L., Amendola, A., Colizzi, V., Davies, P.J.A., Farrace, M.G., Fraziano, M., Gentile, V., Uray, I., Piacentini, M., and Fésüs, L. 1997. Lack of tissue transglutaminase protein cross-linking leads to leakage of macromolecules from dying cells: Relationship to development of autoimmunity in MRLlpr/lpr mice. Cell Death Differ. 4 463–472. [DOI] [PubMed] [Google Scholar]
- Ruopollo, M., Ottu, S., D'Amato, A., Francese, S., Rovero, P., Marino, G., and Esposito, C. 2003. Analysis of transglutaminase protein substrates by functional proteomics. Protein Sci. 12 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, L., Molberg, O., Parrot, I., Hausch, F., Filiz, F., Gray, G.M., Sollid, L.M., and Khosla, C. 2002. Structural basis for gluten intolerance in Celiac Sprue. Science 297 2275–2297. [DOI] [PubMed] [Google Scholar]
- Shimizu, T., Hozumi, K., Horiike, S., Nunomura, K., Ikegami, S., Takao, T., and Shimonishi, Y. 1996. A covalently cross-linked histone. Nature 380 32. [DOI] [PubMed] [Google Scholar]
- Shing, U.S., Kunar, M.T., Kao, Y.-L., and Baker, K.M. 2001. Role of transglutaminase II in retinoic acid-induced activation of RhoA-associated kinase-2. EMBO J. 20 2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura, Y., Hosono, M., Wada, F., Yoshimura, T., Maki, M., and Hitomi, K. 2006. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: Identification of peptide substrates for TGASE 2 and Factor XIIIA. J. Biol. Chem. 281 17699–17706. [DOI] [PubMed] [Google Scholar]
- Szegezdi, E., Szondy, Z., Nagy, L., Nemes, Z., Friis, R.R., Davies, P.J., and Fesus, L. 2000. Apoptosis-linked in vivo regulation of the tissue transglutaminase gene promoter. Cell Death Differ. 7 1225–1233. [DOI] [PubMed] [Google Scholar]
- Thomázy, V. and Fesus, L. 1989. Differential expression of tissue transglutaminase in human cells: An immunohistochemical study. Cell Tissue Res. 255 215–224. [DOI] [PubMed] [Google Scholar]
- Vader, L.W., de Ru, A., van der Wal, Y., Kooy, Y.M.C., Benckhuijsen, W., Meqarin, M.L., Drijfhout, J.W., van Veelen, P., and Koning, F. 2002. Specificity of tissue transglutaminase explains cereal toxicity in Celiac disease. J. Exp. Med. 195 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Lesort, M., Guttman, R.P., and Johnson, G.V.W. 1998. Modulation of the in situ activity of tissue transglutaminase by calcium and GTP. J. Biol. Chem. 273 2288–2295. [DOI] [PubMed] [Google Scholar]