Abstract
Six α-defensins have been found in humans. These small arginine-rich peptides play important roles in various processes related to host defense, being the effectors and regulators of innate immunity as well as enhancers of adoptive immune responses. Four defensins, called neutrophil peptides 1 through 4, are stored primarily in polymorphonuclear leukocytes. Major sites of expression of defensins 5 and 6 are Paneth cells of human small intestine. So far, only one structure of human α-defensin (HNP3) has been reported, and the properties of the intestine defensins 5 and 6 are particularly poorly understood. In this report, we present the high-resolution X-ray structures of three human defensins, 4 through 6, supplemented with studies of their antimicrobial and chemotactic properties. Despite only modest amino acid sequence identity, all three defensins share their tertiary structures with other known α- and β-defensins. Like HNP3 but in contrast to murine or rabbit α-defensins, human defensins 4–6 form characteristic dimers. Whereas antimicrobial and chemotactic activity of HNP4 is somewhat comparable to that of other human neutrophil defensins, neither of the intestinal defensins appears to be chemotactic, and for HD6 also an antimicrobial activity has yet to be observed. The unusual biological inactivity of HD6 may be associated with its structural properties, somewhat standing out when compared with other human α-defensins. The strongest cationic properties and unique distribution of charged residues on the molecular surface of HD5 may be associated with its highest bactericidal activity among human α-defensins.
Keywords: human α-defensins, crystal structures, antimicrobial peptides, chemotaxis
Defensins are small (3–5 kDa), cationic proteins with molecules stabilized by several (usually three) disulfide bonds (Ganz 2002; Lehrer and Ganz 2002). Based primarily on the connectivity of six cysteine residues (CysI, …, CysVI), they are classified into two categories, α- and β-defensins. In α-defensins, three disulfide bridges have topology, CysI–CysVI, CysII–CysIV, CysIII–CysV, whereas the equivalent linkage arrangement of β-defensins is CysI–CysV, CysII–CysIV, CysIII–CysVI (Selsted and Harwig 1989; Tang and Selsted 1993). Despite this difference, the tertiary structures of proteins from both categories are quite similar (Hill et al. 1991; Hoover et al. 2000). So far, six α-defensins have been identified in human (Zasloff 2002), although only five of them originate from specific genes (Sparkes et al. 1989; Linzmeier et al. 1993). Furthermore, defensins 1 through 3 differ only by a single N-terminal residue (Lehrer and Ganz 2002). The first four defensins are often called neutrophile peptides (in this report HNP1–4), as the azurophilic granules of neutrophiles are their primary storage compartments (Lehrer et al. 1983; Selsted et al. 1984; Ganz et al. 1985). Defensins 5 and 6 are frequently referred to as intestinal defensins (here HD5–6), since they are most abundant in the epithelia of the Paneth cells (Jones and Bevins 1992, 1993).
The best known property of defensins is their ability to kill microbial pathogens (Harder et al. 1997; Ganz 2002; Lehrer and Ganz 2002; Zasloff 2002). In addition to their microbicidal properties, other biological activities of these proteins have been observed either in vitro or in vivo (Yang et al. 2004). These activities include chemotaxis, angiogenesis, modulation of adoptive immunological reactions, pro-inflammatory effects, cancer metastasis, etc. Due to the multi-level roles in the defensive, regulatory, and pathological processes, interest in these proteins has been recently rapidly increasing (Yang et al. 2004). Until recently, only the first three human α-defensins (HNP1–3) had been studied relatively extensively (Lehrer et al. 1983; Ganz et al. 1985; Yang et al. 2004), primarily because of the lack of access to sufficient quantities of pure biologically active preparations of the remaining members of this category (Wu et al. 2004). In addition to research focused on the functional (mainly antimicrobial) properties of α-defensins, some studies attempted to rationalize observed biological activities in terms of the structural properties of these proteins (Hill et al. 1991; McManus et al. 2000; Jing et al. 2004). So far, however, not much data is available for human α-defensins, HNP4, HD5, and HD6.
In this report, we present the X-ray structures of three defensins, HNP4, HD5, and HD6, which, together with previous results for HNP3 (Hill et al. 1991), provide a more complete structural description of human α-defensins. Our structural results are complemented with a series of functional assays, and the preliminary analysis of possible relations between structural and functional properties is also presented.
Results
Tertiary structures of α-defensins
The overall fold of the α-defensin monomer is attributed to the three β-strands (for definition, see Fig. 1A), arranged into antiparallel β-sheet, the β-hairpin (T2) formed by strands β2 and β3, seven to eight residues long loop (T1) connecting β1 and β2, and two termini of variable length (one to four amino acids) located close to each other. These architectural elements are restrained in their relative orientations by three disulfide bridges, CysI–CysVI, CysII–CysIV, CysIII–CysV, and one salt bridge formed by the side chains of ArgX and GluY (where I through IV indicate the relative positions in the peptides, and X is 5, 6, or 7, and Y is 13, 14, or 14, in HNP4, HD5, and HD6, respectively). The first two cysteine residues in each sequence are separated by one amino acid, whereas the last two are adjacent. In addition to these eight residues stabilizing the tertiary structure of each protein, two additional Gly residues complete the list of conserved residues.
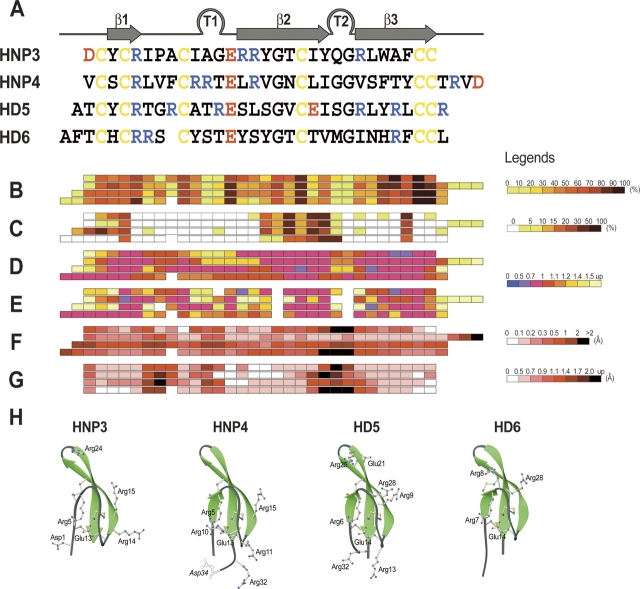
Figure 1.
Structure-related properties of human α-defensins. (A) Alignment of the amino acid sequences. Conserved Cys residues are shown in yellow, positively-charged residues are printed in blue, and anionic residues in red. The distribution of β-strands and turns (T's) common to four α-defensins is shown above the sequence alignment (assignment was prepared with the program DSSP; Kabsch and Sander 1983). (B) Histograms of buried solvent-accessible surfaces for individual residues in folded monomers of defensins. Values of buried surfaces were calculated with program Naccess (www.biochem.ucl.ac.uk/~roman/naccess/naccess.html) and averaged over all crystallographically-independent monomers. The histogram is followed by the legend, describing the color assignment used. (C) Histograms of the solvent-accessible surfaces for individual residues buried upon dimerization. (D) Distributions of the average B-factor values for the main chain atoms of individual residues, divided by the B-factor values averaged over all main chain atoms of each defensin (see also Table 2). (E) As in D, except for the side chain atoms. Values used for the preparation of panels D and E represent the averages over all structurally independent monomers of each defensin. (F) Distributions of the discrepancies (in Å) calculated for the equivalent Cα-atoms of superimposed, structurally-independent monomers of defensins. For each defensin, values represent the averages obtained from all possible superimpositions. (G) Histograms of the discrepancies (in Å) calculated for the equivalent Cα-atoms of superimposed monomers from different defensins. Values used in this figure represent the averages (i.e., each color bar for HNP4 represents an average of 36 equivalent discrepancies). (H) Ribbon diagrams of the monomers of four human α-defensins shown in equivalent orientation. The side chains of all charged and Cys residues are shown in a ball-and-stick representation with labels. For HNP4, the structure of the last residue, Asp34, was not determined due to disorder. For completeness, however, this residue is also drafted in dotted lines and labeled in italics. The panel was prepared with programs Ribbons (Carson 1991) and POV-Ray (http://www.povray.org).
The tertiary structures of HNP4, HD5, and HD6, like those of other α-defensins reported to date (Hill et al. 1991; Xie et al. 2005), are quite similar (Fig. 1H). The root-mean-square (RMS) deviations between the sets of 28–31 equivalent Cα-atoms within the monomers of different defensins vary between 0.3 Å and 1.0 Å. Packed in the core of the defensin's monomer, the 10 conserved residues are significantly buried within the native structure (Fig. 1B) and play a secondary role in the oligomerization of α-defensins (Fig. 1C). Analysis of the histograms shown in Figure 1D–G indicates that the sites of conserved residues coincide with the most rigid and structurally conserved fragments in the defensin molecules. None of these residues appears to be indispensable for the antimicrobial properties of α-defensins (Wu et al. 2003c; Maemoto et al. 2004; Xie et al. 2005). In turn, the proteolytic stability, folding properties, and chemotactic activities of defensins are significantly weakened by substitution of any of these residues (Wu et al. 2003c, 2005; Maemoto et al. 2004; Selsted and Ouellette 2005), supporting their stabilizing role.
Despite a high topological similarity, a significant conformational variability was found for two Paneth cells’ defensins, HD5 and HD6. In the case of each protein, four molecules are present in the asymmetric unit, resulting in eight different crystal environments. In the case of both proteins, we observe two conformationally distinct populations, with the differences between them conferred to the hairpin T2 and conformations of the CysIII–CysIV disulfide bridge. Superposition of monomers accommodating different conformations in HD5 and in HD6 is shown in Figure 2.
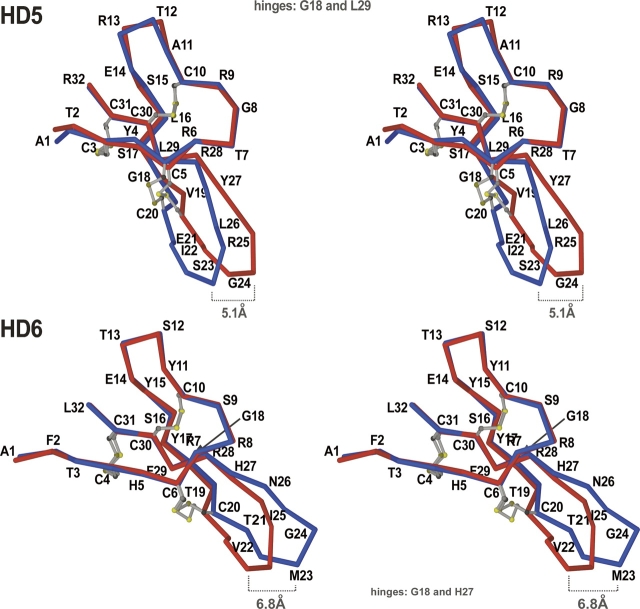
Figure 2.
A superposition of the Cα-traces of the intimate monomers of HD5 (monomers A and C) and HD6 (monomers A and B). Based on the comparison of the equivalent backbone torsion angles for each of the two monomers compared, only the subset of Cα-atoms (1–18 and 29–32 for HD5, 1–18 and 27–32 for HD6) corresponding to the most conformationally-similar residues has been used during alignment. For better readability, all Cα-atoms are labeled, and the disulfide bridges are shown as balls-and-sticks. Two structural differences between the two monomers, common for both defensins, include different conformations of the second disulfide bridge (Cys5–Cys20 in HD5, and Cys6–Cys20 in HD6) and orientations of the β2-hairpins. The latter discrepancy can be measured by the distance between the equivalent atoms at the tip of the hairpin in both monomers and varies from 5 Å (HD5) to nearly 7 Å (HD6). Several residues within this hairpin have different backbone conformations in both monomers, as seen clearly for the fragment Thr21–Asn26 in HD6. In both proteins, the differences are the result of a conformational change of Gly18 (a “hinge” residue). The lesser difference is seen within the loop Ala11–Glu14 in both monomers of HD5. Conformational changes shown in this figure lead to the asymmetry of the dimers (see text for more details).
Analysis of the conformational differences (see the Supplemental Materials) between two backbones of HD5 monomers pinpoints Thr2, Cys3, Arg6, Ala11, Thr12, Glu14, Gly18, Val19, Glu21, and Leu29 as the residues with the most divergent conformations. The cysteines 5 and 20 form the disulfide bridge, which has different conformations in two kinds of monomers. Gly18 and Val19 as well as Leu29 define the ranges of the hairpin T2 region, which accommodates different conformations. Variability in conformations of Thr2 and Cys3 is directly associated with slight flexibility of the N terminus in HD5, whereas similar flexibility of the turn T1 region is manifested by conformational differences found for residues Ala11–Glu14.
In the case of HD6, the conformational differences between two types of monomers are even more pronounced; however, they are localized to the same fragment regions as in HD. The only differences in asymmetry of HD5 versus HD6 are a lower flexibility of the N termini in HD6 monomers as compared with those in HD5, and dramatic differences in the conformation of the turn (Met23 and Gly24) located at the tip of T2. As one of the Cys residues (Cys20) forming the bridge (CysIII–CysIV) in two different conformations is located within the flexible region of the hairpin T2, practically all topological differences observed for the backbones of HD5 or HD6 can be attributed to the movement of the hairpin T2. Interestingly, one of the “hinge” residues in the T2 hairpin is Gly18, a residue conserved in all human defensins. Therefore, the complete conservation of this Gly in α-defensins may also be associated with its role in the hairpin movement.
Human α-defensins form similar dimers
The defensins described here form, in crystals, topologically similar dimers, which were also observed previously for HNP3 (Hill et al. 1991). Figure 1, A and C, highlights the contributions of individual residues (expressed by the loss of solvent-accessible surfaces) to the dimerization and locations of these residues within the structural motifs of the defensins monomers.
In all human α-defensins, an equivalent section of the monomers, spanning over the second half of the strand β2 and a tip of the hairpin T2, is engaged in the intermolecular contacts. In HNP3, HNP4, and HD5, two equivalent residues from this section (Asn18 and Leu20 in HNP4) form two pairs of the >N-H…O= hydrogen bonds, laying approximately in the same plane and related by the pseudo twofold axis of the dimers, perpendicular to this plane. Upon dimerization, two strands (β2 and β3) from either monomer arrange into the four-stranded, antiparallel β-sheet. The overall topology of the dimers is further stabilized by a series of short interactions between the N-terminal residues of both monomers.
These interactions are most extensive in HD5. The pair of equivalent H-bonds, located symmetrically against a dimer's pseudo twofold axis, is formed by the backbone atoms of Cys3 from both monomers. Additional stabilization is provided by asymmetrically distributed (present only on one side of the dimer's pseudo twofold axis) hydrophobic interactions between the side-chains of Cys3 from one monomer and Cys31 from the other. This asymmetry is directly related to the asymmetry of monomers participating in the formation of the HD5 dimer (discussed in the previous paragraph).
The network of intermonomer contacts observed for HD6 is, however, significantly different than that for other human α-defensins; in particular, the arrangement of the hydrogen bonds between two β2 strands is different. At the center of the network are two H-bonds between the N and O backbone atoms of the Thr21 residues from both monomers, approximately perpendicular to the pseudo twofold axis of the dimer. Two residues, Thr19(O) and Met23(N), from each monomer form additional, slightly elongated (3.1–3.5 Å), H-bonds with similar symmetry and orientation against the twofold axis described for other H-bonds.
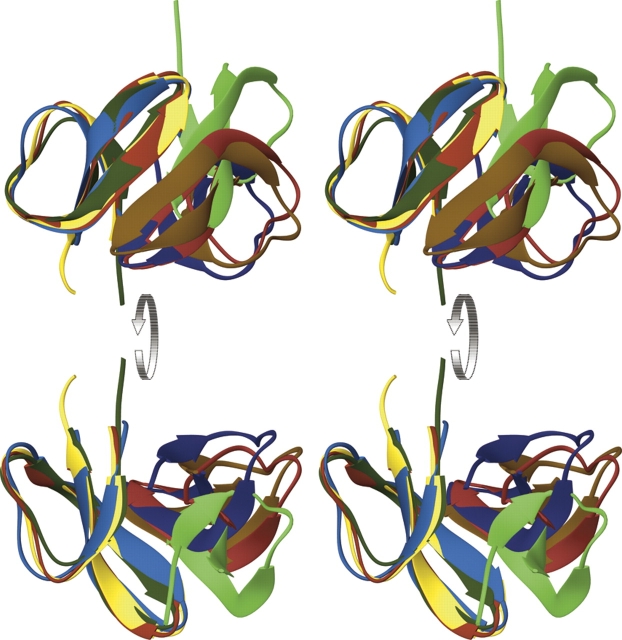
Similar to HD5, dimers of HD6 are asymmetric. Resulting from this asymmetry are subtle changes of the dimer's topology, illustrated in Figure 3. As might be expected, the most similar topology is observed for the dimers of HNP3 and HNP4. In addition to the asymmetry of individual monomers of HD6, two monomers are shifted in relation to each other along their β2 strands, leading to the formation of a different central network of H-bonds. These findings lead us to the interesting questions of (1) whether the specific environment of the crystal packing leads to the asymmetry of the HD5 and HD6 dimers (or monomers), or it is a native feature of these proteins; and (2) how to interpret a relative shift of monomers found in the dimer of HD6. The structural data for seven crystallographically independent monomers of HD5 and HD6, only modest amino acid sequence identity of these proteins (∼38%), different crystallization conditions, and lack of crystal contacts favoring the conformational changes of monomers described for these proteins indicate that ability to accommodate specific yet different conformations by the monomers of α-defensins might be the intrinsic feature of these proteins.
Figure 3.
Dimers of human α-defensins. Dimers of four α-defensins, HNP3 (shown in two shades of red), HNP4 (yellow and brown), HD5 (blue), and HD6 (green), are aligned based on the Cα-atoms from the left (“fixed”) monomer only. The discrepancies between the right (“riding”) monomers illustrate the extent of topological changes within dimers. The largest departure, seen for HD6, is the result of a relative shift of two monomers along their β2 strands.
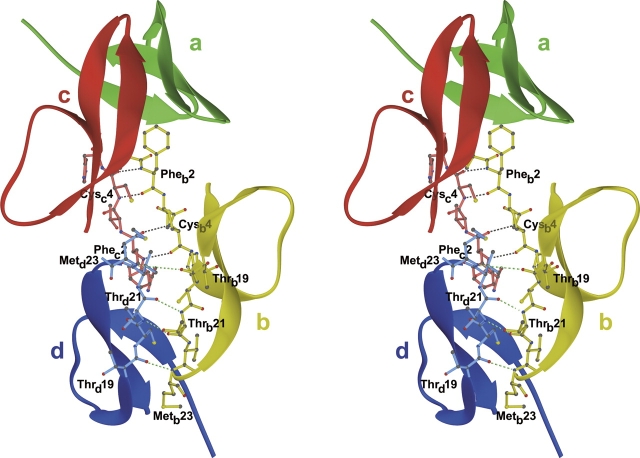
The origin of more significant topological changes observed for HD6 is, however, more ambiguous. In the crystal lattice, each monomer, in addition to the previously described H-bond network contributed by the residues from strand β2, forms another set of H-bonds with adjacent molecules, equally extensive yet utilizing the N-terminal region. This association is shown in Figure 4. As seen, two residues from each of the N termini form a pair of >N-H…O= hydrogen bonds, which increases the overall number of intermonomer H-bonds to eight. This type of interaction in crystals leads to the formation of a continuous chain (elongated helix) of HD6 monomers linked to each other by a network of four backbone–backbone H-bonds. It appears unlikely that such long chains of HD6 monomers are stable in the solution (not analyzed), although a transient occurrence of their shorter variants cannot be excluded. Therefore, the crystal interactions are likely triggering the formation of N termini-associated H-bond networks and partial disruption of the standard H-bond structure contributed by the strands β2 of two monomers.
Figure 4.
Two types of dimerization found in the crystals of human HD6. In addition to four intermonomer H-bonds contributed by the strands β2 and observed for all α-defensins in crystals (here marked with green dashed lines for the monomers b [yellow] and d [blue]), each monomer of HD6 forms the second cluster of four H-bonds (marked with black dashed lines) through the backbone atoms of Phe2 and Cys4. The second network is shown for monomers b (yellow) and c (red). Each mode of dimerization is associated with the formation of four H-bonds and a comparable reduction of the solvent-accessible surface per monomer (584 Å2 and 834 Å2, for β2-strand- and N termini-mediated dimerizations, respectively).
Biological activities of HNP4, HD5, and HD6
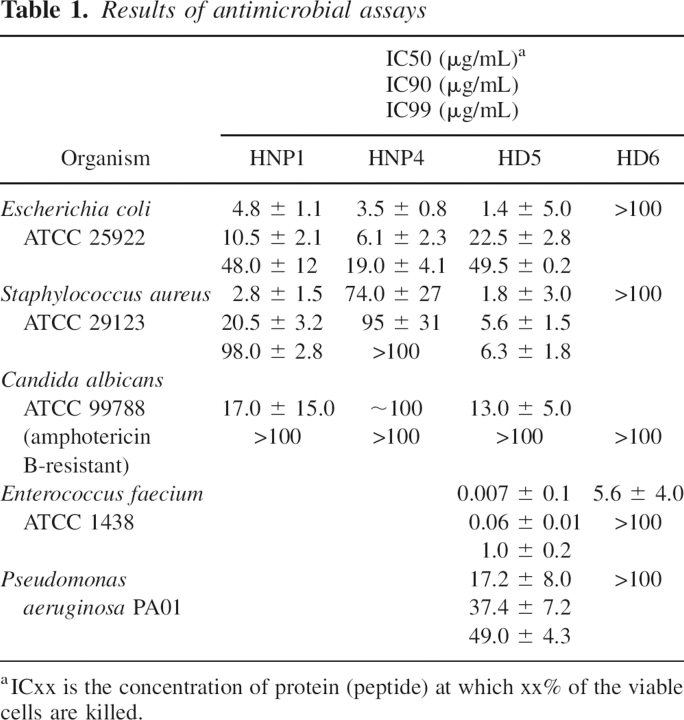
All three defensins studied here were subjected to antimicrobial assays with three pathogenic microorganisms, Gram-negative bacterium Escherichia coli (strain ATCC 25922), Gram-positive Staphylococcus aureus (strain ATCC 29123), and yeast Candida albicans (amphotericin B-resistant strain ATCC 99788). As a reference, we also included HNP1 in these experiments. Furthermore, both intestinal defensins, HD5 and HD6, were assayed against the Gram-positive Enterococcus faecium (strain ATCC 1438), a common pathogen of a digestive tract, and the opportunistic, Gram-negative bacterium, Pseudomonas aeruginosa (strain PA01). In all cases, the highest concentration of defensin used was 100 μg/mL. The results, shown in Table 1, indicate that HNP4 and HNP1 have a comparable antimicrobial activity; however, systematically higher concentrations of the former are needed to achieve complete inhibition of bacterial growth. Additionally, both defensins are rather weak anti-yeast agents, with HNP1 slightly more potent. Comparable activity against three pathogens was observed for HD5. This defensin appeared to be more potent than HNP1 or HNP4 against S. aureus, whereas anti-yeast activity was quite low. Curiously, HD6 was practically inactive. The only modest response to HD6 was observed for E. faecium. In contrast, HD5 was very potent against this digestive tract bacterium, and the complete inhibition was observed at concentrations of defensin <1 μg/mL. Activities of this protein in the assays with P. aeruginosa and E. coli were comparable.
Table 1.
Results of antimicrobial assays
The analysis of the results in Table 1 allows a few general observations. The myeloid defensins, HNP1 and HNP4, appeared more potent against the Gram-negative E. coli than the Gram-positive S. aureus. In turn, HD5 kills Gram-positive S. aureus and E. faecium more efficiently than Gram-negative E. coli or P. aeruginosa. Neither of α-defensins efficiently kills yeast. HD6 showed practically no antimicrobial activity in our assays. Recently, Ericksen et al. (2005) reported similar results for all six human α-defensins. It was reported previously that human neutrophil peptides HNP1–3 induce chemotaxis of T lymphocytes or dendritic cells at nanomolar concentrations (Ericksen et al. 2005), although the cell-surface receptor(s) targeted by the defensins is unknown. No such data are available for HNP4, HD5, or HD6. The chemotactic activity assays for these defensins, as well as for HNP1 and SDF-1α (used as positive references), were completed for human peripheral blood CD4 T cells. Both myeloid defensins showed very comparable chemotactic activity. The maximum migration of cells (doubled over the background) was observed at concentrations between 10 and 100 ng/mL for both proteins. No chemotactic effect, however, was detected in these assays for either of intestinal defensins.
Surface properties of α-defensins
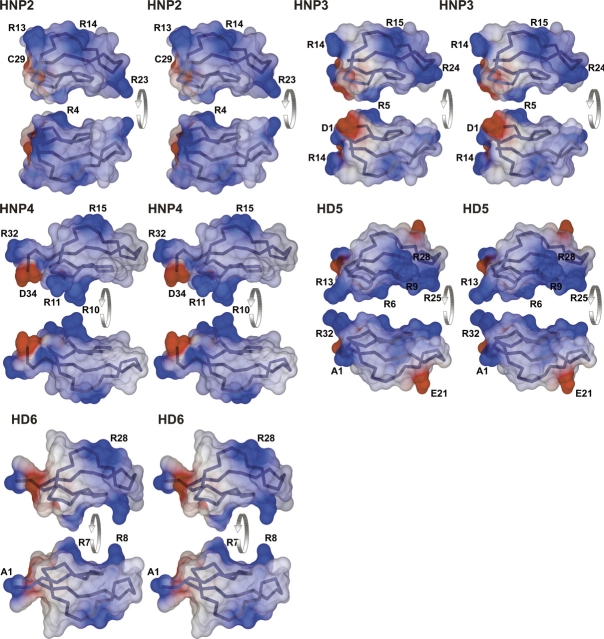
Despite common structural topology and similar amphiphilic properties, α-defensins display divergent antimicrobial properties (Ganz 2002; Yang et al. 2004). Furthermore, we found that neither of the two intestinal defensins is chemotactic for the human peripheral blood CD4 T cells, which respond to the concentration gradient of HNP1–4. Since either of these activities stems from interactions of defensins with components of cellular membranes, the topological and electrostatic properties of the molecular surfaces of their molecules likely play the pivotal role in differential interactions with membranes. Additionally, differences in the oligomerization properties of these proteins may also contribute to their specific activities. In Figure 5, the electrostatic potentials for five human α-defensins are mapped on their solvent-accessible surfaces and colored accordingly. For convenience of analysis, the composite cross-section (see figure legend for definition) assigns five regions of the molecular surface.
Figure 5.
Distribution of the electrostatic potential on solvent-accessible surfaces of human α-defensins. The stereo representations of the solvent-accessible surfaces have been determined with program MSMS (Sanner et al. 1996), using the solvent sphere with radius of 1.4 Å. Surfaces are colored according to the values of Coulomb potential calculated on the basis of the charged residues and the termini. The regions colored in blue are associated with positively-charged groups, red with negative charges, and white-gray areas are electrically neutral. Monomers of all defensins are shown in equivalent orientations and are seen from two opposite views. For convenience, the Cα-representations of the molecules are also shown inside the semitransparent surfaces, and the charged residues are annotated. Drawings for HNP2 and HNP3 are based PDB entries 1ZMI and 1DFN, respectively. The cross-section of the defensin molecule is shown in the lower right panel as a composite of the cross-sections for individual proteins, and it is a flat schematic representation of both faces of the solvent-accessible surfaces. For ease of analysis (see text), solvent-accessible surfaces were arbitrarily divided into five regions (Reg1 through Reg5). Boundaries of the regions were chosen based on careful comparison of electrostatic potentials and shapes of the surfaces, and bear no relevance to the detailed topological features of the proteins. For convenience, areas of the composite cross-sections, charged in at least one defensin, are colored.
Each protein contributes to two structurally-conserved, negatively-charged groups, the C terminus and the Glu residue located four positions past the third cysteine. The effect of the negative charge contributed by the carboxylate group is comparable for all α-defensins as its location is quite conserved (Fig. 5, the red-colored area in region Reg1). The glutamate side chain, in turn, is almost completely buried inside the molecules (see Fig. 1B) and is involved in the salt bridge interaction with the conserved arginine residue. Another common contribution to the electrostatic properties of these proteins is made by their positively-charged N termini located, in all cases, close to the carboxylate of the C termini (region Reg1), leading to mutual attenuation of their contributions to the electrostatic properties of these proteins. Despite the topological similarity of the structural motifs across all human α-defensins, as well as their comparable contributions to electrostatic surface potentials, the region Reg1 may play role in microbicidal and chemotactic specificity of these proteins due to the differences in lengths of the peptide chains determining both the shape and the electrostatic potential of this area.
Another region, defining ∼40%–50% of the molecular surface of α-defensins, is Reg3. This area lacks side chains of charged residues and likely plays a prominent role in the interactions between defensins molecules and hydrophobic components of the membrane, including the transmembrane domains of the receptor(s) coordinating chemotaxis. The size and surface properties of Reg3 are comparable in all human α-defensins. Although the fundamental role of Reg3 for the overall activities of defensins can be easily envisioned, it is harder to reflect this role in terms of the specificities of these proteins.
Whereas in HNP1–3, the surface of Reg2 is positively charged, it is more hydrophobic in HNP4 and HD5, and slightly negatively charged in HD6. In the case of the HNP1–3, Arg14 (HNP3), absent in defensins 4–6, exposes its side chain, defining the charge as well as the shape of this fragment. Although no clear correlation can as yet be identified between the surface properties of this fragment of defensins and their activities, studies of the effects on their biological properties of introducing Arg14 into HD5–6 may provide additional information.
Analysis of Reg4 shows that its properties in HD5 are very different compared with other defensins. In HNP1–4, the electrostatic potential of this fragment of molecular surface is defined by the presence of Arg15. Although this residue is absent in HD6, a similar position is occupied by Arg28, resulting in very similar surface-charge distribution. Equivalent Arg28 in HD5, however, accommodates a quite different conformation, whereas the cationic site of HNP1–4 and HD6 in Reg4 is occupied by the negatively-charged side chain of Glu21 in HD5. The latter residue is found in the amino acid sequence of HD5 only. Therefore, the contribution of Reg4 to the biological properties of human α-defensins 1–4 and 6 is likely comparable, while it may be a determinant for specificity of HD5.
The final section of the molecular surface of defensins, distal to the termini, is Reg5. With the exception of HNP4, this area is defined by one or more positively-charged side chains. One cationic residue, Arg24 in HNP3, contributes to Reg5 in HNP1–3 and HD5. In both intestinal defensins, the side chain of another Arg residue (8 and 9 in HD6 and HD5, respectively), which is absent in HNP1–4, is also located in this area. Consequently, whereas neither of the two arginine residues is found in the sequence of HNP4, both residues are present in HD5, making the electrostatic properties of the Reg5 regions in these defensins opposite.
Discussion
Although human α-defensins became, in recent years, a subject of increased scientific interest (Chang and Klotman 2004; Ericksen et al. 2005; Schneider et al. 2005; Selsted and Ouellette 2005; Buck et al. 2006), only for one member of this family, HNP3, the detail structural studies were described (Hill et al. 1991). Due to the amino acid sequence identity, results obtained for HNP3 are likely quite representative for HNP1 and HNP2; however, the same does not hold for the remaining three human α-defensins. In this report, we present the detailed structural description of these three proteins, supplemented with limited results of antimicrobial and chemotaxis assays. Even though our data confirm the anticipated similarity of the tertiary structures of all α-defensins, significant conformational differences are observed for the specific sections of their molecules. The conformationally most variable fragments are the β-hairpin (T2) formed by the strands β2 and β3, loop (T1) connecting β1 and β2, and the termini. Since these regions have the lowest amino acid sequence identity and are exposed on the molecular surfaces, quite likely they are responsible for the differences in biological activities of different α-defensins. They can also be linked to the oligomerization properties of these proteins. Although at a first glance all human α-defensins form topologically identical dimers, more detailed analysis shows significant differences in the specific intermonomer contacts, stabilizing the dimerization. The effect is most clearly seen for HD6, for which the network of H-bonds relating the strands β2 from two monomers is quite different compared with other human α-defensins, and is associated with a relative mutual shift of two monomers. Another observation is the asymmetry of the dimers in α-defensins, which is most likely their inherent property and not induced by the crystal packing.
The significance of dimerization for biological properties of α-defensins is as yet not completely understood. Several lines of evidence suggest that in vitro formation of dimers or even the particular fold of these proteins may not play a role in their antibacterial and anti-yeast properties (Wu et al. 2003 a,b). Thus, both of these structural features may be important for the in vivo stability of α-defensins. Our recent experiments with chemically modified variants of HNP1 (M. Pazgier, Z. Wu, W. Lu, and J. Lubkowski, in prep.) suggest that for the activation of G-protein-coupled receptor(s) (GPCRs), necessary for chemotaxis, the monomeric α-defensins are sufficient. Therefore, in both activities the ability to form the dimers may eventually play an enhancing role. Very little, however, is known about the mechanisms of interactions between α-defensins and viruses, roles defensins in angiogenesis, or cancer metastasis, therefore a potential role of dimerization for these processes cannot be excluded.
Since the hallmark property of defensins (including the α-family) is their ability to kill or inhibit various pathogens, it was somewhat unexpected to find the lack of activity against the panel of tested microbes for HD6. The amphiphilic character and high content of positively-charged residues are generally associated with microbicidal properties of defensins. Judging by these criteria, HD6 has the lowest number of Arg residues among all six human α-defensins. Additionally, a charge of guanidinium group in Arg7 is effectively “neutralized” within the salt bridge formed with the carboxylate group of Glu14. However, accepting even the connection between the low content of basic residues in HD6 with its lack of antimicrobial properties leaves one significant question: What is the physiological role of this protein? Again, one of possibilities may point to the anti-viral properties, and further studies are needed for some understanding.
In contrast to inactive HD6, the second of the intestinal defensins, HD5, was most potent against the digestive tract bacterium, E. faecium ATCC 1438. This finding correlates well with the site of synthesis in vitro of HD5. Also, the unusually high activity of HD5 against human papillomaviruses was recently reported (Buck et al. 2006).
Neither of intestinal defensins was chemotactic against the cell lines that respond to the gradient of HNPs, distinguishing further these two small subgroups. Since most of the human defensins (from both α- and β-families) were described in the past as chemoattractant, it seems plausible that the subset of cells responsive to the concentration gradient of HD5 and/or HD6 is still to be identified.
Materials and methods
Synthesis, folding, and purification
Details of the synthesis and characterization of α-defensins used in this study were published previously (Wu et al. 2004). Briefly, all peptides were chemically synthesized using Boc solid-phase strategy (references 27 and 28 in Wu et al. 2004) and folded in the presence of reduced and oxidized glutathione and different concentrations of guanidinium chloride. Finally, products were isolated and purified to homogeneity by preparative reverse phase chromatography (RP-HPLC). The electrospray ionization mass spectrometry, analytical RP-HPLC, and activity assays confirmed composition, purity, and activity of synthetic α-defensins.
Crystallization
Crystals of HNP4, HD5, and HD6 were grown using the vapor-diffusion method from the hanging droplets at 20°C. Crystals of HNP4 grew at pH 7.5 (HEPES buffer) in the presence of 2-methyl-2,4-pentanediol (MPD) and PEG400. Crystals of HD5 were also obtained at pH 7.5 from a solution containing lithium sulfate and dioxane. The solution-promoting crystallization of HD6 contained a mixture of sodium acetate trihydrate, MPD, and calcium chloride. For the collection of X-ray data, crystals were frozen at 100 K. The bromide derivatives of HD5 and HD6 were generated by soaking the crystals of native proteins in the solutions obtained by the addition of lithium bromide (0.3–0.6 M final concentration) to the appropriate crystallization solutions. The soaking process extended 30–90 sec. For additional details, see Supplemental Materials.
Data collection and processing
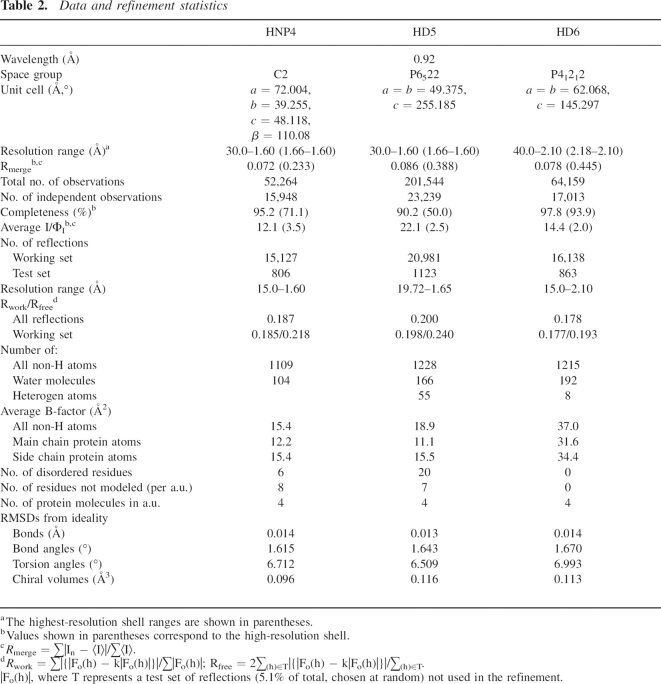
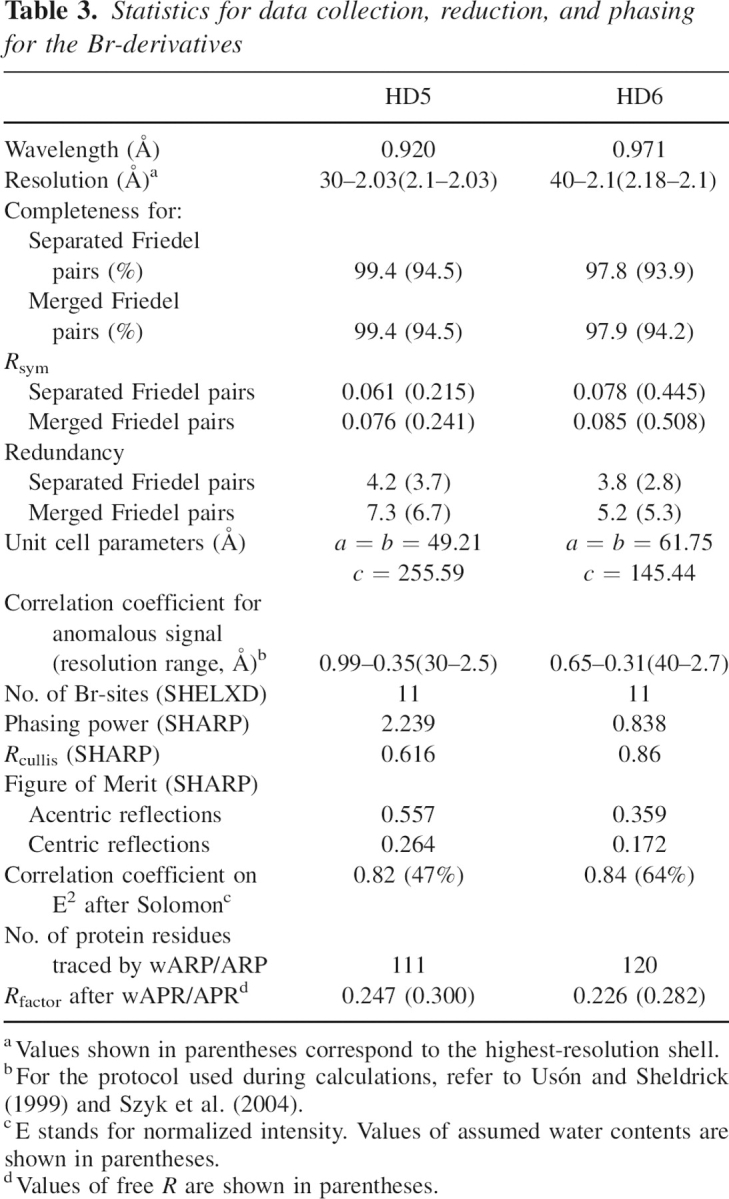
The preliminary X-ray experiments for HNP4 and HD6 were conducted using the conventional radiation source (λCu = 1.5478 Å), passed through Osmic mirrors (Rigaku) originated from the rotating anode mounted on the Rigaku-Ru200 generator operating at 100 V and 50 mA. The diffraction patterns were recorded using the image plate detector, MAR345dtb (MAR Research). Resolution of the datasets extended to ∼2.3 Å and 2.8 Å for HNP4 and HD6, respectively. The final X-ray data for the crystals of native proteins as well as for the bromide derivatives of HD5 and HD6 were collected using the synchrotron radiation (beamline 22BM on the SER-CAT station at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL), with the wavelength adjusted to ∼0.92 Å (absorption edge of Br). The experimental intensities were recorded using the MAR CCD300 detector (MAR Research). The resolution limits of the diffraction patterns for the native proteins varied between 2.1 Å and 1.6 Å and were lower by ∼0.2 Å to 0.3 Å for the bromide derivatives (see also Supplemental Materials). Other properties, such as mosaic spread, radiation sensitivity, or the unit cell parameters, were not affected by the derivatization process. All the X-ray data were processed and scaled using HKL2000 (Otwinowski and Minor 1997). The selected statistics obtained from the reduction of the X-ray data collected for the native and Br-derivative crystals are shown in Tables 2 and 3, respectively.
Table 2.
Data and refinement statistics
Table 3.
Statistics for data collection, reduction, and phasing for the Br-derivatives
Structure solutions and refinements
Initial attempts to solve the structures of three α-defensins by the molecular replacement method were successful only for HNP4. Using programs AMoRe (Navaza 1993) and EPMR (Kissinger et al. 1999), and a model based on the structure of HNP3 (Hill et al. 1991), it was possible to identify clear solution characterized by the correlation coefficient of 0.5, crystallographic R factor of 0.45, and good crystal packing. The asymmetric unit (a.u.) of the HNP4 crystals consists of two nearly identical homodimers, similar to those observed earlier for other human α-defensins (Hill et al. 1991; Xie et al. 2005).
Structures of HD5 and HD6 were solved based on the anomalous signal collected for Br-derivatives. The anomalous signal, evaluated according to previously described strategies (Usón and Sheldrick 1999; Szyk et al. 2004), indicated the specific binding of the Br− ions into the crystal lattice. Based on results obtained from the program SHELXD (Usón and Sheldrick 1999), for each protein, 11 Br sites were selected for further phase refinement with program SHARP (De La Fortelle and Bricogne 1997), followed by phase modification using Salomon (Abrahams and Leslie 1996). The results of these calculations are also characterized in Table 3. In both cases, the initial models were obtained by automatic methods (Lamzin and Wilson 1997).
All three models were first subjected to structural refinement using program CNS (Brünger et al. 1998) and the maximum likelihood as a target function. The structural refinement interspersed by manual model rebuilding lowered the values of the crystallographic R factor and R free to the range of 0.2–0.3. Repetitive refinement and model rebuilding with a gradual extension of the X-ray data resolution allowed modeling of nearly all protein residues. Only in case of HD5, a fragment of one monomer was disordered, and as such could not be modeled throughout the whole refinement process. When the resolution of X-ray data approached the available limits, the well-defined solvent atoms were modeled in the electron density peaks.
At this stage, refinement of all structures was continued with program Refmac 5.0 (Murshudov et al. 1997). In the case of two other defensins, for which X-ray data at higher resolutions were available, we aimed to use the anisotropic B-factor refinement of the sulfur atoms to improve the quality of the modeling of the experimental data. The final stages of refinement consisted of modeling the multiple conformations for several residues, inclusion of small molecules (ions, glycerol, buffer, etc.), and refinement of the individual B-factors for all atoms. The sulfur atoms in HNP4 and HD5 were refined using an anisotropic model for B-factors. In both structures, correctness of this approach was confirmed by lowering of the free R values.
In the final model of HNP4, a single C-terminal residue is missing in monomers B and D, while in each of the monomers, A and C, the last three residues were not modeled due to disorder. While all the protein residues could be modeled in three monomers of HD5, the electron density corresponding to the fragment C10ATRES15 in monomer D could not be interpreted (probably due to the disorder). Two conformations were established for the N terminus (A1TCYCRT7) and the last two C-terminal residues of the monomer D. Six SO4 2− and two Cl− anions, as well as four molecules of glycerol, could be modeled in the electron density peaks. All protein residues could be clearly modeled in the structure of HD6. The complete model also consists of eight Cl− anions. The basic refinement statistics as well as the detail content of a.u. are shown in Table 2. The final coordinates and the experimental structure factors have been deposited with the Protein Data Bank (PDB codes: HNP4, 1zmm; HD5, 1zmp; HD6, 1zmq).
Antimicrobial assay
Three microorganisms, E. coli ATCC 25922, S. aureus ATCC 29213, and C. albicans ATCC 99788 (amphotericin B-resistant), were assayed with all three defensins (and HNP1, used as a reference) studied in this paper. Additionally, microbicidal activity of HD5 and HD6 was tested against E. faecium 1438 and P. aeruginosa PAO1. The microorganisms were grown to mid-logarithmic phase in tryptic soy broth, and then diluted to 1×106 CFU/mL in 10 mM potassium phosphate, 1% tryptic soy broth (pH 7.4); 100 μL of cells were incubated in the presence of different concentrations of peptides for 3 h at 37°C. The cells were then diluted serially in the same buffer, plated on Luria Broth plates, and incubated for 18 h at 33°C, and the colonies were counted. Microbicidal activity was expressed as the ratio of colonies counted to the number of colonies on a control plate. All experiments were completed at least three times, and the average values are reported here. The IC50 is the concentration of protein (peptide) at which 50% of the viable cells are killed.
Chemotactic activity assay
Human peripheral blood mononuclear cells (PBMC) were isolated from leukopacks of healthy blood donors (courtesy of the Transfusion Medicine Department, NIH Clinic Center, Bethesda, MD) by routine Ficoll-Hypaque density gradient centrifugation, with the approval of the National Cancer Institute (NCI) Human Research Committee. Human peripheral blood CD4 T cells were purified from PBMC using human CD4 T cell negative selection columns (R & D Systems) following the manufacturer's recommendation. The purity of T cell subset populations was checked by FACScan analysis, and only cells with a purity of >95% were used.
The migration of CD4 T cells in response to α-defensin(s) was measured by the 48-well microchemotaxis chamber assay as described previously (Yang et al. 2000). In brief, α-defensins or SDF-1α (used as a positive control) diluted in chemotaxis medium (CM, RPMI 1640 containing 1% BSA) were placed in wells of the lower compartment of the chamber (Neuro Probe), and CD4 T cells suspended in CM at 5×106/mL were added to the wells of the upper compartment. The lower and upper compartments were separated by a 5-μm polycarbonate filter (NeuroProbe) precoated overnight at 4°C in RPMI 1640 containing 10 μg/mL fibronectin (Sigma). After incubation at 37°C in humidified air with 5% CO2 for 3 h, the filter was removed, scraped, stained, counted, and presented as the average (mean±SD) number of T cells migrated per high power field (No./HPF) of triplicate wells.
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This project was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Institutes of Health, National Cancer Institute, under Contract NO1-CO-12400.
Footnotes
Reprint requests to: Jacek Lubkowski, National Cancer Institute, Frederick, 539, Frederick, MD 21702, USA; e-mail: jacek@ncifcrf.gov; fax: (301) 846-7101.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062336606.
Supplemental material: see www.proteinscience.org
References
- Abrahams, J.P. and Leslie, A.G.W. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52: 30–42. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunsteleve, R.W., Jiang, J.S., Kuszewski, J., Nigles, M., and Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Buck, C.B., Day, P.M., Thompson, C.D., Lubkowski, J., Lu, W., Lowy, D.R., and Schiller, J. 2006. Human α-defensins block papillomavirus infection. Proc. Natl. Acad. Sci. 103: 1516–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, M. 1991. Ribbons 4.0. J. Appl. Crystallogr. 24: 958–961. [Google Scholar]
- Chang, T.L. and Klotman, M.E. 2004. Defensins: Natural anti-HIV peptides. AIDS Res. 6: 161–168. [PubMed] [Google Scholar]
- De La Fortelle, G.M. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276: 472–494. [DOI] [PubMed] [Google Scholar]
- Ericksen, B., Wu, Z., Lu, W., and Lehrer, R.L. 2005. Antibacterial activity and specificity of the six human α-defensins. Antimicrob. Agents Chemother. 49: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz, T. 2002. Immunology: Versatile defensins. Science 298: 977–979. [DOI] [PubMed] [Google Scholar]
- Ganz, T., Selsted, M.E., Szklarek, D., Harwig, S.S.L., Daher, K., Bainton, D.F., and Lehrer, R.I. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76: 1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder, J., Bartels, J., Christophers, E., and Schröder, J.M. 1997. A peptide antibiotic from human skin. Nature 387: 861. [DOI] [PubMed] [Google Scholar]
- Hill, C.P., Yee, J., Selsted, M.E., and Eisenberg, D. 1991. Crystal structure of defensin HNP-3, an amphiphilic dimer: Mechanisms of membrane permeabilization. Science 251: 1481–1485. [DOI] [PubMed] [Google Scholar]
- Hoover, D.M., Rajashankar, K.R., Blumenthal, R., Puri, A., Oppenheim, J.J., Chertov, O., and Lubkowski, J. 2000. The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275: 32911–32918. [DOI] [PubMed] [Google Scholar]
- Jing, W., Hunter, H.N., Tanabe, H., Ouellette, A.J., and Vogel, H.J. 2004. Solution structure of cryptdin-4, a mouse paneth cell α-defensin. Biochemistry 43: 15759–15766. [DOI] [PubMed] [Google Scholar]
- Jones, D.E. and Bevins, C.L. 1992. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 267: 23216–23225. [PubMed] [Google Scholar]
- Jones, D.E. and Bevins, C.L. 1993. Defensin-6 mRNA in human Paneth cells: Implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 315: 187–192. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kissinger, C.R., Gehlhaar, D.K., and Fogel, D.B. 1999. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 55: 484–491. [DOI] [PubMed] [Google Scholar]
- Lamzin, V.S. and Wilson, K.S. 1997. Automated refinement for protein crystallography. Methods Enzymol. 277: 269–305. [DOI] [PubMed] [Google Scholar]
- Lehrer, R.I. and Ganz, T. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14: 96–102. [DOI] [PubMed] [Google Scholar]
- Lehrer, R.I., Selsted, M.E., Szklarek, D., and Fleischmann, J. 1983. Antibacterial activity of microbicidal cationic proteins 1 and 2, natural peptide antibiotics of rabbit lung macrophages. Infect. Immun. 42: 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzmeier, R., Michaelson, D., Liu, L., and Ganz, T. 1993. The structure of neutrophil defensin genes. FEBS Lett. 321: 267–273. [DOI] [PubMed] [Google Scholar]
- Maemoto, A., Qu, X., Rosengren, K.J., Tanabe, H., Henschen-Edman, A., Craik, D.J., and Ouellette, A.J. 2004. Functional analysis of the α-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279: 44188–44196. [DOI] [PubMed] [Google Scholar]
- McManus, A.M., Dawson, N.F., Wade, J.D., and Craik, D.J. 2000. Three-dimensional structure of RK-1: A novel α-defensin peptide. Biochemistry 39: 15757–15764. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza, J. 1993. On the computation of the fast rotation function. Acta Crystallogr. D Biol. Crystallogr. 49: 588–591. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation model. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Sanner, M.F., Spehner, J.-C., and Olson, A.J. 1996. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 38: 305–320. [DOI] [PubMed] [Google Scholar]
- Schneider, J.J., Unholzer, A., Schaller, M., Schäfer-Korting, M., and Korting, H.C. 2005. Human defensins. J. Mol. Med. 83: 587–595. [DOI] [PubMed] [Google Scholar]
- Selsted, M.E. and Harwig, S.S.L. 1989. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J. Biol. Chem. 264: 4003–4007. [PubMed] [Google Scholar]
- Selsted, M.E. and Ouellette, A.J. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6: 551–557. [DOI] [PubMed] [Google Scholar]
- Selsted, M.E., Szklarek, D., and Lehrer, R.I. 1984. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect. Immun. 45: 150–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes, R.S., Kronenberg, M., Heinzmann, C., Daher, K.A., Klisak, I., Ganz, T., and Mohandas, T. 1989. Assignment of defensin gene(s) to human chromosome 8p23. Genomics 5: 240–244. [DOI] [PubMed] [Google Scholar]
- Szyk, A., Lu, W., Xu, C., and Lubkowski, J. 2004. Structure of the scorpion toxin BmBKTtx1 solved from single wavelength anomalous scattering of sulfur. J. Struct. Biol. 145: 289–294. [DOI] [PubMed] [Google Scholar]
- Tang, Y.-Q. and Selsted, M.E. 1993. Characterization of the disulfide motif in BNBD-12, an antimicrobial β-defensin peptide from bovine neutrophils. J. Biol. Chem. 268: 6649–6653. [PubMed] [Google Scholar]
- Usón, I. and Sheldrick, G.M. 1999. Advances in direct methods for protein crystallography. Curr. Opin. Struct. Biol. 9: 643–648. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Hoover, D.M., Yang, D., Boulegue, C., Santamaria, F., Oppenheim, J.J., Lubkowski, J., and Lu, W. 2003a. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human β-defensin 3. Proc. Natl. Acad. Sci. 100: 8880–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Powell, R., and Lu, W. 2003b. Productive folding of human neutrophil α-defensins in vitro without the pro-peptide. J. Am. Chem. Soc. 125: 2402–2403. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Prahl, A., Powell, R., Ericksen, B., Lubkowski, J., and Lu, W. 2003c. From pro defensins to defensins: Synthesis and characterization of human neutrophil pro α-defensin-1 and its mature domain. J. Pept. Res. 62: 53–62. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Ericksen, B., Tucker, K., Lubkowski, J., and Lu, W. 2004. Synthesis and characterization of human α-defensins 4–6. J. Pept. Res. 64: 118–125. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Li, X., de Leeuw, E., Ericksen, B., and Lu, W. 2005. Why is the Arg5-Glu13 salt bridge conserved in mammalian α-defensins? J. Biol. Chem. 280: 43039–43047. [DOI] [PubMed] [Google Scholar]
- Xie, C., Prahl, A., Ericksen, B., Wu, Z., Zeng, P., Li, X., Lu, W.-Y., Lubkowski, J., and Lu, W. 2005. Reconstruction of the conserved β-bulge in mammalian defensins using D-amino acids. J. Biol. Chem. 280: 32921–32929. [DOI] [PubMed] [Google Scholar]
- Yang, D., Chen, Q., Chertov, O., and Oppenheim, J.J. 2000. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J. Leukoc. Biol. 68: 9–14. [PubMed] [Google Scholar]
- Yang, D., Biragyn, A., Hoover, D.M., Lubkowski, J., and Oppenheim, J.J. 2004. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 22: 181–215. [DOI] [PubMed] [Google Scholar]
- Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415: 389–395. [DOI] [PubMed] [Google Scholar]