Abstract
Deletion of the regulatory N-terminal arms of the AraC protein from its dimerization domain fragments increases the susceptibility of the dimerization domain to form a series of higher order polymers by indefinite self-association. We investigated how the normal presence of the arm inhibits this self-association. One possibility is that arms can act as an entropic bristles to interfere with the approach of other macromolecules, thereby decreasing collision frequencies. We examined the repulsive effect of flexible arms by measuring the rate of trypsin cleavage of a specially constructed ubiquitin-arm protein. Adding an arm to ubiquitin or increasing its length produced only a modest repulsive effect. This suggests that arms such as the N-terminal arm of AraC do not reduce self-association by entropic exclusion. We consequently tested the hypothesis that the arm on AraC reduces self-association by binding to the core of the dimerization domain even in the absence of arabinose. The behaviors of dimerization domain mutants containing deletions or alterations in the N-terminal arms substantiate this hypothesis. Apparently, interactions between the N-terminal arm and the dimerization domain core position the arm to interfere with the protein–protein contacts necessary for self-association.
Keywords: entropic bristle, ubiquitin, velocity sedimentation, fluorescence, trypsin cleavage, AraC, regulation
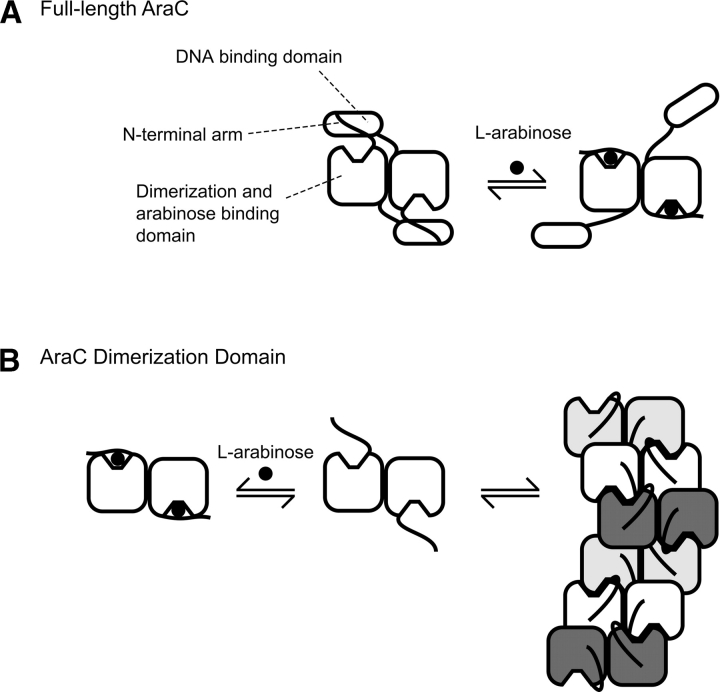
The N-terminal arms, residues 1–15, particularly residues 8–14, of AraC, a transcriptional regulator of the L-arabinose araBAD operon in Escherichia coli, play a critical role in controlling the protein's response to arabinose (Fig. 1A) (Saviola et al. 1998; Seabold and Schleif 1998; Wu and Schleif 2001a,b; Ross et al. 2003; Gryczynski and Schleif 2004). Structure determination (Soisson et al. 1997a) and surface plasmon resonance experiments (Ghosh and Schleif 2001) have shown that in the presence of arabinose, the arms bind to the dimerization domains of AraC over the bound arabinose as illustrated in the right half of Figure 1A. A variety of genetic and physical experiments indicate that in the absence of arabinose, however, the arms bind instead to the DNA-binding domains of AraC (Saviola et al. 1998; Reed and Schleif 1999; Wu and Schleif 2001a). These experiments, plus additional physiological experiments (Harmer et al. 2001), indicate that the interactions between the arms and the DNA-binding domains hold the protein in a conformation that favors DNA looping. Thus, when arabinose is present, the binding of the arms to the dimerization domains frees the DNA-binding domains, allowing them to contact two adjacent direct repeat DNA sites and thereby activate transcription from the araBAD promoter.
Figure 1.
Behavior of the full-length AraC protein (A) and the dimerization domain fragment of AraC (B) in the presence and absence of the ligand L-arabinose. For full-length AraC protein in the absence of arabinose, binding of the N-terminal arms to the DNA binding domains helps hold the domains fixed. As explained in the cited references, in vivo this favors DNA looping and repression of the nearby ara genes. When arabinose is present, the arms bind more tightly to the dimerization domains, thereby freeing the DNA binding domains, which ultimately results in induction of the nearby ara genes. AraC protein lacking the DNA binding domain; that is, dimerization domain in the absence of ligand forms indefinite-length oligomers through interactions involving the ligand-binding pocket as indicated.
This study addresses unanswered questions about the allosteric mechanism of AraC. First, do the N-terminal arms perform a function other than stiffening the protein in the absence of arabinose through interactions with the DNA-binding domains? Second, is binding of the N-terminal arms to the dimerization domains an all or none phenomenon controlled by arabinose, or do the arms have significant affinity for the dimerization domains in the absence of arabinose? The experiments described in this article originated from an observation that the tendency of the AraC dimerization domain to form insoluble aggregates, first described by Soisson et al. (1997a), is strongly enhanced when their N-terminal arms are deleted (N. Moore, J. Weldon, and R. Schleif, unpubl.). The structural basis of the aggregation appears to be understood. In the absence of arabinose, a solvent-exposed tyrosine residue from one dimerization domain monomer binds into the arabinose-binding pocket of a dimerization domain from another dimer. This joins two dimers in a mode that can continue indefinitely (Fig. 1B). Because deleting the arms accentuates the aggregation of dimerization domain, the arms apparently interfere with the indefinite self-association process. In this article, we document that deleting the arms strongly increases the self-association of AraC dimerization domain. We then test possible explanations for the solubilizing effect of the arms.
We envision three potential explanations for how the N-terminal arms of AraC inhibit aggregation of dimerization domain. First, the arms may act as entropic bristles to resist the approach of other macromolecules in solution. Second, the arms may not be completely free in the absence of ligand and may bind to the core of the dimerization domain, obscuring contacts necessary for self-association. Third, the arms may not interfere with the specific interactions that lead to indefinite self-association but may act as a “solubility tag” to enhance the solubility of the dimerization domain by virtue of the arms’ intrinsic solubility.
Entropic bristles are flexible polypeptide regions in which thermal motion generates a time-averaged “domain” that tends to exclude other macromolecules because restricting the movement of the bristle is entropically unfavorable (Hoh 1998). The intrusion of another macromolecule into a bristle domain essentially requires compression of the bristle “gas,” and hence is resisted by the bristle's thermal motion. The arms of AraC are attached to the cores of the dimerization domain immediately adjacent to the ligand-binding pockets and the interface involved in self-association (Soisson et al. 1997a). Thus their location is compatible with the arms acting as entropic bristles.
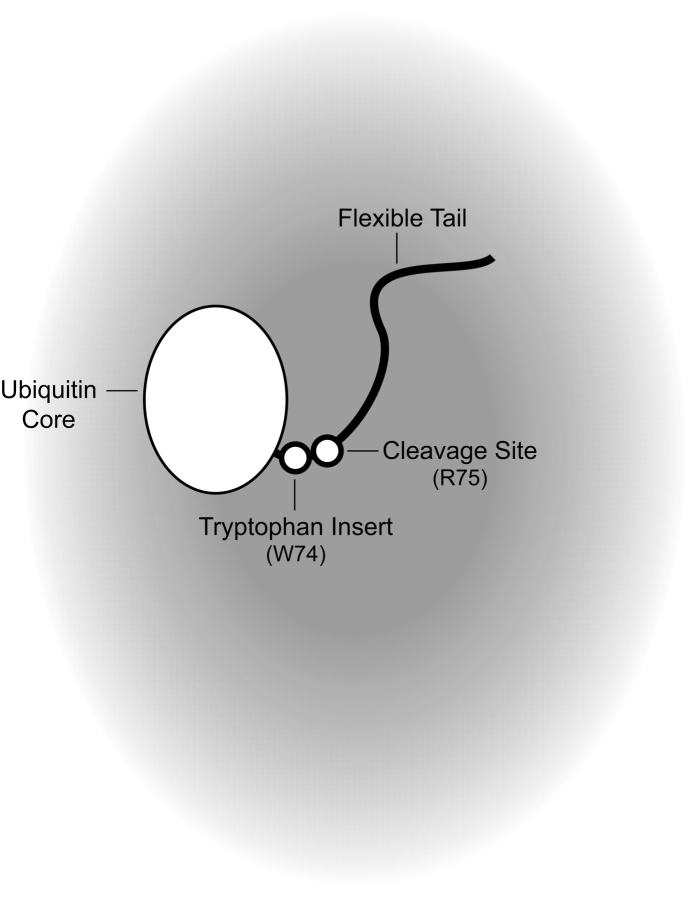
We developed a real-time fluorescence-based assay suitable for measuring the ability of a peptide similar in size to the arm of AraC to exclude another macromolecule from its vicinity. The assay measures the rate of trypsin cleavage near the base of a polypeptide arm introduced at the C terminus of a ubiquitin protein core (Fig. 2). While the results of this assay show a small decrease in the cleavage rate as the length of the flexible polypeptide arm is increased from two to 18 residues, the change is insufficient to explain the difference in solubility between dimerization domain containing the N-terminal arm and dimerization domain lacking the arm.
Figure 2.
Design of an entropic bristle test construct. Shading represents the effective concentration of the entropic bristle.
Since short peptides the length of the N-terminal arm of AraC do not possess a strong entropic bristle effect, the arms more likely inhibit oligomerization by a mechanism other than entropic exclusion. Therefore, we tested the possibility that, even in the absence of arabinose, the arms bind significantly to the dimerization domains, and that this interferes with the interactions necessary to self-associate and form insoluble oligomers. Altering or deleting the functionally important residues (Ross et al. 2003), 8–14, of the arm yields a dimerization domain whose properties of self-association are almost identical to those of protein deleted of residues 2–14. Since the identities of residues in the arms are vital to the inhibition of indefinite self-association, we conclude that specific interactions between the arm and dimerization domain are required to reduce self-association of the AraC dimerization domain. These data also suggest that the arms are not acting as “solubility tags” because there is no significant change in charge density or hydrophilicity of the protein when residues 8–14 are mutated.
Results
The N-terminal arm of AraC reduces self-association of the dimerization domain
Deletion of the N-terminal arms from the AraC dimerization domain yields protein that is much less soluble than intact dimerization domain (N. Moore, J. Weldon, and R. Schleif, unpubl.). To verify and examine more quantitatively the role of the arms on the solubility of the dimerization domain, we compared the sedimentation properties of wild-type to mutant AraC dimerization domain deleted of residues 2–14–Δ(2–14). Deletions beyond residue 14 yield a protein that is too poorly soluble to purify and concentrate to useful concentrations. Velocity sedimentation experiments have previously shown that the wild-type dimerization domain sediments as a single species in the presence of L-arabinose and as a distribution of higher-order species in the absence of L-arabinose (Soisson et al. 1997a). We expected that the deletion of residues 2–14 would enhance self-association of the dimerization domain, which would lead to the formation of larger dimerization domain assemblies in the absence of L-arabinose. We also expected that the addition of L-arabinose would decrease the extent of self-association for both proteins.
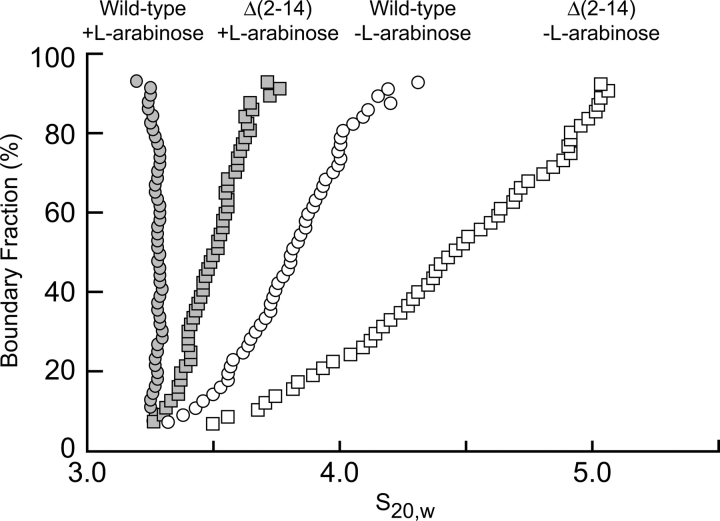
Figure 3 presents the results of an analysis of velocity sedimentation data of both wild-type and Δ(2–14) AraC dimerization domain in the presence and absence of L-arabinose. A homogeneous noninteracting species will sediment with a single sedimentation coefficient S, whereas an indefinitely self-associating species will sediment with a distribution of S values. As expected, the protein with the deletion of residues 2–14, sediments with a considerably broader range of S values than wild-type protein, both in the presence and in the absence of arabinose, and arabinose decreases the range of S values for both proteins (Fig. 3). This demonstrates that the deletion of residues 2–14 allows the dimerization domain to self-associate with a greater affinity than wild-type protein. These results are consistent with the indefinite self-association as described (Soisson et al. 1997a), in which tyrosine competes with L-arabinose for access to the ligand-binding pocket of AraC. Although L-arabinose reduces the formation of higher-order species of the Δ(2–14) construct, it does not entirely eliminate their formation. This behavior is as expected for competition between arabinose binding and self-association if the Δ(2–14) construct has both an increased propensity to oligomerize and a decreased affinity for L-arabinose. Indeed, we found that arabinose binds more weakly to the Δ(2–14) protein, K D ≈ 2.2 ± 0.1 mM, than to wild-type protein, K D ≈ 0.6 ± 0.1 mM, as measured by the fluorescence assay described in the Materials and Methods section.
Figure 3.
Extrapolation plot from a van Holde-Weischet analysis of velocity sedimentation data from the wild-type and Δ(2–14) AraC dimerization domain constructs in the presence and absence of L-arabinose.
We imagine three potential explanations for the increase in indefinite self-association displayed by the protein with the deletion of residues 2–14 of the N-terminal arm. As discussed above, the N-terminal arm of AraC could be acting like an entropic bristle to restrict the spatial proximity of dimerization domain molecules, thereby inhibiting self-association. A second possibility is that the arm is not disordered in solution but instead makes specific interactions with the core of the dimerization domain that inhibit the formation of contacts necessary for self-association. Last, the arm could be acting as a “solubility tag” to enhance the solubility of the core protein by virtue of its own exceptional solubility. We first tested the hypothesis that the arm is acting as an entropic bristle.
Design and construction of an entropic bristle test system
We designed and constructed a system to evaluate entropic bristle behavior of short polypeptide arms. A short polypeptide chain extending from a protein might act like an entropic bristle and restrict macromolecular access to a nearby binding site. Our entropic bristle test system (Fig. 2) utilizes ubiquitin, a 76-residue compact, tryptophan-free, globular protein that is used physiologically as a signal for protein degradation (for review, see Hershko 2005). When folded, ubiquitin contains a single naturally occurring trypsin cleavage site near the C terminus, just after Arg74, which is followed by a Gly-Gly sequence. We monitored cleavage at this site by measuring the fluorescence of a tryptophan residue introduced immediately before Arg74. Cleavage at the arginine changes the environment of the only tryptophan in the construct and hence changes its intrinsic fluorescence, thus allowing a real-time measurement of the kinetics of cleavage. We introduced additional residues at the C terminus of the protein to form an arm that is likely to be unstructured and flexible. We then measured the rate of trypsin cleavage as the arm was incrementally extended from two to 10 to 18 residues.
It is important that the residues added to the protein be flexible and preferably unstructured. To maximize the chances that the arm be flexible, an eight-residue sequence was chosen based on the repeated unit of the entropic bristle in neurofilaments (Brown and Hoh 1997). Subsequent analysis of these proteins using programs designed to predict disordered regions of polypeptide chains (VL2, VL3, VL3H, and VL3E accessed through DisProt [Vucetic et al. 2005], http://www.disprot.org) predict that, indeed, the added residues are unstructured (Obradovic et al. 2003; Vucetic et al. 2003; Peng et al. 2005).
Real-time observation of trypsin cleavage
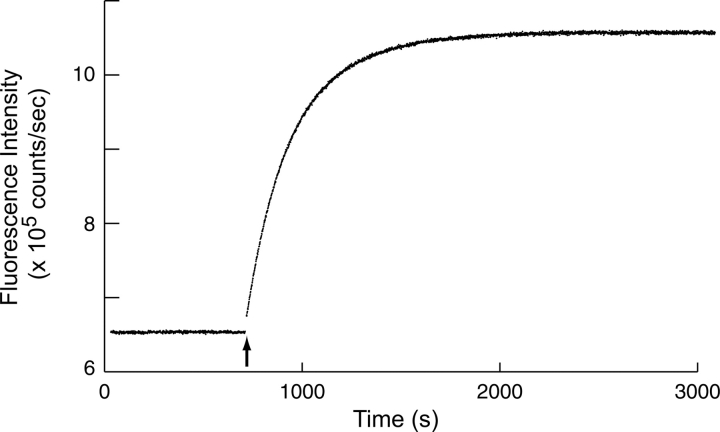
A time course of cleavage for ubiquitin constructs with two-residue, 10-residue, and 18-residue arms was observed by monitoring the increase in intrinsic tryptophan fluorescence following the addition of trypsin. A typical time course is shown in Figure 4. Rate constants from an exponential rate function fitted to the data are presented in Table 1. We see a modest decrease of ∼16% in the rate constant of the cleavage reaction as the arm is lengthened from two to 18 residues. This result suggests the presence of a very small entropic bristle effect. Although a quantitative relationship between an entropic bristle effect and self-association is likely to be somewhat obscure, it seems highly unlikely that the small bristle effects we measured for short bristles could be the major reason why deleting the N-terminal arm of AraC so dramatically decreases its solubility. We therefore concluded that some other effect is a more likely explanation for the reduction in self-association promoted by the arms on the AraC dimerization domain.
Figure 4.
Fluorescence intensity time course of the UbW74-A0 construct before and after the addition of trypsin. The arrow indicates the point of trypsin addition.
Table 1.
Trypsin cleavage rate constants for ubiquitin-tail constructs
The effects of mutations in the N-terminal arm of the AraC dimerization domain
The results presented in the previous section suggest that the N-terminal arms of AraC may interfere with self-association of the dimerization domain in a way other than by entropic exclusion. Perhaps, even in the absence of arabinose, the arm interacts with the dimerization domain core in a manner that competes with the formation of contacts necessary for self-association. For example, residues 8–14 of the arm, which normally bind to the domain core in the presence of arabinose (Soisson et al. 1997a) and which play a key role in the protein's response to arabinose (Ross et al. 2003), might retain a degree of affinity for binding over the ligand-binding pocket even in the absence of interactions with ligand. To test this hypothesis, we compared the sedimentation properties of the Δ(2–14) dimerization domain construct with the properties of a dimerization domain construct deleted of residues 8–14, Δ(8–14), and a construct that replaced residues 8–14 with a peptide sequence different from wild-type protein, Mut(8–14). The arm sequences of these constructs are described in Table 2.
Table 2.
Mutations in the N-terminal arm of the AraC dimerization domain
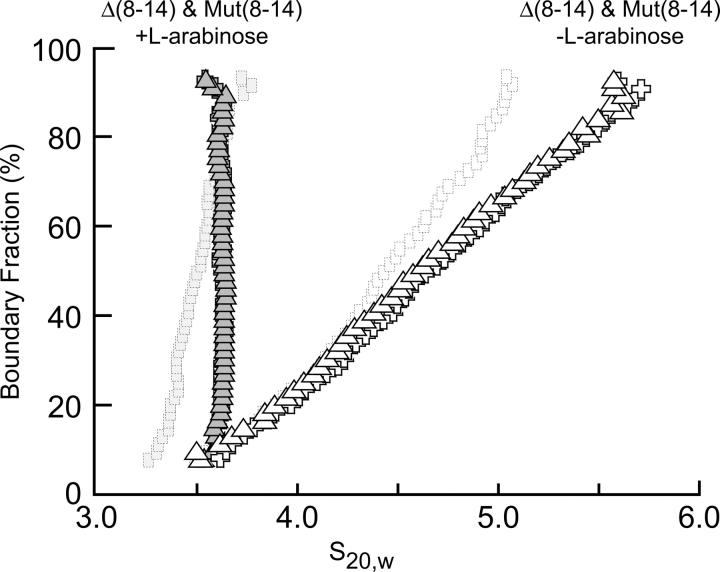
We used velocity sedimentation in the presence and absence of L-arabinose to compare the self-association of the deletion constructs Δ(2–14) and Δ(8–14) with the construct whose arm is altered in sequence, but not in length, Mut(8–14) (Fig. 5). All three proteins behave in a similar manner, which indicates that a property of specific residues in positions 8–14 of the arm must reduce self-association. In addition, these results indicate that altering residues 8–14 is equivalent to deleting them. Thus, the identity of residues 8–14, not just the presence of any residue at those positions, affects the extent of oligomerization.
Figure 5.
Extrapolation plot from a van Holde-Weischet analysis of velocity sedimentation data from the Δ(8–14) and Mut(8–14) AraC dimerization domain constructs in the presence and absence of L-arabinose. The behavior of the Δ(2–14) dimerization domain mutant (Fig. 1) is presented in the background for reference.
The fact that specific residues in the arm are required to inhibit self-association of the dimerization domain is most easily explained if the wild-type residues in the arm are capable of significant interactions with the core of the protein in the absence of arabinose. This conclusion is consistent with our previous finding that the entropic bristle effects of short arms on the approach of other proteins appears to be very small. In addition, these data suggest that the arms are not acting as “solubility tags” because altering the identities of residues 8–14 without generating appreciable change in the charge density or hydrophilicity of the dimerization domain still enhanced self-association of the mutant protein.
Discussion
The experiments reported here confirm that the N-terminal arm of the dimerization domain of AraC protein substantially hinders indefinite self-association of the core dimerization domain. To learn the basis for this hindrance, we tested whether the inhibition is a result of the arm acting as an entropic bristle that repels other proteins from its vicinity. Not only would this effect be important for the behavior of AraC, but it would also be important for understanding other arm-domain proteins, as well as systems into which an arm has been engineered. Arm effects could also have significant implications for protein–protein interactions in general because many proteins possess unstructured peptide arms with lengths up to 15 residues or so at their N and/or C termini.
By engineering an arm system onto the compact, tryptophan-free, and easily purified protein ubiquitin, it proved relatively simple to measure directly the effect of short arms on the exclusion of other proteins from the arm's vicinity. We added a tryptophan residue to the arm engineered onto ubiquitin in such a position that cleavage of the arm with trypsin would change the environment, and hence the fluorescence, of the tryptophan. Because the fluorescence could be accurately measured in real time, it was possible to make precise measurements of the rate of cleavage of the arm as a function of the arm length.
Although we did observe a decrease in the cleavage rate as the arm increased in length from two to 10 to 18 residues, the decrease in cleavage rate was rather modest, only 16% overall. Thus, while it was possible to detect an entropic bristle effect for the short arms we studied, its magnitude is insufficient to explain the strong effect of the 18-residue arm on AraC protein in inhibiting indefinite self-association of the dimerization domain. Even though we took precautions to reduce the possibility that the arms all possessed stable, rigid, and compact structures, that possibility cannot be excluded. Overall, we consider entropic bristle effect to be an unlikely explanation for the self-association properties of AraC.
When residues eight through 14 of the N-terminal arm on the dimerization domain of AraC were altered, the protein formed higher-order species as extensively as when residues two through 14 were deleted. Because altering residues eight to 14 does not significantly alter the charge density or the hydrophilicity of the arms, or the protein as a whole, it seems unlikely that N-terminal arms on the dimerization domain act to keep the protein soluble by functioning as a “solubility tag.” Instead, it seems that the native sequence of the N-terminal arm binds specifically, that is, in a residue-dependent manner, to the dimerization domain and it blocks formation of the interaction through the ligand-binding pocket that leads to indefinite polymerization of the protein.
An interaction between the arm and dimerization domain that inhibits self-association could easily be similar to the one that forms in the presence of arabinose (Soisson et al. 1997a). Arabinose may simply provide additional interactions necessary to strengthen the contact between the arm and dimerization domain to the level needed to shift the equilibrium position of the arm from predominantly bound to DNA-binding domain to largely bound to dimerization domain.
In principle, structural studies could determine the arm's position in the absence of arabinose, but in practice, this has not yet been possible. The crystal structure of the dimerization domain in the absence of ligand shows the tyrosine-mediated indefinite self-association (Soisson et al. 1997a) that necessitates displacement of the arm from its arabinose-bound position. The high concentration of dimerization domain required for crystallization could easily have shifted the equilibrium between arm binding and self-association to favor indefinite self-association. In addition, crystallization of this form was possible only at pH 9. Since structural evidence suggests that low pH strengthens binding between the arm and the core of the protein by protonating a histidine (Soisson et al. 1997b), the use of a high pH may have sufficiently weakened arm binding that displacement of the arm and crystallization became possible. Due to the relatively high molecular weight of the dimerization domain, NMR characterization of the freedom of the N-terminal arm would also be a technically challenging problem.
In summary, we have demonstrated that the repulsive effects of peptide arms acting as entropic bristles can be observed, but arms of the length and sequences we studied, two to 18 residues, did not produce dramatic entropic exclusion. Our measurements of self-association, evaluation of entropic exclusion, and the effects of deleting or altering arm sequence, as well as structures of the arabinose-bound and apo-AraC dimerization domain, all indicate that the presence of the arms directly interferes with self-association of the dimerization domain. This likely results from specific interactions between the arm and the core of the dimerization domain that exists even in the absence of arabinose.
Materials and methods
Generation and purification of ubiquitin constructs
Plasmid pET16b-ubiquitin consists of the full-length 231-nucleotide ubiquitin reading frame cloned between the NdeI and BamHI sites of expression vector pET16b (ampr). The resulting vector expresses full-length ubiquitin protein (residues 1–76) preceded by an N-terminal 10-His tag that is connected to the protein with a nine-residue linker containing a Factor Xa cleavage site (You et al. 1999). Oligonucleotide mutagenesis primers were obtained from Integrated DNA Technologies. Plasmid DNA was isolated and purified from cells using the Wizard Plus Miniprep DNA purification system from Promega. Mutations and insertions were introduced into the pET16b-ubiquitin plasmid using the QuikChange Site-directed Mutagenesis kit from Stratagene and were confirmed by DNA sequencing.
We deleted DNA coding for the N-terminal 10-His tag and the linker containing the Factor Xa cleavage site from the pET16b-ubiquitin expression vector. The resulting plasmid, termed pUb, expresses only the full-length ubiquitin construct (residues 1–76). Next, a codon for a single tryptophan residue was inserted immediately prior to the codon for residue R74. The resulting construct, pUbW74-A0, expresses ubiquitin that contains a two-residue (GG) arm following residues W74 and R75. Three DNA sequences coding for eight-residue segments were then added in succession to the end of the gene on pUbW74 to generate expression vectors that produce ubiquitin constructs with a 10-residue (pUbW74-A1; GGASAPTSPA), an 18-residue (pUbW74-A2; GGASAPTSPAPSTAPASA), or a 26-residue (pUbW74-A3; GGASAPTSPAPSTAPASAGGTAPGSA) flexible arm immediately subsequent to R75. The sequence of the eight-residue inserts was based on the repeated unit of the entropic bristle in neurofilaments (Brown and Hoh 1997), but with lysine residues removed. Subsequent analysis of these proteins using programs designed to predict disordered regions of polypeptide chains (VL2, VL3, VL3H, and VL3E accessed through DisProt [Vucetic et al. 2005], www.disprot.org) indicates that the added residues are unstructured (Obradovic et al. 2003; Vucetic et al. 2003; Peng et al. 2005).
Mutant constructs derived from ubiquitin were expressed using the pET16b-ubiquitin plasmid in the expression-optimized E. coli strain BL21 (DE3) from Stratagene and purified as follows. Cells were grown with shaking at 37°C in YT medium (Schleif and Wensink 1981) to ∼1 × 108 cells/mL, induced by the addition of IPTG to 1 mM, and grown for an additional 3 h; then they were harvested by sedimentation at 6000g for 10 min at 4°C. The cell pellet was resuspended in 20 mM HEPES (pH 7.5), 0.1 M NaCl, 5% glycerol, 10 mM MgCl2, 1 mM PMSF, with 10 μg/mL each DNase I and RNase A added immediately before use, and lysed by a French press. Debris was removed by centrifugation at 10,000g for 15 min at 4°C. Acetic acid (50%) was added drop-wise until the pH dropped to ∼4.5–5.0. The extract was then centrifuged at 10,000g for 20 min at 4°C, and the pH of the supernatant was raised to >5 with one small pellet of NaOH. The solution was then dialyzed against 10- to 100-fold excess of 20 mM NH4-acetate (pH 5.1) at 4°C. The dialyzed sample was filtered through a 0.22-μm PVDF low-protein-binding syringe filter from Millipore; bound to an Amersham Pharmacia Biotech HiTrap SP HP, 1 mL volume, cation exchange column; and eluted at 100–200 mM in a 40-mL gradient from 20 to 500 mM NH4-acetate (pH 5.1) and 0.1 mM NaN3. Fractions containing ubiquitin (the only major elution peak) were dialyzed against buffer containing 20 mM HEPES (pH 7.7) and 100 mM NaCl.
Generation and purification of AraC dimerization domain constructs
Plasmid AraCTF (LaRonde-LeBlanc and Wolberger 2000), encoding the arm and dimerization domain of AraC, was derived from a pET21b (ampr) expression vector by inserting nucleotides 1–546 of AraC between the NdeI and AdeI sites of the plasmid. The resulting vector expresses residues 1–182 of AraC followed by a C-terminal Leu-Glu linker and a 6-His tag. Mutations were introduced and verified as above. Wild-type dimerization domain protein and mutant constructs were expressed as above and purified by methods modified from those previously described (LaRonde-LeBlanc and Wolberger 2000). Cells grown, induced, and harvested as above were resuspended in 15 mM Tris-Cl (pH 8.0), 0.1 M NaCl, 5% glycerol, 10 mM MgCl2, and 50 mM L-arabinose with 10 μg/mL each DNase I and RNase A added immediately before use, lysed by a French press, and centrifuged at 10,000g for 15 min at 4°C. The supernatant was incubated at 4°C, rocking gently for a minimum of 2 h with 1 mL Ni-NTA agarose beads from QIAGEN per estimated 10 mg target protein. Typically, 5 mL of beads were incubated with the material from 1 L of cell growth medium. The beads were rinsed once in bulk and then packed into a column and washed with 15 mM Tris-Cl (pH 8.0), 0.1 M NaCl, 50 mM L-arabinose, and 10 mM imidazole until the OD280nm of the flow-through was <0.05. Bound protein was eluted with two column volumes of buffer containing 15 mM Tris-Cl (pH 8.0), 10 mM NaCl, 50 mM L-arabinose, and 1 M imidazole. Trypsin cleavage was performed overnight at 4°C using 1 μg trypsin per 1 mg estimated protein to remove the 6-His tag, followed by binding to a Pharmacia Mono-Q HR 5/5, 1 mL volume, anion exchange column and elution in 15 mM Tris-Cl (pH 8.0) and 50 mM L-arabinose using a 40 mL gradient from 10 mM to 1 M NaCl.
Fluorescence assay of trypsin cleavage rate
A 2.5-mL sample of protein at a concentration of 13.5 μM was stirred continuously in a 10-mm quartz fluorescence cuvette. Samples were excited at 295 nm with slit widths adjusted for a 1-nm spectral bandwidth. Emission was detected at 340 nm with slit widths adjusted for a 5-nm spectral bandwidth. After establishing an initial baseline, 1 μL of trypsin at a concentration of 3.5 mg/mL was added to the sample, and data were collected until the cleavage reaction was essentially completed. As the kinetics of trypsin cleave should be first order, we fit the fluorescence data to a standard exponential increase rate equation. Preliminary experiments showed that the amplitude of the fluorescence change of the ubiquitin construct with a 26-residue arm (UbW74-A3) was only one quarter the change of the other three constructs. We did not further use this protein because of the discrepancy.
Velocity sedimentation
Samples of purified dimerization domain were dialyzed overnight into 75 mM KCl and 15 mM Tris-Cl (pH 8.0), with or without 0.2% (w/v) L-arabinose, ∼13 mM. Samples were loaded into ultracentrifugation cells with 12-mm path length double-sector charcoal-filled Epon centerpieces and sapphire windows. A loading concentration of 0.5 mg/mL, corresponding to an OD280nm of ∼0.8 for a 12-mm cell, was used for all samples. Cells were centrifuged at 20°C in a Beckman XLI analytical ultracentrifuge at 50,000 rpm in an An-50Ti rotor. Absorbance scans at 280 nm were taken in continuous scan mode at ∼2.5-min intervals without replicates. A nominal radial step size of 0.003 cm was used. Data were analyzed by the van Holde-Weischet method (Van Holde and Weischet 1978), as implemented in the Ultrascan v6.0 software (Demeler et al. 1997). The partial specific volume of each construct was calculated from amino acid composition by the program Sednterp v1.07 (Laue et al. 1992). Because the Sednterp database lacked an entry for arabinose, the same molar concentration of glucose was used in place of arabinose to calculate the density of the buffer from its composition. Density was taken to be 1.00221 g/cm3 without L-arabinose and 1.00309 g/cm3 with 0.2% (w/v) L-arabinose.
Fluorescence measurements of arabinose binding
Purified wild-type and Δ(2–14) AraC dimerization domain were dialyzed extensively into 15 mM Tris-Cl (pH 8.0) and 75 mM KCl and then diluted into the final dialysis buffer to reach a concentration of 10 μg/mL, ∼475 nM. Samples were measured in triplicate as above with excitation at 295 nm and emission data collected from 320 to 370 nm. L-Arabinose stock solutions, dissolved in the final dialysis buffer at concentrations of 1 M, 0.1 M, and 10 mM, were added to the sample to produce final concentrations from 0 to 50 mM. Data were collected at 1 point/sec at 1-nm intervals. Equilibrium appeared to be reached by 1 min. An average emission wavelength at each arabinose concentration was calculated with a spreadsheet and plotted with KaleidaGraph. The resulting curve was fit to a standard ligand-binding isotherm to extract ligand affinity.
Acknowledgments
This work was supported by a National Institutes of Health grant GM18277 to R.F.S.
Footnotes
Reprint requests to: Robert F. Schleif, Department of Biology, Johns Hopkins University, Mudd Hall, 3400 North Charles Street, Baltimore, MD 21218, USA; e-mail: schleif@jhu.edu; fax: (410) 516-5213.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062327506.
References
- Brown, H.G. and Hoh, J.H. 1997. Entropic exclusion by neurofilament sidearms: A mechanism for maintaining interfilament spacing. Biochemistry 36: 15035–15040. [DOI] [PubMed] [Google Scholar]
- Demeler, B., Saber, H., and Hansen, J.C. 1997. Identification and interpretation of complexity in sedimentation velocity boundaries. Biophys. J. 72: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, M. and Schleif, R.F. 2001. Biophysical evidence of arm-domain interactions in AraC. Anal. Biochem. 295: 107–112. [DOI] [PubMed] [Google Scholar]
- Gryczynski, U. and Schleif, R. 2004. A portable allosteric mechanism. Proteins. 57: 9–11. [DOI] [PubMed] [Google Scholar]
- Harmer, T., Wu, M., and Schleif, R. 2001. Role of rigidity in DNA looping-unlooping by AraC. Proc. Natl. Acad. Sci. 98: 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A. 2005. The ubiquitin system for protein degradation and some of its roles in the control of cell division cycle. Cell Death Differ. 12: 1191–1197. [DOI] [PubMed] [Google Scholar]
- Hoh, J.H. 1998. Functional protein domains from the thermally driven motion of polypeptide chains: A proposal. Proteins. 32: 223–228. [PubMed] [Google Scholar]
- LaRonde-LeBlanc, N. and Wolberger, C. 2000. Characterization of the oligomeric states of wild type and mutant AraC. Biochemistry. 39: 11593–11601. [DOI] [PubMed] [Google Scholar]
- Laue, T.M., Shah, B.D., Ridgeway, T.M., and Pelletier, S.L. 1992. Computer aided interpretation of analytical sedimentation data for proteins. In Analytical ultracentrifugation in biochemistry and polymer sciene (eds. S.E. Harding et al.), pp. 90–125. The Royal Society of Chemistry, Cambridge, UK.
- Obradovic, Z., Peng, K., Vucetic, S., Radivojac, P., Brown, C., and Dunker, A.K. 2003. Predicting intrinsic disorder from amino acid sequence. Proteins. 53: 566–572. [DOI] [PubMed] [Google Scholar]
- Peng, K., Vucetic, S., Radivojac, P., Brown, C., Dunker, A.K., and Obradovic, Z. 2005. Optimizing intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 3: 35–60. [DOI] [PubMed] [Google Scholar]
- Reed, W.L. and Schleif, R.F. 1999. Hemiplegic mutations in AraC protein. J. Mol. Biol. 294: 417–425. [DOI] [PubMed] [Google Scholar]
- Ross, J.J., Gryczynski, U., and Schleif, R. 2003. Mutational analysis of residue roles in AraC function. J. Mol. Biol. 328: 85–93. [DOI] [PubMed] [Google Scholar]
- Saviola, B., Seabold, R., and Schleif, R.F. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278: 539–548. [DOI] [PubMed] [Google Scholar]
- Schleif, R.F. and Wensink, P.C. 1981. Practical methods in molecular bioloy. Springer-Verlag, New York.
- Seabold, R.R. and Schleif, R.F. 1998. Apo-AraC actively seeks to loop. J. Mol. Biol. 278: 529–538. [DOI] [PubMed] [Google Scholar]
- Soisson, S.M., MacDougall-Shackleton, B., Schleif, R., and Wolberger, C. 1997a. Structural basis for ligand-regulated oligomerization of AraC. Science. 276: 421–425. [DOI] [PubMed] [Google Scholar]
- Soisson, S.M., MacDougall-Shackleton, B., Schleif, R., and Wolberger, C. 1997b. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with D-fucose. J. Mol. Biol. 273: 226–237. [DOI] [PubMed] [Google Scholar]
- Van Holde, K.E. and Weischet, W.O. 1978. Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers. 17: 1387–1403. [Google Scholar]
- Vucetic, S., Brown, C.J., Dunker, A.K., and Obradovic, Z. 2003. Flavors of protein disorder. Proteins. 52: 573–584. [DOI] [PubMed] [Google Scholar]
- Vucetic, S., Obradovic, Z., Vacic, V., Radivojac, P., Peng, K., Iakoucheva, L.M., Cortese, M.S., Lawson, J.D., Brown, C.J., and Sikes, J.G., et al. 2005. DisProt: A database of protein disorder. Bioinformatics. 21: 137–140. [DOI] [PubMed] [Google Scholar]
- Wu, M. and Schleif, R. 2001a. Mapping arm-DNA-binding domain interactions in AraC. J. Mol. Biol. 307: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Wu, M. and Schleif, R.F. 2001b. Strengthened arm-dimerization domain interactions in AraC. J. Biol. Chem. 276: 2562–2564. [DOI] [PubMed] [Google Scholar]
- You, J., Cohen, R.E., and Pickart, C.M. 1999. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli . Biotechniques. 27: 950–954. [DOI] [PubMed] [Google Scholar]