Abstract
PH0459, from the hyperthermophilic archaeon Pyrococcus horikoshii OT3, is a probable haloacid dehalogenase with a molecular mass of 26,725 Da. Here, we report the 2.0 Å crystal structure of PH0459 (PDB ID: 1X42) determined by the multiwavelength anomalous dispersion method. The core domain has an α/β structure formed by a six-stranded parallel β-sheet flanked by six α-helices and three 310-helices. One disulfide bond, Cys186–Cys212, forms a bridge between an α-helix and a 310-helix, although PH0459 seems to be an intracellular protein. The subdomain inserted into the core domain has a four-helix bundle structure. The crystal packing suggests that PH0459 exists as a monomer. A structural homology search revealed that PH0459 resembles the l-2-haloacid dehalogenases l-DEX YL from Pseudomonas sp. YL and DhlB from Xanthobacter autotrophicus GJ10. A comparison of the active sites suggested that PH0459 probably has haloacid dehalogenase activity, but its substrate specificity may be different. In addition, the disulfide bond in PH0459 probably facilitates the structural stabilization of the neighboring region in the monomeric form, although the corresponding regions in l-DEX YL and DhlB may be stabilized by dimerization. Since heat-stable dehalogenases can be used for the detoxification of halogenated aliphatic compounds, PH0459 will be a useful target for biotechnological research.
Keywords: haloacid dehalogenase, HAD superfamily, hydrolase, detoxification, intracellular disulfide bond, hyperthermophilic archaeon, structural genomics
PH0459, from the hyperthermophilic archaeon Pyrococcus horikoshii OT3, is a hypothetical protein with a molecular mass of 26,725 Da (232 amino acids) (Kawarabayasi et al. 1998). PH0459 homologs are conserved among several prokaryotes (Fig. 1A). The PH0459 protein shares 77% and 60% identities to PF0463 from Pyrococcus furiosus DSM 3638 (Maeder et al. 1999) and TK0058 from Thermococcus kodakarensis KOD1 (Fukui et al. 2005), respectively. PH0459 belongs to a large superfamily of hydrolases, the haloacid dehalogenase (HAD) superfamily (Pfam, PF00702) (Koonin and Tatusov 1994; Aravind et al. 1998). Based on their three conserved sequence motifs, the l-2-haloacid dehalogenases, epoxide hydrolases, P-type ATPases, and a variety of phosphatases are recognized as members of this superfamily. The X-ray structures of two l-2-haloacid dehalogenases (EC 3.8.1.2), l-DEX YL from Pseudomonas sp. YL (Hisano et al. 1996; Li et al. 1998), and DhlB from Xanthobacter autotrophicus GJ10 (Ridder et al. 1997, 1999), were reported. Haloacid dehalogenase catalyzes the hydrolytic dehalogenation of a haloalkanoate to the corresponding hydroxyalkanoate. The PH0459 protein shares 16% and 20% identities to l-DEX YL and DhlB, respectively. Here, we report the crystal structure of PH0459 at 2.0 Å resolution and discuss its structural aspects.
Figure 1.
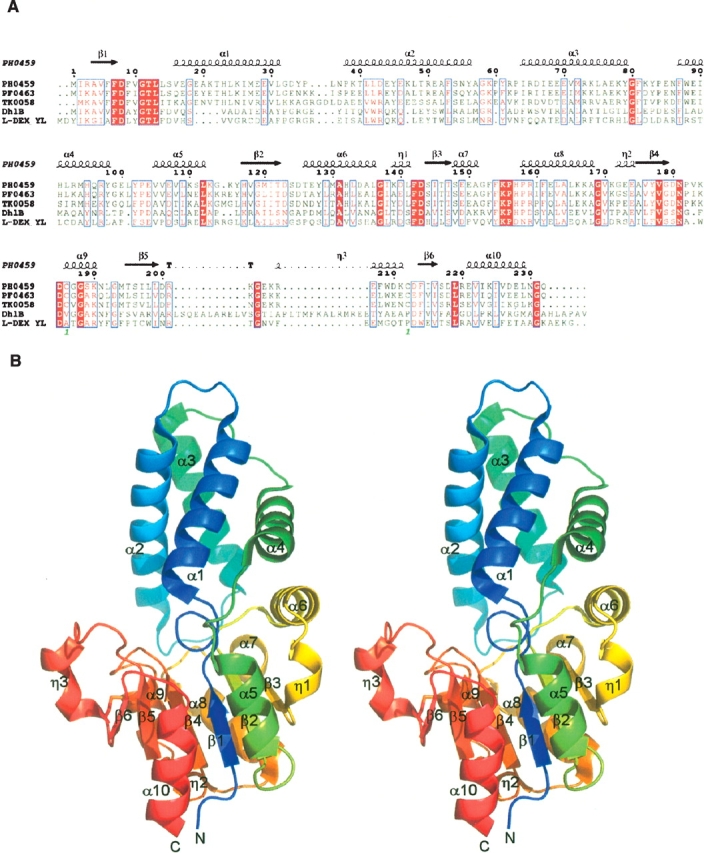
(A) Sequence alignment of homologs of the PH0459 protein. The alignment was generated by CLUSTALW (Thompson et al. 1994) and ESPript (Gouet et al. 1999). The secondary structures of PH0459, as determined by DSSP (Kabsch and Sander 1983), are shown above the sequences (α, α-helix; β, β-strand; η, 310-helix; T, β-turn). (B) Ribbon representations of the PH0459 structure (stereo view). (Color gradient [blue, green, yellow, red] from N terminus to C terminus.) The disulfide bond, Cys186–Cys212, is shown in a stick model.
Results and Discussion
The PH0459 crystal belongs to the monoclinic space group P21, with unit cell constants of a = 34.11 Å, b = 71.27 Å, c = 53.62 Å, α = 90.00°, β = 93.51°, γ = 90.00°, and contains one protein molecule per asymmetric unit. The structure was refined to 2.0 Å by the multiwavelength anomalous dispersion (MAD) method (Hendrickson 1991). The crystallographic data are summarized in Table 1. The final model includes 230 amino acid residues and 150 water molecules in the asymmetric unit. The two C-terminal residues are invisible due to disorder. PH0459 consists of the core domain and the subdomain (Fig. 1B). The core domain has an α/β structure formed by a six-stranded parallel β-sheet flanked by six α-helices and three 310-helices. The subdomain inserted into the core domain has a four-helix bundle structure. There is an active site cavity between the two domains.
Table 1.
X-ray data collection, phasing, and refinement statistics
| Peak | Edge | Remote | |
| Data collection | |||
| Wavelength (Å) | 0.97890 | 0.97930 | 0.96000 |
| Resolution (Å) | 50–2.0 | 50–2.0 | 50–2.0 |
| Total reflections | 62,653 | 62,667 | 62,818 |
| Unique reflections | 17,044 | 17,066 | 17,071 |
| Redundancy | 3.7 (3.6) | 3.7 (3.6) | 3.7 (3.6) |
| Completeness (%) | 99.3 (98.8) | 99.2 (98.7) | 99.3 (98.8) |
| I/σ(I) | 12.0 (5.00) | 14.3 (4.92) | 13.3 (5.02) |
| Rsyma (%) | 8.9 (27.2) | 8.2 (27.3) | 8.2 (27.4) |
| MAD analysis | |||
| Resolution (Å) | 50–2.0 | ||
| No. of Se sitesb | 6 | ||
| FOMMADc | 0.54 | ||
| FOMRESOLVEd | 0.74 | ||
| Refinement | |||
| Resolution (Å) | 42.8–2.0 | ||
| No. of reflections | 16,891 | ||
| No. of protein atoms | 1869 | ||
| No. of water molecules | 150 | ||
| Rwork (%) | 16.6 | ||
| Rfree (%)e | 20.7 | ||
| RMSD bond length (Å) | 0.014 | ||
| RMSD bond angles (°) | 1.6 | ||
| Average B factor (Å2) | 22.5 | ||
| Ramachandran plot | |||
| Most favored regions (%) | 96.0 | ||
| Additional allowed regions (%) | 4.0 | ||
| Generously allowed regions (%) | 0.0 | ||
| Disallowed regions (%) | 0.0 | ||
All numbers in parentheses represent last outer shell (2.07–2.00 Å) statistics.
aRsym = ∑|Ii − Iavg|/∑Ii, where Ii is the observed intensity and Iavg is the average intensity.
bNumber of selenium sites located with SOLVE.
cFigure of merit after SOLVE phasing.
dFigure of merit after RESOLVE.
eRfree is calculated for 10% of randomly selected reflections excluded from refinement.
A DALI (Holm and Sander 1993) structural homology search revealed that PH0459 resembles two l-2-haloacid dehalogenases, l-DEX YL from Pseudomonas sp. YL (Hisano et al. 1996; Li et al. 1998) (PDB ID: 1JUD, Z = 22.3, RMSD = 2.6 Å over 213 Cα atoms) and DhlB from X. autotrophicus GJ10 (Ridder et al. 1997, 1999) (PDB ID: 1QQ5, Z = 21.5, RMSD = 2.6 Å over 210 Cα atoms), and other HAD superfamily hydrolase proteins. The overall structures of PH0459, l-DEX YL, and DhlB overlap considerably, especially in the core domain (Fig. 2A). The major difference between PH0459 and DhlB is the extra-domain of DhlB, with two antiparallel helices involved in dimer formation. The crystal packing suggests that PH0459 exists as a monomer, whereas l-DEX YL and DhlB form homodimers. In PH0459, one disulfide bond, Cys186–Cys212, forms a bridge between α9 and η3 (Fig. 1), although PH0459 seems to be an intracellular protein. These cysteines are conserved in the P. furiosus PF0463 and T. kodakarensis TK0058 proteins from hyperthermophilic archaeons (Fig. 1A). Recently, genomic evidence indicated a critical role for disulfide bonds in the structural stabilization of intracellular proteins from thermophiles (Mallick et al. 2002; Beeby et al. 2005), suggesting that the disulfide bond in PH0459 contributes to the structural stability. The disulfide bond probably facilitates the structural stabilization of the neighboring region in the monomeric form, although the corresponding regions in l-DEX YL and DhlB may be stabilized by dimerization (Fig. 2B).
Figure 2.
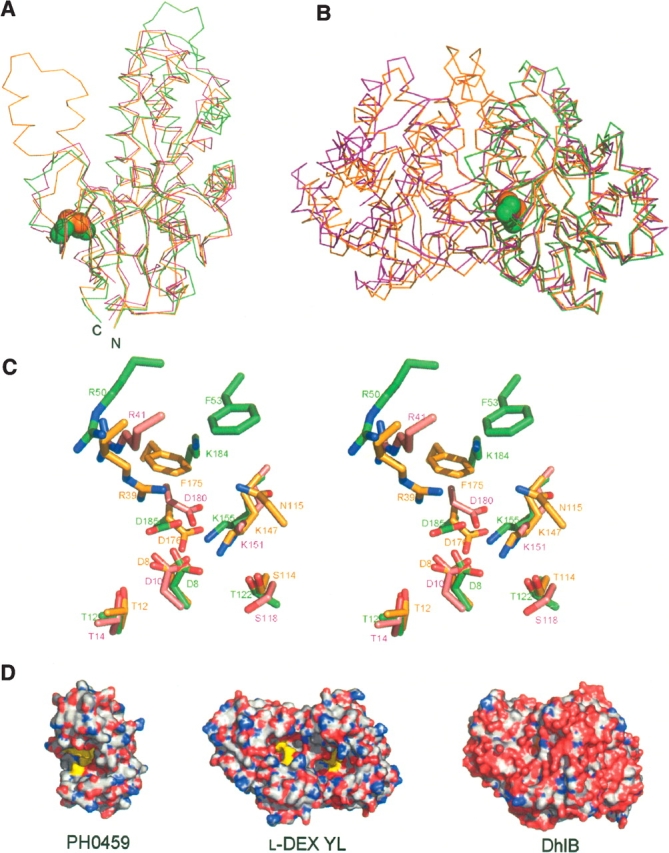
(A) The superimposition of the main-chain structures of PH0459 (green), l-DEX YL (magenta) (PDB ID: 1JUD), and DhlB (orange) (PDB ID: 1QQ5) monomers. The disulfide bond in PH0459 is shown in a sphere model. (B) The superimposition of the main-chain structures of the PH0459 (green) monomer, the l-DEX YL (magenta), and DhlB (orange) dimers. The disulfide bond in PH0459 is shown in a sphere model. (C) The superimposition of the active sites of PH0459 (green), l-DEX YL (magenta), and DhlB (orange) (stereo view). (D) Surface representations of PH0459, l-DEX YL, and DhlB. The active site residues are colored yellow. The structures are oriented in the same direction.
Most of the essential active site residues, Asp8, Thr12, Thr122, Lys155, Asn181, and Asp185 in PH0459, are well conserved (Asp10, Thr14, Ser118, Lys151, Asn177, and Asp180 in l-DEX YL, and Asp8, Thr12, Ser114, Lys147, Asn173, and Asp176 in DhlB, respectively) (Fig. 1A), suggesting that Asp8 works as the nucleophile in the enzymatic reaction in the same way as in the catalysis by l-DEX YL (Li et al. 1998) and DhlB (Ridder et al. 1999). The salt bridge to the positively charged Lys155 probably reduces the pKa of Asp8, thereby increasing its nucleophilicity. In l-DEX YL, Arg41 functions as the halogen abstraction residue (Li et al. 1998), and in DhlB, Arg39, Asn115, and Phe175 form a halide-stabilizing cradle (Ridder et al. 1999). In the case of PH0459, Arg50 probably functions as the halogen abstraction residue, and Arg50, Phe53, and Lys184 may form a halide-stabilizing cradle (Fig. 2C). However, their locations are slightly different from those of other proteins. In addition, the active site cavities of PH0459 and l-DEX YL are open to the solvent, whereas in DhlB it is shielded from the solvent (Fig. 2D). The active site cavity of PH0459 is more open to the solvent than that of l-DEX YL, due to the monomeric form of PH0459. These results suggest that PH0459 probably has haloacid dehalogenase activity, but the substrate specificity may be somewhat different from those of other proteins. DhlB can only efficiently degrade short substrates up to the size of l-2-propionate (van der Ploeg et al. 1991), whereas l-DEX YL can degrade larger substrates, e.g., l-2-bromohexadecanoic acid (Liu et al. 1994). PH0459 may have broad substrate specificity, because its active site cavity is widely open to the solvent. Since heat-stable dehalogenases can be used in biotechnological applications to detoxify environmentally damaging halogenated aliphatic compounds, PH0459 will be a useful target for applied research.
Materials and methods
Protein expression, purification, and crystallization
The gene encoding full-length PH0459 was cloned into the plasmid vector pET11a (Novagen, EMD Biosciences). The selenomethionine-substituted PH0459 protein was expressed in Escherichia coli BL21-CodonPlus(DE3)-RIL-X (Stratagene). The E. coli lysate was heated at 90°C for 11.5 min, and the proteins were purified by a series of TOYOPEARL SuperQ-650M (Tosoh), RESOURCE Q (Amersham Biosciences, GE Healthcare), CHT5-I (Bio-Rad), and HiLoad 16/60 Superdex 75 pg (Amersham Biosciences) column chromatography steps (Amersham Biosciences). Initial screening for crystallization was performed using the TERA automatic crystallization system (Sugahara and Miyano 2002). The crystals of PH0459 were obtained in drops composed of 1 μL of 18.5 mg/mL protein and 1 μL of precipitant solution (0.1 M Tris-HCl buffer at pH 7.0 containing 0.88 M tri-sodium citrate) by microbatch crystallization under Al’s oil (Hampton Research). Some clusters of plate-like crystals were obtained within a few months.
Data collection, structure determination, and refinement
A single crystal segment (~50 × 50 × 5 μm3) was isolated from the crystal cluster and was used for data collection. The data collection was carried out at 100 K, with the reservoir solution containing 20% glycerol as a cryoprotectant. The MAD data were collected at three different wavelengths at BL26B1 (Yamamoto et al. 2002), SPring-8 (Hyogo, Japan), and were recorded on a Jupiter 210 CCD detector (Rigaku). All diffraction data were processed with HKL2000 (Otwinowski and Minor 1997). SOLVE (Terwilliger and Berendzen 1999) was used to locate the selenium sites and to calculate the phases, and RESOLVE (Terwilliger 2002) was used for the density modification and partial model building. The rest of the model was built with O (Jones et al. 1991) and the model was refined with Refmac5 (Murshudov et al. 1997) and CNS (Brunger et al. 1998). The quality of the model was inspected by the program PROCHECK (Laskowski et al. 1993). Graphic figures were created using PyMol (DeLano Scientific). The atomic coordinates and the structure factors have been deposited in the Protein Data Bank, with the accession code 1X42.
Acknowledgments
We thank Ms. Y. Fujimoto for protein preparations, Dr. M. Sugahara for crystallization screening, Dr. M. Yamamoto for data collection at the RIKEN Structural Genomics beamline, and Ms. K. Yajima, Ms. A. Ishii, and Ms. T. Nakayama for clerical assistance. This work was supported by the RIKEN Structural Genomics/Proteomics Initiative (RSGI), the National Project on Protein Structural and Functional Analyses, the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051922406.
References
- Aravind, L., Galperin, M.Y., and Koonin, E.V. 1998. The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biochem. Sci. 23: 127–129. [DOI] [PubMed] [Google Scholar]
- Beeby, M., O’Connor, B.D., Ryttersgaard, C., Boutz, D.R., Perry, L.J., and Yeates, T.O. 2005. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 3: e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Fukui, T., Atomi, H., Kanai, T., Matsumi, R., Fujiwara, S., and Imanaka, T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15: 352–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D.I., and Metoz, F. 1999. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A. 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254: 51–58. [DOI] [PubMed] [Google Scholar]
- Hisano, T., Hata, Y., Fujii, T., Liu, J.Q., Kurihara, T., Esaki, N., and Soda, K. 1996. Crystal structure of l-2-haloacid dehalogenase from Pseudomonas sp. YL. An α/β hydrolase structure that is different from the α/β hydrolase fold. J. Biol. Chem. 271: 20322–20330. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi, Y., Sawada, M., Horikawa, H., Haikawa, Y., Hino, Y., Yamamoto, S., Sekine, M., Baba, S., Kosugi, H., Hosoyama, A., et al. 1998. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5: 55–76. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. and Tatusov, R.L. 1994. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 244: 125–132. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Macarthur, M.W., Moss, D.S., and Thornton, J.M. 1993. Procheck—A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Li, Y.F., Hata, Y., Fujii, T., Hisano, T., Nishihara, M., Kurihara, T., and Esaki, N. 1998. Crystal structures of reaction intermediates of l-2-haloacid dehalogenase and implications for the reaction mechanism. J. Biol. Chem. 273: 15035–15044. [DOI] [PubMed] [Google Scholar]
- Liu, J.Q., Kurihara, T., Hasan, A.K., Nardi-Dei, V., Koshikawa, H., Esaki, N., and Soda, K. 1994. Purification and characterization of thermostable and nonthermostable 2-haloacid dehalogenases with different stereospecificities from Pseudomonas sp. strain YL. Appl. Environ. Microbiol. 60: 2389–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, D.L., Weiss, R.B., Dunn, D.M., Cherry, J.L., Gonzalez, J.M., DiRuggiero, J., and Robb, F.T. 1999. Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics 152: 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick, P., Boutz, D.R., Eisenberg, D., and Yeates, T.O. 2002. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl. Acad. Sci. 99: 9679–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Ridder, I.S., Rozeboom, H.J., Kalk, K.H., Janssen, D.B., and Dijkstra, B.W. 1997. Three-dimensional structure of l-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate. J. Biol. Chem. 272: 33015–33022. [DOI] [PubMed] [Google Scholar]
- Ridder, I.S., Rozeboom, H.J., Kalk, K.H., and Dijkstra, B.W. 1999. Crystal structures of intermediates in the dehalogenation of haloalk-anoates by l-2-haloacid dehalogenase. J. Biol. Chem. 274: 30672–30678. [DOI] [PubMed] [Google Scholar]
- Sugahara, M. and Miyano, M. 2002. Development of high-throughput automatic protein crystallization and observation system. Tanpakushitsu Kakusan Koso 47: 1026–1032. [PubMed] [Google Scholar]
- Terwilliger, T.C. 2002. Automated structure solution, density modification and model building. Acta Crystallogr. D 58: 1937–1940. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D 55: 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg, J., van Hall, G., and Janssen, D.B. 1991. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J. Bacteriol. 173: 7925–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., Kumasaka, T., Ueno, G., Ida, K., Kanda, H., Miyano, M., and Ishikawa, T. 2002. RIKEN structural genomics beamlines at SPring-8. Acta Crystallogr. A 58: C302. [Google Scholar]


