Abstract
We have optimized the expression level of 20 mammalian G protein-coupled receptors (GPCRs) in the methylotrophic yeast Pichia pastoris. We found that altering expression parameters, including growth temperature, and supplementation of the culture medium with specific GPCR ligands, histidine, and DMSO increased the amount of functional receptor, as assessed by ligand binding, by more than eightfold over standard expression conditions. Unexpectedly, we found that the overall amount of GPCR proteins expressed, in most cases, varied only marginally between standard and optimized expression conditions. Accordingly, the optimized expression conditions resulted in a marked fractional increase in the ratio of ligand binding-competent receptor to total expressed receptor. The results of this study suggest a general approach for increasing yields of functional mammalian GPCRs severalfold over standard expression conditions by using a set of optimized expression condition parameters that we have characterized for the Pichia expression system. Overall, we have more than doubled the number of GPCR targets that can be produced in our laboratories in sufficient amounts for structural studies.
Keywords: G protein-coupled receptor, GPCR, expression screening, binding assay, optimization, Pichia pastoris
Members of the G protein-coupled receptor (GPCR) superfamily play a pivotal role in eukaryotic signal transduction (Bockaert and Pin 1999). They are involved in almost all physiological regulatory mechanisms in the human body. For this reason, GPCRs represent one of the largest classes of pharmacological therapeutic targets. Currently, half of all pharmaceuticals primarily target members of this membrane protein family (Klabunde and Hessler 2002). Understanding drug-target interaction requires detailed structural and functional knowledge of the target receptors (Kuhn et al. 2002).
Nevertheless, structural investigations of membrane proteins are typically limited by several fundamental experimental barriers including adequate protein expression, solubilization, purification, and crystallization. To date, only a few dozen structures of membrane proteins have been reported (http://www.rcsb.org/pdb/; http://www.mpibp-frankfurt.mpg.de/michel/public/memprotstruct.html). Among them, only one is a member of the GPCR family (bovine rhodopsin) (Palczewski et al. 2000; Li et al. 2004). Because of its high natural abundance in native tissue, this protein was successfully purified and crystallized, demonstrating that obtaining a three-dimensional structure of a GPCR is feasible provided that enough pure and functional protein is available. However, since most GPCRs are naturally produced in low abundance only, heterologous expression systems are inevitably required to obtain adequate amounts of protein for structural and functional studies. Despite the remarkable advances that have been realized in recombinant protein production (Tate 2001), expression and purification success rates are still low for membrane intrinsic proteins. In light of this, it is highly desired to find universal systems to produce and evaluate GPCRs. This point is particularly well illustrated by recent publications reporting successful GPCR heterologous expression (Drew et al. 2003; Lundstrom 2003; Massotte 2003; Sarramegna et al. 2003; Akermoun et al. 2005; Reinhart and Krettler 2006). The quantity of expressed active receptors has been observed to vary dramatically, in fact >100-fold in some cases, depending on the receptor sequence, its source organism, type of expression system, promoter and fusion sequences, gene dosage, and expression conditions. These results led us to the idea that a small subset of expression condition parameters might be chiefly responsible for optimizing overall expression levels in any heterologous expression system.
MePNet (Membrane Protein Network), an academic consortium comprising five European laboratories, is a structural genomics program focusing on membrane proteins (Lundstrom 2005). The general approach used by the consortium was to produce 100 GPCRs in three expression systems (Semliki Forest Virus, Pichia pastoris, Escherichia coli). The specific aim was to increase yields of functional GPCRs and also to create general methods to study their structure and function.
Based on our previous investigations (Weiss et al. 1998a; Schiller et al. 2000; Grünewald et al. 2004; Reinhart and Krettler 2006), two groups within the consortium decided to focus on GPCR production in P. pastoris. First, we generated a collection of P. pastoris clones expressing 100 different GPCRs. Based on previous observations, we optimized each step of the expression procedure, including lowering temperature during induction (N. André and C. Prual, unpubl.) and adding specific ligands to the growth/induction media (Sizmann et al. 1996; Weiss et al. 1998a; Grünewald et al. 2004), in addition to other compounds such as histidine and dimethyl sulfoxide (DMSO) (Ivanovic 2001). It was previously reported that DMSO can modify membrane physical properties and alter the regulation of intracellular biochemical pathways (Murata et al. 2003). The present study represents the first systematic investigation of optimizing expression parameters affecting functional protein yield and has resulted in uniform increases in functional GPCR yield for most of the 20 GPCRs we chose to investigate initially. This suggests that the culture condition refinements reported here may be generally applicable to increase yields of functional GPCRs.
Results
The goal of this work was to define a simple, universal method to increase the production level of functional GPCRs in P. pastoris to reach receptor quantity and quality required for structural studies. Twenty GPCRs were selected from the 100 receptors studied within the MePNet program (Table 1) and analyzed for both yield and activity in a systematic way. Among the selection criteria, the availability of proper compounds and protocols for ligand binding measurements was the most important. These selected receptors varied in their initial production levels according to preliminary immunodetection experiments. Different GPCR subfamilies and subtypes were represented, and, in a few cases, receptor orthologs from various organisms were studied.
Table 1.
Binding conditions for 20 GPCRs expressed in P. pastoris
Based on a derivative of the pPIC9K vector (Weiss et al. 1998a; C. Reinhart, C. Krettler, H. Reiländer, and H. Michel, in prep.), we designed a new construct for the large-scale production of GPCRs in P. pastoris (Fig. 1). The expression cassette comprises an N-terminal α-factor signal sequence from Saccharomyces cerevisiae followed by a Flag-tag and a decahistidine-tag. The multiple cloning site is flanked by two tobacco etch virus (TEV) protease sites. The biotinylation domain from Propionibacterium shermanii is placed at the C terminus following the protein to be expressed. In total, 20 receptor cDNAs were ligated into the expression vector via the BamHI and SpeI sites.
Figure 1.
Schematic drawing of the P. pastoris expression vector for heterologous production of 20 G protein-coupled receptors. (AOX1P) Alcohol oxidase 1 gene promoter, (α-F) coding region for the prepropeptide of the S. cerevisiae mating type factor α, (Flag) coding region for the FLAG-tag, (H10) His-tag consisting of 10 consecutive histidine codons, (TEV) coding region for the tobacco etch virus protease cleavage site, (Bio) coding region for the biotinylation domain of the transcarboxylase from Propionibacterium shermanii.
Transformation of the protease-deficient Pichia strain SMD1163 with these plasmids resulted in clones containing multiple copies of the gene expression cassette. These clones were selected for relative expression cassette copy number by antibiotic resistance screening (G418) and further analyzed. It appeared that the highest expression evaluated by immunodetection was not always obtained from a clone growing on the highest antibiotic concentration and thus not containing the greatest expression cassette copy number. Similar effects were observed for the 5HT5 receptor produced in P. pastoris (Weiss et al. 1998a).
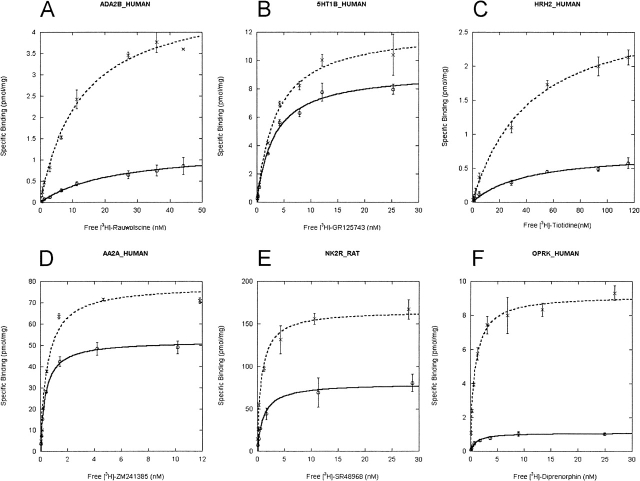
Saturation ligand binding experiments were performed on crude membrane preparations for all receptors (Table 1). Ligand binding was saturable and specific for each of the 20 receptors enabling the determination of total ligand binding sites (Bmax) and affinity (Kd). Representative binding curves for a few receptor/ligand combinations are depicted in Figure 2.
Figure 2.
Radioligand binding studies on membranes of P. pastoris cells producing the following receptors: (A) ADA2B_HUMAN, (B) 5HT1B_HUMAN, (C) HRH2_HUMAN, (D) AA2A_HUMAN, (E) NK2R_RAT, (F) OPRK_HUMAN. Yeast cells were induced as described in Materials and Methods. Binding assays were performed under the conditions outlined in Table 1. Binding assays were performed as outlined in Materials and Methods. The results shown are from three independent experiments. (Solid line) Membranes from yeast cells grown under standard conditions, (broken line) membranes from yeast cells grown under optimized conditions.
Expression of the different receptors under standard expression conditions (BMMY medium, 30°C, 18-h induction time) yielded production levels ranging from 120 femtomoles (fmol) to 71 picomoles (pmol) bound ligand per milligram total membrane protein (i.e., expression level of 20 pmol/mg membrane of a 50-kDa receptor corresponds to 0.38 mg in 1 L of Pichia culture). Nine receptors were expressed at low level (<2 pmol/mg); eight, at mid-range level (2–20 pmol/mg); and three, at high level (>20 pmol/mg). Notably, the expression level of histamine receptor was low. Members of the group of opioid and serotonin receptors were found either in the low or the mid-range of expression. Furthermore, all muscarinic acetylcholine receptors (ACM1_Mouse > ACM1_Human > ACM2_Pig > ACM2_Human) and all adrenergic receptors (ADA1B_Human > ADA2B_Human) were expressed at a particularly low level, whereas the dopamine receptors and the NK receptors were expressed at a medium or a high level.
The receptors heterologously expressed in P. pastoris revealed high affinity binding for their radioligand ranging from 50 pM to 38 nM (Fig. 1).
In our binding experiments, comparing NK2 receptors from various organisms, that from rat exhibited a 10 times higher affinity for the ligand [3H]SR48968 than that of human. Similar findings were obtained for κ opioid receptors in Pichia: That from mouse bound the respective ligand with four times higher affinity than that from human. However, no significant ligand binding difference was found between D2DR_Human and D2DR_Mouse.
According to this classification, 45% of the GPCRs were expressed at low level, 40% at medium level, and 15% at high level. Since we considered only receptors expressed in the abundance that could be used for structural studies, only three targets were found to be of use for further structural study by the MePNet project (NK2R_HUMAN, AA2A_HUMAN, and NK2R_RAT). As a consequence, expression levels of the 17 other GPCRs needed to be significantly improved before planned structural studies could be realized.
Impact of various expression parameters on GPCR production
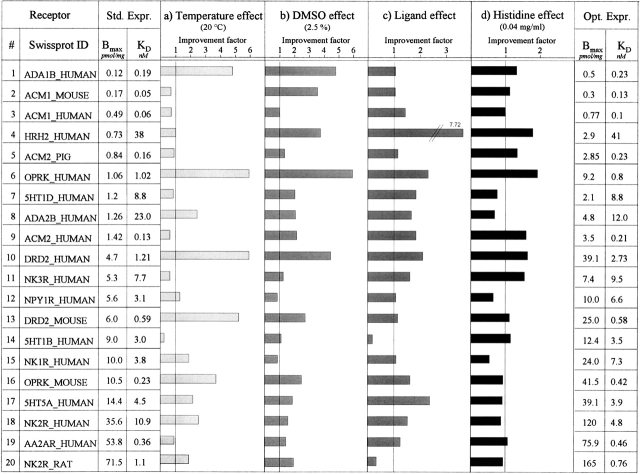
We focused in our study on the impact of four different parameters affecting protein expression. First, the effect of varying each parameter (temperature, DMSO, ligand, histidine) was assessed separately in order to design a screen that would yield information for all 20 receptors (data not shown). This prescreening revealed that the optimal expression improvement due to adjusting each parameter individually was tightly clustered for all receptors studied; we thus chose the mean value of each optimized parameter with respect to all receptors. These expression parameter adjustments included (1) temperature lowering from 30° to 20°C and (2) supplementing the medium with a specific ligand at a concentration close to 100-fold Kd or with other additives such as (3) 2.5% DMSO or (4) 0.04 mg/mL histidine.
In Figure 3, values in the column “Std. Expr.” represent binding data (Bmax and Kd) for each receptor under standard conditions (30°C, 18-h induction time, BMMY). The following four columns depict changes of the standard expression level for each receptor upon changing each of the four parameters one at a time. Control ligand binding experiments with Pichia cells transformed with empty vector (pPIC9K) revealed no specific binding for any of the tested conditions (data not shown).
Figure 3.
Production level and its improvement factor for 20 GPCRs in various conditions. Cells were grown in BMMY at 30°C, 225 rpm, 22 h. For a, temperature was shifted to 20°C during induction; for b, BMMY medium was supplemented with 2.5% DMSO; for c, medium was supplemented with antagonist or agonist (100× Kd); for d, medium was supplemented with histidine (0.04 mg/mL). The binding values for standard expression conditions (Std. Expr.) and optimized expression conditions (Opt. Expr.) were calculated from three independent saturation curves. The improvement factor represents the Bmax value of the changed condition divided by the Bmax value of the standard conditions from a, b, c, or d. Bmax values from a, b, c, or d were calculated from single point measurements using 5–10× Kd for the radioactive ligand. For the optimized conditions, only parameters showing a positive effect (improvement factors >1) were combined.
The improvement factors were calculated as the ratio of the binding value obtained for modified condition over the value obtained for standard conditions.
The temperature change from 30° to 20°C during induction resulted in a Bmax increase for 12 receptors. For eight receptors, the expression level was enhanced by a factor of at least 2, and among them, four showed a fourfold Bmax increase. Nevertheless, for seven receptors, this temperature shift led to no significant change. For one receptor (5H1B_HUMAN), the expression was reduced significantly.
Upon addition of 2.5% DMSO to induction medium, 16 receptors revealed an increased expression level. The Bmax of nine of them more than doubled, with a maximum improvement of sixfold (OPRK_HUMAN). The Bmax of four receptors were not affected by the addition of DMSO.
When 100× Kd ligand concentration was present in the induction medium, the production level of functional protein increased for 18 receptors. The Bmax values for four of them showed more than a twofold increase and even a sevenfold increase for the Histamine receptor. Two of them (5HT1B_Human, NK2R_Rat) revealed a decreased Bmax.
When 0.04% histidine was present during induction, 12 receptors revealed a higher expression yield, and one receptor's production level increased more than twofold. There was no change for eight receptors.
Optimization
The parameters that had a positive effect were combined to create an optimized condition for each receptor. For all the studied GPCRs, the Bmax values were always higher than the ones obtained in standard conditions, validating this optimization procedure. Under optimized condition, the production level doubled for 13 receptors. Seven receptors had their Bmax increased by a factor of at least 4 with a maximum of 8.7 for the OPRK_HUMAN. No significant changes in affinity could be observed under optimized conditions.
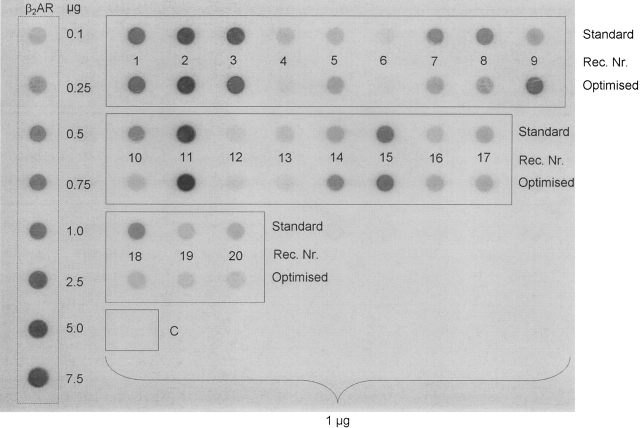
Determining the total amount of receptor by dot-blot analysis
In addition to investigating the amount of functional receptor (ligand binding), we determined the total receptor production. Spots for each receptor for standard and optimized conditions were analyzed and expressed relative to each other. For the standard curve, membranes containing the β2AR (C. Reinhart, C. Krettler, H. Reiländer, and H. Michel, in prep.) were used. Each spot corresponds to 1 μg of crude Pichia membrane. Total receptor production was revealed by immunodetection method using the Flag-tag present on each receptor.
The dot-blot revealed that the levels of expressed GPCRs were varying considerably (Fig. 4). Only four receptors (HRH2R_HUMAN, ACM2_PIG, ACM2_HUMAN, NK3R_HUMAN) revealed a stronger dot-blot signal after optimization, whereas for most of them no significant change or a slight decrease was observed.
Figure 4.
Dot-blot experiments on yeast membranes producing the 20 GPCRs. (β2AR) Pichia membrane containing human β2 adrenergic receptor (25 pmol/mg), (1–20) receptor number according to Table 1, (C) control cells transformed with empty vector pPIC9K. One microgram (1 μg) of Pichia membrane was applied to PVDF membrane. The top row represents the results obtained from standard conditions; the bottom row, results from optimized conditions. Immunoblot analysis was performed using the anti-FLAG M2 antibody as described in Materials and Methods.
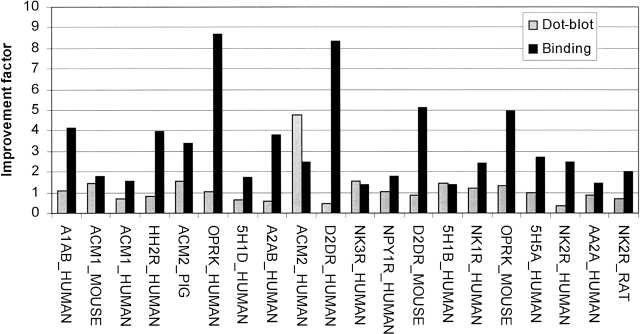
To further analyze, we compared the improvement factor in dot-blot experiments with the one of the binding experiments. Interestingly, they did not correlate with each other (Fig. 5). D2DR_Human revealed a greater than eightfold increase in the Bmax, whereas the signal from the dot-blot experiment decreased by a factor of 2. For the ACM2_Human, the total amount of receptor increased almost five times (Fig. 5), whereas the Bmax only doubled. This is the only target where the ratio was shifted toward nonfunctional receptor.
Figure 5.
Dot-blot vs. ligand binding. Correlation between total and functional receptor. (Shaded bars) Improvement factor for total receptor production (dot-blot signal optimized condition divided by dot-blot signal standard conditions), (solid bars) improvement factor for functional receptor production (Bmax value optimized condition divided by Bmax value standard condition).
Discussion
As part of a European membrane protein network (MePNet), our aim was to express 100 GPCRs in P. pastoris (see http://www.mepnet.org) and use the targets with the highest yields of functional protein for structural studies. Since no such systematic study of large-scale production of GPCRs in P. pastoris has been conducted to date, we wanted to develop, evaluate, and improve new tools for the expression, solubilization, and purification of these targets with the aim of increasing the number of candidates available for structural studies.
Thus, as a starting point, we exploited the latest vector and yeast strain optimization for GPCR expression. The 20 recombinant clones investigated in the present study were selected as a representative panel (in terms of family and origin) of the total GPCR expressed in MePNet project. When placed in standard expression conditions, these clones showed significant specific ligand binding properties (Bmax), but only a few receptors (NK2R_HUMAN, AA2A_HUMAN, and NK2R_RAT) were considered expressed at sufficient level to be compatible with structural studies.
The ligand affinity (Kd) of HRH2R_HUMAN, NPY1R_HUMAN, A2AB_HUMAN, and NK3R_HUMAN receptors was 3-, 6-, 13-, and 38-fold higher, respectively, in Pichia membranes (Fig. 3) relative to the levels previously described from native tissue (Leurs et al. 1994; Oury-Donat et al. 1995; Parker et al. 1998; Millan et al. 2000). As reported for other GPCRs, this could be explained by the differential lipid composition (nature and proportion) of yeast and mammalian membranes (e.g., ergosterol vs. cholesterol) (Opekarová and Tanner 2003).
In order to improve the expression levels observed for the 20 receptors, we optimized the parameters that influence expression conditions. For soluble proteins, parameters including the balance of salts and nutrients in the media were also shown to be influential in improving yields of soluble proteins in Pichia (Boettner et al. 2002). However, we found that the same parameters had little effect on the production of membrane proteins including GPCRs (data not shown). Similarly, some studies described that optimizing pH values during expression could be valuable for the expression of soluble protein (Koganesawa et al. 2002). For the μ-opioid receptor (Sarramegna et al. 2002) and other GPCRs (data not shown), a similar observation was made. In the present study, we showed that a change in temperature or the addition of receptor-specific ligands, DMSO, or histidine in the medium resulted in significantly increased levels of recombinant protein.
Temperature effect on expression level
In previous work, it was shown that lowering temperature during expression influences the yield of recombinant proteins. This has been observed with soluble proteins (Hong et al. 2002; Jahic et al. 2003b), but also with the μ-opioid receptor (Sarramegna et al. 2002). Previous unpublished work revealed that a temperature range of 18°–24°C during expression was optimal for various receptors, as measured by ligand binding. In this study, at 20°C, we found for 10 targets (ADA1B_HUMAN, OPRK_HUMAN, D2DR_HUMAN, D2DR_MOUSE, NK1R_HUMAN, OPRK_MOUSE, 5HT5_HUMAN, NK2R_HUMAN, and NK2R_RAT) that lowered expression temperature increased active receptor yield up to sixfold (Fig. 3). Interestingly, receptors from the muscarinic acetylcholine family (ACM1_HUMAN, ACM1_MOUSE, ACM2_HUMAN, ACM2_PIG) exhibited slightly decreased Bmax values upon temperature shift from 30° to 20°C, whereas two targets from the α 1B adrenergic receptor family, two from the D2 dopamine receptor family, and two from the κ opioid family revealed two- to sixfold increased Bmax for the same temperature difference. This shows that the temperature effect is strongly receptor-specific.
Possible explanations for the temperature effect include slowing down protein production and not overloading the translocation machinery, protein processing, or intracellular trafficking. Lowering temperature could reduce proteolytic activities (Jahic et al. 2003a, b) or up-regulate cold-shock proteins such as chaperones.
Chaperones are proteins responsible for correct folding and assembly of polypeptide chains and are present in all virtually living organisms (Walter and Buchner 2002). Previous study showed that certain cold-shock proteins, including chaperones, undergo induction following temperature shift from 30° to 10°C in S. cerevisiae (for review, see Inouye and Phadtare 2004). To our knowledge, not much is known about the induction of cold-shock proteins in P. pastoris, but it could be possible that similar effects increase the level of chaperones after lowering the temperature to 20°C in this yeast species. Furthermore, an increased chaperone level could lead to greater amounts of properly folded recombinant receptors in yeast.
In addition to chaperone induction, it was also reported that S. cerevisiae undergo up- and down-regulation of many other genes upon cold shock. These are chiefly genes involved in metabolism, protein folding, protein synthesis, transport, and localization, as well as several uncharacterized proteins (Homma et al. 2003). Assuming that similar effects take place in P. pastoris after shifting to lower temperature, there might be a combination of effects such as chaperone induction and slow protein synthesis leading to increased amounts of functional receptor under these growth conditions.
Ligand effect on recombinant receptor yield
Over 10 years ago, it was found that adding specific ligand to S. cerevisiae expression medium enhanced the relative fraction of receptors that specifically binds ligand (King et al. 1990). We previously reported similar effects for the 5HT5 receptor, the β2 adrenergic receptor (Weiss et al. 1998a), and the dopamine D2 receptor (Grünewald et al. 2004). In the present study, we investigated how using ligands in P. pastoris culture media could form the basis for a general method to increase the overall yield of receptor binding sites for this system.
Interestingly, it was possible to increase the expression level for all receptors except for two (5H1B_HUMAN, NK2R_RAT) (see Fig. 3) by adding ligand to the expression medium. For some targets like the OPRK_HUMAN, DRD2_HUMAN, and the 5HT5A_HUMAN, the level of functional receptor could be increased by more than twofold compared with standard conditions, and for the HRH2R_HUMAN receptor the improvement factor was even more than 7.
There is evidence that a specific ligand can serve as a molecular chaperone during protein folding (Petäjä-Repo et al. 2002; Bernier et al. 2004; Grünewald et al. 2004).
DMSO effect on expression level
To our knowledge, this study is the first to report the effect of DMSO on GPCR production in yeast. The production yield (Bmax) of 16 out of 20 tested receptors was increased up to sixfold compared with the standard conditions by adding 2.5% (v/v) DMSO to the expression medium (Fig. 3). Four receptors revealed almost no effect or slightly reduced production yield (ACM1_HUMAN, 5HT1D_HUMAN, NPY1R_HUMAN, and NK1R_HUMAN).
It has been reported that DMSO in the growth medium dramatically alters the expression pattern of yeast (Zhang et al. 2003). Genes involved in lipid synthesis were up-regulated (Murata et al. 2003) and could have positive effects on membrane proteins in yeasts. Furthermore, it was shown that DMSO alters the membrane properties of several organisms dramatically (Yu and Quinn 1994), leading to increased permeability of the membrane. This could explain the additive effect for the combination ligand/DMSO (e.g., DRD2_Mouse) where the ligand can cross the membrane more easily and reach therefore the receptor population in internal membrane compartments.
Histidine effect
Twelve of the tested receptors revealed increased expression level upon supplementing the medium with additional histidine. Here the improvement factor was not as strong as for the other parameters (<2). There have been no reports about the effect of histidine-enriched expression media on recombinant membrane protein production. One scenario could be that histidine has a nutrition-mediated effect on protein expression rather than a direct interaction on the receptor. This would help for the recombinant protein production or protect the cells from toxic side effects of the recombinant protein. It was reported that histidine rather than other amino acids can act as a physiological “anti-oxidant” (Murakami et al. 1997). However, no evident data are available to explain this measurable effect and actually no enhanced growth could be detected upon addition of histidine (data not shown).
Optimized condition
By combining all positively acting parameters in an optimized condition (temperature to 20°C, DMSO to 2.5%, specific ligand, 0.04% histidine), all clones revealed higher ligand binding values (Bmax) compared with the standard condition. Strikingly, eight out of 20 receptors revealed high Bmax values (>20 pmol/mg) after optimization. Several receptors tested (DRD2_HUMAN, NK2R_HUMAN,…) exhibited higher Bmax values than those reported for expression in mammalian or insect cell lines (Sarramegna et al. 2003).
Even for receptors where each separate effect was very little or negative (e.g., NPY1R_HUMAN), the Bmax value was almost doubled (5.6–10 pmol/mg) by combining all conditions with a positive effect. Contrastingly, for some receptors where significant improvement factors were observed for all parameters, the final Bmax value was not consistent with an additive effect indicating that the maximum optimization level was reached. From this observation, we then proposed that, depending on the target, a certain amount of receptor can be produced in a functional or nonfunctional form, and, dependent on the expression condition, the equilibrium can be shifted toward the functional form.
Correlation between ligand binding data and total receptor yield
In our study, we sought to optimize the amount of functional receptor as measured by ligand binding. Additionally, the total amount of receptor was semiquantitatively evaluated by virtue of M2 antibody binding to its Flag-tag. Because we have quantitative data for active receptor but only semiquantitative data for total receptor, we cannot simply subtract to determine the amount of inactive receptor.
Unexpectedly, for all 20 receptors we studied, the amount of active receptor increased by applying optimized conditions, whereas concerning the total amount of receptor, applying optimized conditions did not lead to systematic increases shown by our dot-blot results (Fig. 2). For example, ADA1B_HUMAN, ACM1_MOUSE, and ACM1_HUMAN revealed strong signal in the dot-blot but little receptor binding in radioactive receptor binding assay (Bmax < 1 pmol/mg). Similarly, the receptor NK3R_HUMAN revealed a strong signal in dot-blot and a Bmax of 7.4 pmol/mg only. The receptors NK2R_HUMAN, AA2AR_HUMAN, and NK2R_RAT revealed high ligand binding (Bmax > 75 pmol/mg), whereas the total amount of expressed receptor was low. This led to the conclusion that the amount of functional receptor (in terms of ligand binding) does not scale with the total amount of receptor produced. Consistent results were previously obtained for the μ-opioid receptor also expressed in Pichia (Sarramegna et al. 2002): The total amount of GFP-tagged receptor (revealed by fluorescence measurement) was compared with ligand binding, determining a ratio of functional receptor to total receptor of 1:20 under standard conditions. In the same study, this ratio was increased to 4:18 after optimization, indicating that the improvements were acting on the functionality rather than on the total amount of the receptor produced. This was also the case for 14 receptors of the present study since the immunodetected signals were not changed after optimization while the total number of binding sites (Bmax) were increased from 1.3- to more than eightfold. For the six residual GPCRs, even if the total amount of detected protein was slightly increased, it was still less than the Bmax improvements. These data are all in agreement with the idea that GPCRs are expressed in Pichia under a functional/nonfunctional equilibrium that can be modulated with the expression conditions used.
Improving GPCR expression in P. pastoris
Finally, we created a valuable tool for the optimization of the GPCR production in P. pastoris. This toolbox consists of a protease-deficient yeast strain, an optimized vector, and a set of expression conditions to screen (temperature, DMSO, ligand, and histidine). Even if each separate condition was not producing a positive effect with every GPCR tested, at least one of the four was always beneficial to expression levels. Further, the combination of all positive parameters increased the Bmax in every case, with often some significant cooperative effects observed. For some receptors, additional experiments improved further the number of active receptors in subtly refining the temperature, the type, and the concentration of ligand or the DMSO concentration (data not shown). Overall, eight out of 20 receptors were produced in Pichia with expression yields >20 pmol/mg after optimization and therefore providing sufficient amounts for structural studies.
As a result, this kind of generic approach could be of great interest to rapidly and efficiently screen for rough optimization of one given GPCR expressed in P. pastoris. On this basis, further optimization may then be realized in refining each parameter as a second screening.
Materials and methods
Materials
For molecular biology experiments, the restriction, modification, and ligation enzymes were from Fermentas Life Science. Recombinant plasmids were propagated using either XL-1 Blue (Stratagene GmbH) or TOP10 (Invitrogen) E. coli strains.
The protease-deficient P. pastoris strain SMD1163 (his4, pep4, pbr) (Invitrogen) was used for transformation and subsequent protein production. Yeast nitrogen base and yeast extract were purchased from Difco (Beckton Dickinson). Peptone was obtained from Sigma. G418 was from Calbiochem or from Invitrogen. L-Histidine was from Fluka or Sigma. DMSO was purchased from Roth or Acros Organics.
Glass beads (0.5 mm) were from Euler Prozesstechnik or from Sigma and BCA reagents were from Pierce.
[3H]-GR125743, [3H]-SR142801, and [3H]-NPY were purchased from Amersham. [3H]-Diprenorphine, [3H]-LSD, [3H]-Spiperone, [3H]-SR48968, [3H]-Prazozine, [3H]-Rauwolscine, [3H]-QNB, [3H]-Tiotidine, and [3H]-SR140333 were from PerkinElmer. GR159897, [3H]-ZM241385, GR127935, Yohimbine, Prazozine, Cimetidine, SB222200, and GR231118 were from Tocris Bioscience. Scintillation cocktail was purchased from Roth (Rotiszint eco plus) or PerkinElmer (cocktail Ultima Gold MW). PVDF membranes (Immobilon transfer membranes) and 96-filter plates (MultiScreen HTS FB) were from Millipore. Ninety-six-well GF/B filter plates (96 unifilter GF/B) were from PerkinElmer. Glass filters GF/F or GF/B were from Whatman. All other chemicals were obtained from Sigma.
Construction of expression plasmids
Twenty GPCR-encoding cDNAs were selected for this study (see Fig. 3). All of them were obtained from the MePNet consortium. A common plasmid collection was set up whereby >100 GPCR genes were cloned as ORFs without stop codon into a pCR4Blunt-TOPO vector before being sequence-verified. Depending on the clone, a type IIS restriction endonuclease digestion (Esp3I or BpiI) generated an oriented insert, which was ligated into the expression vector via BamHI/SpeI. For the present study, the 20 selected cDNAs were subcloned into the plasmid pPIC9KFlagMCSTEVBio.
This plasmid was designed for this study and was derived from the pPIC9KFlagHis10ΔGβ2ARBio vector (C. Reinhart, C. Krettler, H. Reiländer, and H. Michel, in prep.). For the new construct, a BamHI/EcoRI fragment of the plasmid pPIC9KFlagHis10ΔGβ2ARBio was removed and replaced by annealed oligonucleotides TEVfor (5′-GATCCCGCGGCCGGACCGGCCCACGTGGAATTCGAAAACCTGTACTTTCAAGGTC-3′) and TEVrev (5′-AATTGACCTTGAAAGTACAGGTTTTCGAATTCCACGTGGGCCGGTCCGGCCGCGG-3′). Subsequently, a second TEV site was introduced by replacing a BamHI/EcoRI fragment by the annealed oligonucleotides TEVIIfor (5′-GATCGAGAAAACCTGTACTTTCAAGGTGGATCCGCTACTAGTG-3′) and TEVIIrev (5′-AATTCACTAGTAGCGGATCCACCTTGAAAGTACAGGTTTTCTC-3′).
Yeast culture
The 20 different integrative expression vectors were linearized using the restriction enzyme PmeI and transformed by electroporation (1500 V, 25 μF, and 600 Ω) using a Gene Pulser I (Bio-Rad) into the strain SMD1163. Clone selection was performed as previously described (Scorer et al. 1994; Weiss et al. 1998b). In brief, recombinant His+ clones were selected on MD agar plates (1.34% yeast nitrogen base without amino acids, 2% dextrose, 0.00004% biotin, 1.5% agar). In a second step aimed at selecting for multicopy transformants, His+ clones were grown on YPD agar plates containing various concentrations of G418 (1% yeast extract, 2% peptone, 2% dextrose, 2% agar, 0.05–0.5 mg/mL G418). Representative clones exhibiting resistance to various G418 concentrations were tested for recombinant protein production by dot-blot analysis using anti-M2 antibody (Sigma).
For receptor production, cells were precultured in BMGY medium (1% yeast extract, 2% peptone, 1.34% yeast nitrogen base without amino acid, 0.00004% biotin, 1% glycerol, 0.1 M phosphate buffer at pH 6) at 30°C and 250 rpm until an OD600 of 2–6 was reached. As standard condition, induction was achieved in BMMY medium (same as BMGY with 0.5% methanol instead of glycerol) at 30°C from an initial OD600 of 1. Depending on the different induction conditions evaluated in this study, several parameters were then separately or jointly modified. After 18-h induction, cells were pelleted at 3000g for 5 min and subsequently used for membrane preparation.
Membrane preparation
All work was carried out at 0°–4°C. Cells were washed once with ice-cold breaking buffer (50 mM sodium-phosphate buffer at pH 7.4, 100 mM NaCl, 5% glycerol, 2 mM EDTA, 1 mM PMSF) and resuspended to 30% wet weight. After 0.5-mm glass beads were added to the cell suspension, yeast cells were broken by vigorous vortexing at 4°C for 10 min. Breaking efficiency was inspected with a light microscope and was usually >80%. Intact cells and cell debris were separated from the membrane suspension by a low-speed centrifugation (3000g, 5 min, 4°C). Membranes were then pelleted using an ultracentrifuge (100,000g, 45 min, 4°C), resuspended in membrane buffer (50 mM Tris at pH 8.0, 120 mM NaCl, 20% glycerol, 1 mM PMSF) using a dounce homogenizer. Membrane proteins were quantified following the bicinchoninic acid (BCA) method (Pierce), using bovine serum albumin as a standard. Membranes were snap-frozen in liquid nitrogen and stored at −80°C.
Radioligand binding assays
For each tested receptor, specific ligand binding assays were realized following the protocols detailed in Table 1. At a fixed temperature, membrane proteins were incubated in triplicate with specific radioligands until equilibrium was reached. For nonspecific binding determination, similar incubations were performed in parallel in the presence of an excess of unlabeled specific ligand. Bound and free ligands were separated by rapid filtration (Whatman GF/B or GF/F filter, or Perkin-Elmer GF/B 96-unifilter) presoaked in 0.3% polyethylenimine. After the filters were washed three times, the retained radioactivity was measured by liquid-scintillation counting. Single point binding assays were performed at one nonsaturating radioligand concentration. Saturation curves were analyzed by nonlinear regression using Kaleidagraph (Synergy Software) allowing determination of Bmax and Kd.
Dot-blot immunodetection
Pichia membranes were diluted in TBS buffer with 0.2% SDS. For each sample, 1 μg of total membrane protein was applied to a wet PVDF membrane (0.45 μm; Millipore) previously washed with methanol and subsequently with 38 mM Glycine, 10 mM Tris, 20% methanol using a 96-well microfiltration apparatus (Bio-Dot, Bio-Rad). PVDF membrane was then blocked with 5% lowfat milk powder in TBS for 1 h at room temperature. Further, PVDF membrane was washed three times for 1 min with TBS, and subsequently incubated with alkaline phosphatase conjugated anti-FLAG M2 antibody (Sigma). After three 15-min washing steps, the blot was developed in AP-buffer (100 mM Tris at pH 9.5, 100 mM NaCl, 5 mM MgCl2) with the addition of 330 μg/mL 5-bromo-4-chloro-3-indolylphosphate p-toluidinium salt and 165 μg/mL nitroblue tetrazolium chloride. A standard curve was done with Pichia membranes containing the β2 adrenergic receptor with specific ligand binding of 25 pmol/mg (C. Reinhart, C. Krettler, H. Reiländer, and H. Michel, in prep.).
Acknowledgments
We thank Dr. C. Bevans and Dr. D. Parcej for fruitful discussions and critically reading the manuscript, and Gabriele Maul for excellent technical assistance. We are grateful to Angela Finch for the cDNA encoding ADA1B_HUMAN, Susanna Cotecchia for ADA2B_HUMAN, Neil Nathanson for ACM1_MOUSE, and Tatsuya Haga for ACM2_PIG. This work was supported by the Max-Planck-Gesellschaft, Fonds der chemischen Industrie, and the Membrane Protein Network (MePNet).
We dedicate this article in memory of Helmut Reiländer (1955–2004), who pioneered expression of GPCRs in various heterologous expression systems.
Footnotes
Reprint requests to: Christoph Reinhart, Max-Planck-Institute of Biophysics, Max-von-Laue-Str. 3, D-60438 Frankfurt am Main, Germany; e-mail: christoph.reinhart@mpibp-frankfurt.mpg.de; fax: +49-69-6303-1002.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062098206.
References
- Akermoun M., Koglin M., Zvalova-Iooss D., Folschweiller N., Dowell S.J., Gearing K.L. 2005. Characterization of 16 human G protein-coupled receptors expressed in baculovirus-infected insect cells Protein Expr. Purif. 44: 65–74. [DOI] [PubMed] [Google Scholar]
- Audinot V., Newman-Tancredi A., Cussac D., Milla N.M.J. 2001. Inverse agonist properties of antipsychotic agents at cloned, human (h) serotonin (5-HT)(1B) and h5-HT(1D) receptors Neuropsychopharmacology 25: 410–422. [DOI] [PubMed] [Google Scholar]
- Befort K., Tabbara L., Kling D., Maigret B., Kieffer B.L. 1996. Role of aromatic transmembrane residues of the δ-opioid receptor in ligand recognition J. Biol. Chem. 271: 10161–10168. [DOI] [PubMed] [Google Scholar]
- Bernier V., Bichet D.G., Bouvier M. 2004. Pharmacological chaperon action on G-protein-coupled receptors Curr. Opin. Pharmacol. 4: 528–533. [DOI] [PubMed] [Google Scholar]
- Bockaert J. and Pin J.P. 1999. Molecular tinkering of G protein-coupled receptors: An evolutionary success EMBO J. 18: 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner M., Prinz B., Holz C., Stahl U., Lang C. 2002. High-throughput screening for expression of heterologous proteins in the yeast Pichia pastoris J. Biotechnol. 99: 51–62. [DOI] [PubMed] [Google Scholar]
- Cabrele C., Wieland H.A., Langer M., Stidsen C.E., Beck-Sickinger A.G. 2001. Y-receptor affinity modulation by the design of pancreatic polypeptide/neuropeptide Y chimera led to Y(5)-receptor ligands with picomolar affinity Peptides 22: 365–378. [DOI] [PubMed] [Google Scholar]
- Domenech T., Beleta J., Palacios J.M. 1997. Characterization of human serotonin 1D and 1B receptors using [3H]-GR-125743, a novel radiolabelled serotonin 5HT1D/1B receptor antagonist Naunyn Schmiedebergs Arch. Pharmacol. 356: 328–334. [DOI] [PubMed] [Google Scholar]
- Drew D., Froderberg L., Baars L., de Gier J.W. 2003. Assembly and overexpression of membrane proteins in Escherichia coli Biochim. Biophys. Acta 1610: 3–10. [DOI] [PubMed] [Google Scholar]
- Grisshammer R., Little J., Aharony D. 1994. Expression of rat NK-2 (neurokinin A) receptor in E. coli Receptors Channels 2: 295–302. [PubMed] [Google Scholar]
- Grünewald S., Haase W., Molsberger E., Michel H., Reiländer H. 2004. Production of the human D2S receptor in the methylotrophic yeast P. pastoris. Recept. Channels 10: 37–50. [DOI] [PubMed] [Google Scholar]
- Hill P., Lai Y., Hnilo J., Lin C.C., Karla M., Bounds S., Herz J., Mitchell R. 1996. Cloning, expression, and comparison of the binding characteristics of the known human dopamine receptors Adv. Neurol. 69: 41–52. [PubMed] [Google Scholar]
- Homma T., Iwahashi H., Komatsu Y. 2003. Yeast gene expression during growth at low temperature Cryobiology 46: 230–237. [DOI] [PubMed] [Google Scholar]
- Hong F., Meinander N.Q., Jonsson L.J. 2002. Fermentation strategies for improved heterologous expression of laccase in Pichia pastoris Biotechnol. Bioeng. 79: 438–449. [DOI] [PubMed] [Google Scholar]
- Inouye M. and Phadtare S. 2004. Cold shock response and adaptation at near-freezing temperature in microorganisms Sci. STKE 237: pe26. [DOI] [PubMed]
- Ivanovic A. “Klonierung rekombinanter Histamin H1-rezeptoren und deren charakterisierung; solubilisierung und reinigung nach heterologer produktion.” Ph.D. thesis 2001. Johann-Wolfgang-Goethe-Universität, Frankfurt am Main, Germany.
- Jahic M., Gustavsson M., Jansen A.K., Martinelle M., Enfors S.O. 2003a. Analysis and control of proteolysis of a fusion protein in Pichia pastoris fed-batch processes J. Biotechnol. 102: 45–53. [DOI] [PubMed] [Google Scholar]
- Jahic M., Wallberg F., Bollok M., Garcia P., Enfors S.O. 2003b. Temperature limited fed-batch technique for control of proteolysis in Pichia pastoris bioreactor cultures Microb. Cell Fact. 18: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K., Dohlman H.G., Thorner J., Caron M.G., Lefkowitz R.J. 1990. Control of yeast mating signal transduction by a mammalian β 2-adrenergic receptor and Gs α subunit Science 250: 121–123 [Erratum: Science 1991 251: 144]. [DOI] [PubMed] [Google Scholar]
- Klabunde T. and Hessler G. 2002. Drug design strategies for targeting G-protein-coupled receptors ChemBioChem 3: 928–944. [DOI] [PubMed] [Google Scholar]
- Koganesawa N., Aizawa T., Shimojo H., Miura K., Ohnishi A., Demura M., Hayakawa Y., Nitta K., Kawano K. 2002. Expression and purification of a small cytokine growth-blocking peptide from armyworm Pseudaletia separata by an optimized fermentation method using the methylotrophic yeast Pichia pastoris Protein Expr. Purif. 25: 416–425. [DOI] [PubMed] [Google Scholar]
- Kuhn P., Wilson K., Patch M.G., Stevens R.C. 2002. The genesis of high-throughput structure-based drug discovery using protein crystallography Curr. Opin. Chem. Biol. 6: 704–710. [DOI] [PubMed] [Google Scholar]
- Leurs R., Smit M.J., Menge W.M., Timmerman H. 1994. Pharmacological characterization of the human histamine H2 receptor stably expressed in Chinese hamster ovary cells Br. J. Pharmacol. 112: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Edwards P.C., Burghammer M., Villa C., Schertler G.F. 2004. Structure of bovine rhodopsin in a trigonal crystal form J. Mol. Biol. 343: 1409–1438. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. 2003. Semliki Forest virus vectors for rapid and high-level expression of integral membrane proteins Biochim. Biophys. Acta 1610: 90–96. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. 2005. Structural genomics of GPCRs Trends Biotechnol. 23: 103–108. [DOI] [PubMed] [Google Scholar]
- Lundstrom K., Hawcock A.B., Vargas A., Ward P., Thomas P., Naylor A. 1997. Effect of single point mutations of the human tachykinin NK1 receptor on antagonist affinity Eur. J. Pharmacol. 337: 73–81. [DOI] [PubMed] [Google Scholar]
- Massotte D. 2003. G protein-coupled receptor overexpression with the baculovirus-insect cell system: A tool for structural and functional studies Biochim. Biophys. Acta 1610: 77–89. [DOI] [PubMed] [Google Scholar]
- Millan M.J., Newman-Tancredi A., Audinot V., Cussac D., Lejeune F., Nicolas J.P., Coge F., Galizzi J.P., Boutin J.A., Rivet J.M.et al. 2000. Agonist and antagonist actions of yohimbine as compared to fluparoxan at α(2)-adrenergic receptors (AR)s, serotonin (5-HT)(1A), 5-HT(1B), 5-HT(1D) and dopamine D(2) and D(3) receptors. Significance for the modulation of frontocortical monoaminergic transmission and depressive states Synapse 35: 79–95. [DOI] [PubMed] [Google Scholar]
- Murakami K., Onoda Y., Kimura J., Yoshino M. 1997. Protection by histidine against oxidative inactivation of AMP deaminase in yeast Biochem. Mol. Biol. Int. 42: 1063–1069. [DOI] [PubMed] [Google Scholar]
- Murata Y., Watanabe T., Sato M., Momose Y., Nakahara T., Oka S., Iwahashi H. 2003. Dimethyl sulfoxide exposure facilitates phospholipid biosynthesis and cellular membrane proliferation in yeast cells J. Biol. Chem. 278: 33185–33193. [DOI] [PubMed] [Google Scholar]
- Opekarová M. and Tanner W. 2003. Specific lipid requirements of membrane proteins—a putative bottleneck in heterologous expression Biochim. Biophys. Acta 1160: 11–22. [DOI] [PubMed] [Google Scholar]
- Oury-Donat F., Carayon P., Thurneyssen O., Pailhon V., Emonds-Alt X., Soubrie P., Le Fur G. 1995. Functional characterization of the nonpeptide neurokinin3 (NK3) receptor antagonist, SR142801 on the human NK3 receptor expressed in Chinese hamster ovary cells J. Pharmacol. Exp. Ther. 274: 148–154. [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E.et al. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor Science 289: 739–745. [DOI] [PubMed] [Google Scholar]
- Parker E.M., Babij C.K., Balasubramaniam A., Burrier R.E., Guzzi M., Hamud F., Mukhopadhyay G., Rudinski M.S., Tao Z., Tice M.et al. 1998. GR231118 (1229U91) and other analogues of the C-terminus of neuropeptide Y are potent neuropeptide Y Y1 receptor antagonists and neuropeptide Y Y4 receptor agonists Eur. J. Pharmacol. 349: 97–105. [DOI] [PubMed] [Google Scholar]
- Peralta E.G., Ashkenazi A., Winslow J.W., Smith D.H., Ramachandran J., Capon D.J. 1987. Distinct primary structures, ligand-binding properties and tissue-specific expression of four human muscarinic acetylcholine receptors EMBO J. 6: 3923–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petäjä-Repo U.E., Hogu E.M., Bhalla S., Laperriere A., Morello J.-P., Bouvier M. 2002. Ligands act as pharmacological chaperones and increase the efficiency of δ opioid receptor maturation EMBO J. 21: 1628–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees S., den Daas I., Foord S., Goodson S., Bull D., Kilpatrick G., Lee M. 1994. Cloning and characterisation of the human 5-HT5A serotonin receptor FEBS Lett. 355: 242–246. [DOI] [PubMed] [Google Scholar]
- Reinhart C. and Krettler C. 2006. Expression of membrane proteins in yeasts In Structural genomics and rational drug design in membrane proteins (ed. Lundstrom K.) . pp. 115–152. Marcel Dekker, New York (in press).
- Sarau H.M., Griswold D.E., Bush B., Potts W., Sandhu P., Lundberg D., Foley J.J., Schmidt D.B., Webb E.F., Martin L.D.et al. 2000. Nonpeptide tachykinin receptor antagonists. II. Pharmacological and pharmacokinetic profile of SB-222200, a central nervous system penetrant, potent and selective NK-3 receptor antagonist J. Pharmacol. Exp. Ther. 295: 373–381. [PubMed] [Google Scholar]
- Sarramegna V., Demange P., Milon A., Talmont F. 2002. Optimizing functional versus total expression of the human μ-opioid receptor in Pichia pastoris Protein Expr. Purif. 24: 212–220. [DOI] [PubMed] [Google Scholar]
- Sarramegna V., Talmont F., Demange P., Milon A. 2003. Heterologous expression of G-protein-coupled receptors: Comparison of expression systems from the standpoint of large-scale production and purification Cell. Mol. Life Sci. 60: 1529–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller H., Haase W., Molsberger E., Janssen P., Michel H., Reiländer H. 2000. The human ET(B) endothelin receptor heterologously produced in the methylotrophic yeast Pichia pastoris shows high-affinity binding and induction of stacked membranes Recept. Channels 7: 93–107. [PubMed] [Google Scholar]
- Scorer C.A., Clare J.J., McCombie W.R., Romanos M.A., Sreekrishna K. 1994. Rapid selection using G418 of high copy number transformants of Pichia pastoris for high-level foreign gene expression Biotechnology (N.Y.) 12: 181–184. [DOI] [PubMed] [Google Scholar]
- Sizmann D., Kuusinen H., Keranen S., Lomasney J., Caron M.G., Lefkowitz R.J., Keinanen K. 1996. Production of adrenergic receptors in yeast Recept. Channels 4: 197–203. [PubMed] [Google Scholar]
- Tate C.G. 2001. Overexpression of mammalian integral membrane proteins for structural studies FEBS Lett. 504: 94–98. [DOI] [PubMed] [Google Scholar]
- Walter S. and Buchner J. 2002. Molecular chaperons—Cellular machines for protein folding Angew. Chem. Int. Ed. Engl. 41: 1098–1113. [DOI] [PubMed] [Google Scholar]
- Weiss H.M., Haase W., Michel H., Reiländer H. 1998a. Comparative biochemical and pharmacological characterization of the mouse 5HT5A 5-hydroxytryptamine receptor and the human β2-adrenergic receptor produced in the methylotrophic yeast Pichia pastoris Biochem. J. 330: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss H.M., Haase W., Reiländer H. 1998b. Expression of an integral membrane protein, the 5HT5A receptor Methods Mol. Biol. 103: 227–239. [DOI] [PubMed] [Google Scholar]
- Weiss H.M. and Grisshammer R. 2002. Purification and characterization of the human adenosine A2a receptor functionally expressed in Escherichia coli Eur. J. Biochem. 269: 82–92. [DOI] [PubMed] [Google Scholar]
- Yu Z.W. and Quinn P.J. 1994. Dimethyl sulphoxide: A review of its applications in cell biology Biosci. Rep. 14: 259–281. [DOI] [PubMed] [Google Scholar]
- Zhang W., Needham D.L., Coffin M., Rooker A., Hurban P., Tanzer M.M., Shuster J.R. 2003. Microarray analyses of the metabolic responses of Saccharomyces cerevisiae to organic solvent dimethyl sulfoxide J. Ind. Microbiol. Biotechnol. 30: 57–69. [DOI] [PubMed] [Google Scholar]