Abstract
Tetratricopeptide repeat (TPR) domains bind specific peptide ligands and are thought to mediate protein–protein interactions in a variety of biological systems. Here we compare peptide ligand-binding by several different TPR domains. We present specific examples that demonstrate that TPR domains typically undergo little or no structural rearrangement upon ligand binding. Our data suggest that, contrary to a recent proposal, coupled folding and binding is not the common mechanism of ligand recognition by TPR domains.
Keywords: tetratricopeptide repeat (TPR), Hsp90, protein phosphatase 5 (PP5), Vpu-binding protein/small glutamine-rich protein (UBP/SGT), ligand binding
The tetratricopeptide repeat (TPR) is a 34-amino-acid repeat motif that is found in many diverse proteins in all organisms. Different numbers of tandem repeats, from 3 to >16, occur in different proteins (Lamb et al. 1995; D'Andrea and Regan 2003). TPR proteins are thought to function as protein–protein interaction domains, although in the vast majority of cases, the identity of a particular TPR domain's cognate ligand has not yet been identified. There are several examples in which a TPR domain has been produced in isolation from the rest of the protein and has been shown to exhibit the same binding characteristics as it displays in the context of the full-length protein (Brinker et al. 2002; Yang et al. 2005). It is of interest, in the context of a general consideration of protein–ligand interactions, to analyze whether there are significant structural rearrangements associated with TPR–ligand interactions. Such information is important both for our understanding of TPR function within the cell and for efforts to predict TPR binding specificities from sequence comparisons. Here we present several lines of evidence supporting a mechanism of recognition in which the folded TPR module interacts with its cognate ligand with little, if any, structural rearrangements of the TPR fold.
The X-ray crystal structure of the TPR domain of PP5, which contains three tandem TPR repeats, was the first high-resolution TPR structure determined (Das et al. 1998). This structure revealed that each individual 34-amino-acid repeat forms a helix-turn-helix structure. Tandem repetition of this motif, with near-identical inter-repeat angles and interactions, generates a structure, with an overall “super coil.” Within the context of the full-length protein, the conformation of the TPR domain of PP5 is the same as in the isolated domain with backbone RMSD values of 0.6 Å, only the last helix of the domain that connects the TPR with the phosphatase domain is rotated by ∼5° (Yang et al. 2005). Since the original PP5 TPR domain structure was determined, several more structures of both natural and designed TPR domains, with and without ligand bound, have been solved (Das et al. 1998; Scheufler et al. 2000; Taylor et al. 2001; Main et al. 2003; Sinars et al. 2003). There are no significant differences between the structures of TPRs without ligand bound and those with ligand bound. More explicitly, all the free TPR domains are folded and structured in the absence of ligand. Recently, however, Cliff et al. (2005) presented evidence that a particular TPR construct, from the protein PP5, was largely unfolded in absence of ligand and was stabilized when bound to ligand. The investigators proposed that folding coupled to binding might be a common mechanism of ligand recognition by TPR domains. The results we present in this paper, which compare stability, structure, and ligand-binding data for several natural and designed TPR proteins (Table 1), suggest, however, that coupled folding and binding is a relatively uncommon recognition mechanism for TPR domains.
Table 1.
Protein constructs
All of the TPR domains were cloned into pProEx-HTA vector (Invitrogen), and the constructs encode the desired TPR domains as N-terminal hexa-histidine-tagged fusion proteins. All proteins were overexpressed in Escherichia coli BL21(DE3) and purified by affinity chromatography on Talon resin (BD Biosciences, Clontech) according to manufacturer's protocol. The His-tag was cleaved using rTEV protease and removed by metal-affinity chromatography. All proteins were soluble with purification yields between 10 and 40 mg/L.
a The protein from which the TPR fragment was abstracted.
b The TPR is named after the protein source unless >1 TPR domain is present in the same protein.
c The residues (numbering from the N terminus of the full-length protein) that comprise each TPR construct studied.
Results
High-resolution crystal structures of six 3-TPR domains have been reported: two in complex with their peptide ligand (Scheufler et al. 2000) and four without ligand bound (Das et al. 1998; Taylor et al. 2001; Main et al. 2003; Sinars et al. 2003). We have compared these structures in detail and find that they are all completely superimposable, with backbone RMSD values that vary from 1.1 to 1.9 Å for different pairwise structural alignments. Moreover, we find no consistent differences between the structures of free TPR domains and the structures of those in complex with a peptide ligand. Figure 1A shows ribbon representations of the crystal structures of an unliganded 3-TPR domain, the designed protein CTPR3 (Main et al. 2003). Also shown in Figure 1B is the superimposition of two unliganded 3-TPR domains, CTPR3 and the TPR domain of the natural protein PP5 (Das et al. 1998), and the TPR-peptide cocrystal structures of TPR1 and TPR2A from Hsp-organizing protein (HOP) (Scheufler et al. 2000).
Figure 1.
Ribbon representation of crystal structures of TPR domains. (A) CTPR3 domain, in yellow is shown the first TPR repeat (helix-turn-helix motif) (PDB ID 1NA0; Main et al. 2003). (B) Structural alignment of TPR domains: (Blue) CTPR3 (PDB ID 1NA0; Main et al. 2003); (orange) TPR domain (residues 19–177) of PP5 (PDB ID 1A17; Das et al. 1998); (green) TPR2A (PDB ID 1ELR; Scheufler et al. 2000); and (yellow) TPR1 (PDB ID 1ELW; Scheufler et al. 2000). TPR2A and TPR1 domains of Hop, from their co-crystal structures with the C-terminal peptides of Hsp90 and Hsp70, respectively.
Because it has been proposed that the TPR domain of PP5 is unfolded in solution (Cliff et al. 2005), we considered it essential to demonstrate that the folded structure of this domain is not induced by the crystallization conditions nor by crystal packing interactions. In the crystal structure of the TPR domain of PP5 that we study, there are crystal packing interactions between the C-terminal helix and the concave surface of a neighbor molecule forming a crystallographic dimer (Das et al. 1998). We therefore investigated the oligomerization state of this TPR domain in solution, using analytical gel filtration chromatography. We found no indication of any species other than the monomer. We also detected a single, monomeric species for all the other TPR domains that we have characterized (data not shown). Figure 2A shows the CD spectrum of the TPR domain of PP5 in aqueous solution (again, the same construct for which the crystal structure was determined). It is clear from this spectrum that the TPR domain is not unfolded but rather exhibits a highly helical CD spectrum, consistent with its folded, helical structure. Moreover, thermal denaturation studies of the TPR domain of PP5 show that it is folded up to a temperature of ∼40°C; it then undergoes a cooperative, reversible, unfolding transition (Fig. 2B).
Figure 2.
Thermodynamic stability and binding activity of TPR domain of PP5. (A) Far-UV CD spectrum of the TPR domain of PP5 (residues 24–177) at 12 μM protein concentration in a 0.1-cm pathlength cuvette. The CD spectrum was recorded with a bandwidth of 1 nm at 1-nm increments and 10-sec average time. All CD experiments were performed using an AVIV Model 215 CD spectrophotometer (AVIV Instruments) in 150 mM NaCl, 50 mM phosphate buffer (pH 6.5) at 25°C. (B) Thermal denaturation of PP5 TPR domain. Thermal denaturation was monitored at 6 μM protein concentration following the ellipticity at 222 nm from 15°C to 95°C and in the reverse direction from 95°C to 15°C in a 0.1-cm path-length cuvette. The temperature ramp was performed in 1°C steps with an equilibration time at each temperature of 1 min. (C) Interaction of the TPR domain of PP5 with the 24-mer C-terminal peptides of Hsp90 (•) and Hsp70 (○) measured by surface plasmon resonance (SPR) in HBS-EP buffer (150 mM NaCl, 3 mM EDTA, 0.005% [v/v] polysorbate 20, 10 mM Hepes at pH 7.5). SPR measurements were performed using a BIACORE 3000 (BIACORE AB) as described previously (Cortajarena et al. 2004). Equilibrium response units were plotted vs. the protein concentration. The data were fit to a 1:1 binding model to calculate the dissociation constant (KD TPR-PP5/Hsp90 = 660 nM).
These results clearly demonstrate that the TPR domain of PP5 is folded in solution. In addition, we have demonstrated that this folded protein is active in ligand binding. Figure 2C shows a binding curve measured by surface plasmon resonance for the TPR domain of PP5 interacting with its cognate ligand, namely the C-terminal peptide of Hsp90. The experimental data are well fit to a binding curve for a 1:1 TPR–peptide interaction, with a dissociation constant of 0.66 μM. Moreover, the binding is specific for the cognate ligand, as evidenced by the extremely low binding observed between the TPR of PP5 and a different but related peptide, namely the C-terminal peptide of Hsp70 (Fig. 2C).
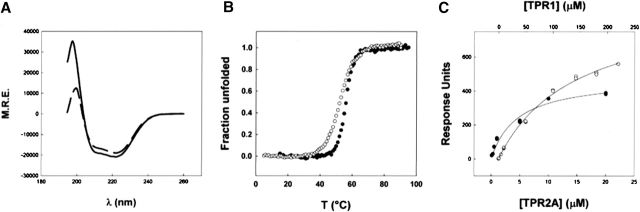
Particularly well-characterized examples of TPR domains are the TPR1 and TPR2A domains of HOP, which bind to the C-terminal peptides of Hsp70 and Hsp90, respectively. It has been shown that the affinity of the isolated TPR domains for their peptide ligands matches their affinity in the context of full-length HOP (Scheufler et al. 2000; Brinker et al. 2002). Furthermore, the crystal structures of the TPR1–Hsp70 and TPR2A–Hsp90 complexes have been solved at high resolution and reveal the nature of the TPR–peptide interactions in atomic detail (Scheufler et al. 2000). Figure 3A shows CD spectra of TPR1 and TPR2A in the absence of ligand. The spectra demonstrate that both domains are highly helical in solution. Again, both the TPR1 and TPR2A domains undergo cooperative unfolding transitions induced by either heat or chemical denaturants, as shown in Figure 3B. They show tight and specific binding to their cognate ligands in solution, as shown in Figure 3C. Both TPR1 and TPR2A are active and completely folded in solution in the absence of ligand, with no indication of ligand-induced folding or conformational change upon ligand binding.
Figure 3.
Thermodynamic stability and binding activity of TPR2A and TPR1 domains of Hop. (A) Far-UV spectra of TPR2A domain (residues 223–349) (solid line) and TPR1 domain (residues 1–115) (dashed line) at 12 μM protein concentration in 150 mM NaCl, 50 mM phosphate buffer (pH 6.5) at 25°C. (B) Thermal denaturation curves of TPR2A (•) and TPR1 (○) domains monitored by the change in CD signal at 222 nm as described in Figure 2. (C) Binding of TPR2A (•) and TPR1 (○) domains of Hop to their cognate ligands, C-terminal peptide of Hsp90 and Hsp70, respectively. Equilibrium response units measured by surface plasmon resonance were plotted vs. protein concentration, and the data were fit to a simple one-site binding mode to obtain the dissociation binding constants (KD TPR2A/Hsp90 = 5 μM; KD TPR1/Hsp70 = 100 μM).
The final example of a natural 3-TPR domain, whose characterization strongly supports the proposition that TPR domains do not undergo dramatic conformational changes upon ligand binding, is the 3-TPR domain of the protein UBP (Vpu binding protein), also called SGT (small glutamine-rich protein) (Callahan et al. 1998; Cziepluch et al. 1998). The CD spectrum of the TPR domain of UBP is shown in Figure 4A and is again consistent with a fully helical protein. The thermal denaturation curve of UBP is shown in Figure 4B. It has similar stability to the TPR domain of PP5, being folded up to ∼40°, then undergoing a cooperative denaturation transition. UBP binds to the C-terminal peptide of Hsp70, with a dissociation constant of 225 μM as shown in Figure 4C. The NMR spectrum of the TPR domain of UBP is completely assigned (Pai et al. 2003). Thus, we are able to use two-dimensional NMR methods to compare in residue-specific detail the structures of the TPR domain of UBP, with and without ligand bound. Figure 5A shows the heteronuclear single quantum coherence (HSQC) spectrum of the TPR domain of UBP in the absence of ligand. The spectrum displays well-dispersed peaks, which is consistent with the TPR domain adopting a folded, unique structure. Figure 5B shows the HSQC spectrum of the same protein, but in the presence of 2 molar equivalents of the C-terminal peptide of Hsp70. It is clear that there is not a dramatic structural change upon ligand binding. Formation of a TPR–ligand complex is occurring under these conditions because localized changes in the chemical shifts of certain residues are observed. These shifts can be mapped onto a model for the UBP structure, allowing us to confirm the ligand-binding site, as illustrated in Figure 5C.
Figure 4.
Thermodynamic stability and binding activity of the TPR domain of UBP. (A) Far-UV CD spectrum of the TPR domain of UBP (residues 88–208) at 12 μM protein concentration in 150 mM NaCl, 50 mM phosphate buffer (pH 6.5) at 25°C. (B) Thermal denaturation curve of UBP TPR domain monitored by the change in ellipticity signal at 222 nm as described in Figure 2. (C) Binding activity of UBP TPR domain to C-terminal 24-mer peptide of Hsp70 measured by SPR. The response at the equilibrium data at increasing protein concentration was fit to a one-site saturation model to calculate the dissociation binding constant (KD UBP-TPR/Hsp70 = 225 μM).
Figure 5.
1H–15N HSQC spectra of the TPR domain of UBP. (A) 1H–15N HSQC spectrum of TPR domain of UBP collected at 1 mM protein concentration in 100 mM NaCl, 10 mM NaN3, and 0.1 mM EDTA, 50 mM K2HPO4 (pH 6.5), 10% D2O at 25°C. (B) 1H–15N HSQC perturbation map of UBP TPR domain upon binding the C-terminal peptide of Hsp70. Overlay 1H–15N HSQC spectra of the TPR domain of UBP in the absence (red) and in the presence (blue) of 2 molar equivalents of 20-mer C-terminal peptide of Hsp70 15N-glycine labeled (GGGAPPSGGASSGPTIEEVD-COOH). No further changes in the chemical shifts were observed between the addition of 1 and 2 molar equivalents of peptide. The new peaks observed upon the addition of the peptide located around 110 p.p.m. at ω1-15N and 8.4 p.p.m. at ω2-1H are the chemical shifts of the 15N-labeled glycines from the Hsp70 peptide. All NMR spectra were recorded at 25°C on a Varian Inova (Varian Inc.) 800-MHz spectrophotometer, processed with NMRPipe (Delaglio et al. 1995), and analyzed with Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco). (C) Map of ligand binding site of UBP TPR domain. Surface representations of the NH chemical shift changes observed upon binding the C-terminal 24-mer peptide of Hsp70 onto the front/concave face of the modeled structure of the TPR domain of UBP. The model was generated using Swiss-Model by automated homology modeling (http://swissmodel.expasy.org//SWISS-MODEL.html) (Schwede et al. 2003). The chemical shift data were mapped onto the modeled structure running a script generated by Protskin (http://www.mcgnmr.ca/ProtSkin/) on the molecular surface created using GRASP (http://trantor.bioc.columbia.edu/grasp/) (Nicholls et al. 1993). The color range from white to deep blue corresponds to the range in values of chemical shifts perturbation from 0 to 1.5 p.p.m.
A consensus 3-TPR protein has also been designed and characterized (Main et al. 2003). Ligand-binding activity has been introduced onto the designed protein (Cortajarena et al. 2004) to create a series of TPR domains that bind their peptide ligand with affinities ranging from 200 μM to <1 μM. All these proteins, in the absence of ligand, are fully folded and undergo chemically and thermally induced cooperative thermal denaturation transitions. Moreover, when ligand binding is monitored using NMR to follow the changes in chemical shifts that occur upon interaction with ligand, it is clear the protein is folded to start with and remains folded when peptide ligand binds (Cortajarena et al. 2004). The only residues to shift are those that come in close contact with the ligand.
Discussion
We have investigated the details of TPR–ligand interactions for four different 3-TPR domains. In all these examples, the TPR domain is shown to fold cooperatively as an independent unit. We also find that the TPR domain is folded with and without ligand bound and that there are no substantial changes in protein conformation upon ligand binding. These results lead us to conclude that the TPR domain is a versatile binding motif that presents a common, folded framework onto which specific ligand-binding residues are “grafted.”
Acknowledgments
A.L.C. was the recipient of a postdoctoral fellowship from Spanish Ministry of Education, Culture, and Sports. We thank Weilan Pan and Chris Wilson for preliminary studies with the PP5 TPR domain. We thank Tommi Kajander, Tina Liu, Irina Pozdnyakova, Dorina Saro, and Fang Yi for critical reading of the manuscript.
Footnotes
Reprint requests to: Lynne Regan, Department of Chemistry, Yale University, 266 Whitney Avenue, New Haven, CT 06520, USA; e-mail: lynne.regan@yale.edu; fax: (203) 432-5175.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062092506.
Abbreviations: TPR, tetratricopeptide repeat; RMSD, root mean square deviation; CD, circular dichroism spectroscopy; HSQC, heteronuclear single quantum coherence; PP5, protein phosphatase 5; UBP, Vpu-binding protein; SGT, small glutamine-rich protein; MRE, mean residue ellipticity; Hop, Hsp-organizing protein
References
- Brinker A., Scheufler C., Von Der Mulbe F., Fleckenstein B., Herrmann C., Jung G., Moarefi I., Hartl F.U. 2002. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70·Hop·Hsp90 complexes J. Biol. Chem. 277: 19265–19275. [DOI] [PubMed] [Google Scholar]
- Callahan M., Handley M., Lee Y., Talbot K., Harper J., Panganiban A. 1998. Functional interaction of human immunodeficiency virus type 1 Vpu and Gag with a novel member of the tetratricopeptide repeat protein family J. Virol. 72: 5189–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff M., Williams M., Brooke-Smith J., Barford D., Ladbury J. 2005. Molecular recognition via coupled folding and binding in a TPR domain J. Mol. Biol. 346: 717–732. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Kajander T., Pan W., Cocco M.J., Regan L. 2004. Protein design to understand peptide ligand recognition by tetratricopeptide repeat proteins Protein Eng. Des. Sel. 17: 399–409. [DOI] [PubMed] [Google Scholar]
- Cziepluch C., Kordes E., Poirey R., Grewenig A., Rommelaere J., Jauniaux J. 1998. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1 J. Virol. 72: 4149–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea L. and Regan L. 2003. TPR proteins: The versatile helix Trends Biochem. Sci. 28: 655–662. [DOI] [PubMed] [Google Scholar]
- Das A.K., Cohen P.W., Barford D. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein–protein interactions EMBO J. 17: 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F.G.S., Vuister G.W., Zhu G., Pfeifer J.B.A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich S., Hieter P. 1995. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem. Sci. 20: 257–259. [DOI] [PubMed] [Google Scholar]
- Main E.R.G., Xiong Y., Cocco M.J., D'Andrea L., Regan L. 2003. Design of stable α-helical arrays from an idealized TPR motif Structure 11: 497–508. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj R., Honig B. 1993. GRASP—graphical representationand analysis of surface properties Biophys. J. 64: A166. [Google Scholar]
- Pai M.T., Yang C.S., Tzeng S.R., Wang C., Cheng J.W. 2003. 1H, 15N and 13C resonance assignments of the tetratricopeptide repeat (TPR) domain of hSGT J. Biomol. NMR 26: 381–382. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. 2000. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine Cell 101: 199–210. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. 2003. SWISS-MODEL: An automated protein homology-modeling server Nucleic Acids Res. 31: 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinars C.R., Cheung-Flynn J., Rimerman R.A., Scammell J.G., Smith D.F., Clardy J. 2003. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes Proc. Natl. Acad. Sci. 100: 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P., Dornan J., Carrello A., Minchin R.F., Ratajczak T., Walkinshaw M.D. 2001. Two structures of cyclophilin 40: Folding and fidelity in the TPR domains Structure 9: 431–438. [DOI] [PubMed] [Google Scholar]
- Yang J., Roe S., Cliff M., Williams M., Ladbury J., Cohen P., Barford D. 2005. Molecular basis for TPR domain-mediated regulation of protein phosphatase 5 EMBO J. 24: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]