Abstract
An abundant enzyme of liver cytosol, 10-formyltetrahydrofolate dehydrogenase (FDH), is an interesting example of a multidomain protein. It consists of two functionally unrelated domains, an aldehyde dehydrogenase-homologous domain and a folate-binding hydrolase domain, which are connected by an ~100-residue linker. The amino-terminal hydrolase domain of FDH (Nt-FDH) is a homolog of formyl transferase enzymes that utilize 10-formyl-THF as a formyl donor. Interestingly, the concerted action of all three domains of FDH produces a new catalytic activity, NADP+-dependent oxidation of 10-formyltetrahydrofolate (10-formyl-THF) to THF and CO2. The present studies had two objectives: First, to explore the modular organization of FDH through the production of hybrid enzymes by domain replacement with methionyl-tRNA formyltransferase (FMT), an enzyme homologous to the hydrolase domain of FDH. The second was to explore the molecular basis for the distinct catalytic mechanisms of Nt-FDH and related 10-formyl-THF utilizing enzymes. Our studies revealed that FMT cannot substitute for the hydrolase domain of FDH in order to catalyze the dehydrogenase reaction. It is apparently due to inability of FMT to catalyze the hydrolysis of 10-formyl-THF in the absence of the cosubstrate of the transferase reaction despite the high similarity of the catalytic centers of the two enzymes. Our results further imply that Ile in place of Asn in the FDH hydrolase catalytic center is an important determinant for hydrolase catalysis as opposed to transferase catalysis.
Keywords: FDH, folate, domain replacement, hydrolase catalysis, transferase, multidomain enzymes
Domain organization is an intrinsic element of protein structure. The majority of proteins consist of distinctive domains that can act independently or cooperatively to achieve a unique function (Vogel et al. 2004; Bornberg-Bauer et al. 2005). The interaction between functional modules within a single polypeptide determines mechanisms by which multidomain proteins operate. An interesting example of a multidomain protein is the chimeric folate enzyme, 10-formyltetrahydrofolate dehydrogenase (FDH). FDH consists of two distinct catalytic domains, the amino-terminal and carboxy-terminal, connected by an intermediate linker (Fig. 1) (Cook et al. 1991; Krupenko et al. 1997a). The carboxy-terminal domain of FDH (Ct-FDH), an aldehyde dehydrogenase (ALDH) homolog (Cook et al. 1991), is capable of catalyzing an NADP+-dependent aldehyde dehydrogenase reaction utilizing short-chain aldehydes as a substrate (Fig. 1) (Krupenko et al. 1997b). The amino-terminal domain (Nt-FDH) is a homolog of the formyl transferase enzymes L-methionyl-tRNA formyltransferase (FMT) and glycinamide ribonucleotide formyltransferase (GARFT) (Cook et al. 1991). Similar to FDH, these enzymes utilize 10-formyltetrahydrofolate (10-formyl-THF) as a substrate. Nt-FDH, however, does not possess transferase activity but instead hydrolyzes 10-formyl-THF to yield formate and THF (Fig. 1) (Cook and Wagner 1995). The intermediate domain of FDH does not have significant homology with any known protein, nor does it have any known catalytic activity (Cook et al. 1991).
Figure 1.
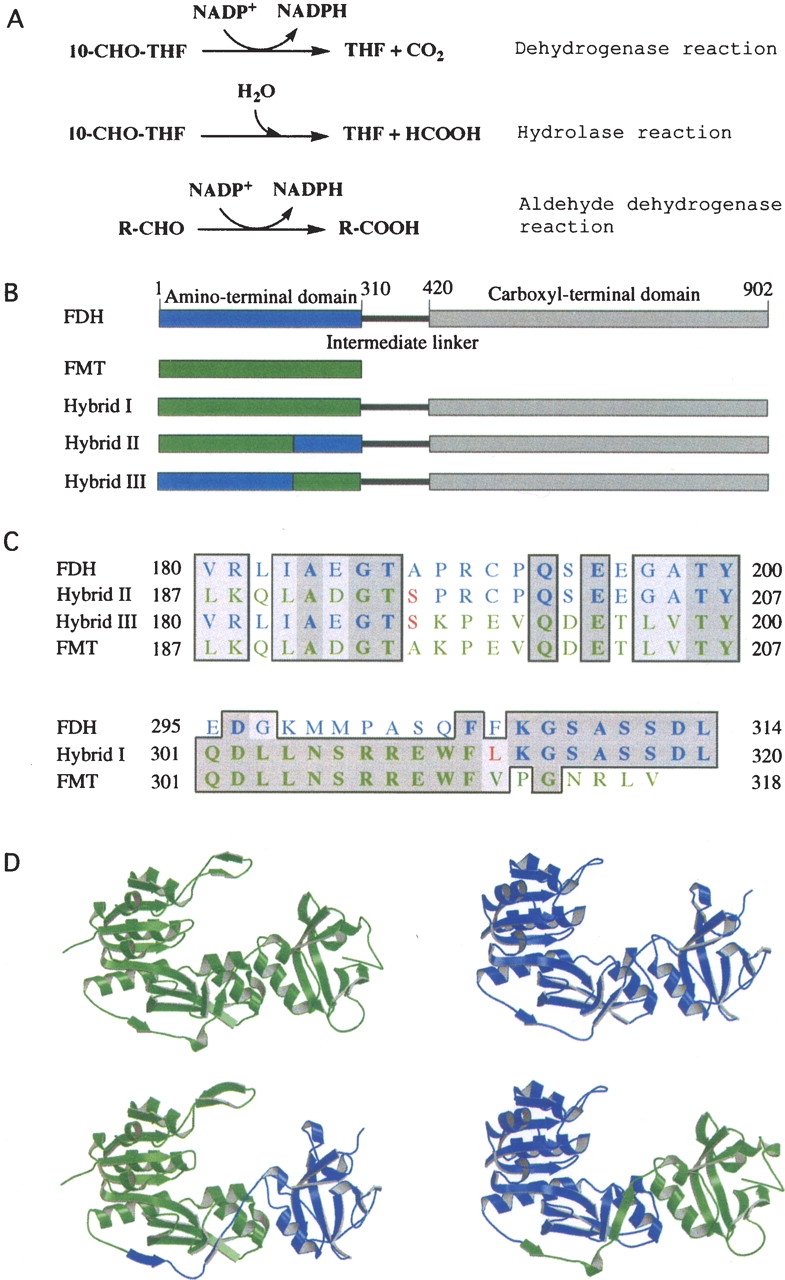
(A) Reactions catalyzed by FDH (the full-length FDH catalyzes all three reactions; the amino-terminal domain catalyzes the hydrolase reaction; the carboxy-terminal domain catalyzes the aldehyde dehydrogenase reaction). (B) Schematic diagram of FDH monomer (blue, the amino-terminal domain; gray, the carboxy-terminal domain; black, the intermediate linker) and FMT/FDH hybrid proteins, showing portions of Nt-FDH (blue) and FMT (green) within each hybrid. (C) Partial sequence alignments of FDH, FMT, and hybrid proteins showing the amino-terminal splice junction (top) and the carboxy-terminal splice junction (bottom). Dark-shaded residues are identical, whereas light-shaded residues are similar. Residues originating from FDH are shown in blue and those from FMT in green. Unique residues created by the insertion of restriction sites are in red. Note that hybrid III has a carboxy-terminal splice junction identical to that of hybrid I. The alignment was prepared using MacVector. (D) Ribbon presentation of crystal structures of FMT (upper left) (PDB ID 2FMT) and Nt-FDH (upper right) (PDB ID 1S3I) along with models of the Nt-FDH homologous region of hybrid II (lower left) and hybrid III (lower right). Hybrid models were prepared using SYBYL 7.0. FMT and Nt-FDH structures were superimposed, and relevant portions of each protein were merged together in a separate area. Hybrid structures were generated by creating bonds between the chains. The resulting hybrid structures were energy-minimized. Visual depictions were generated using MOLSCRIPT (Kraulis 1991).
The two catalytic domains of FDH, connected by an intermediate linker, work in concert to create a new enzymatic activity: The full-length FDH converts 10-formyl-THF to tetrahydrofolate (THF) in an NADP+-dependent dehydrogenase reaction in which the formyl group is oxidized to CO2 (Min et al. 1988). Of note, within the full-length FDH, both catalytic domains are still capable of independently catalyzing their intrinsic reactions (Cook et al. 1991). It is not known, however, whether these reactions have any physiological significance. On the other hand, the main FDH activity, which requires the concerted action of both catalytic domains, can have a strong impact on the cell. Recent studies from this laboratory have shown that expression of FDH in FDH-deficient cancer cells inhibits proliferation, leading to apoptosis (Krupenko and Oleinik 2002; Oleinik and Krupenko 2003). Importantly, these effects can be produced only by catalytically functional, full-length FDH (Krupenko and Oleinik 2002). Therefore, understanding how the two domains act together to create a new enzymatic mechanism is crucial for understanding the biological role of FDH.
The shuffling of pre-existing functional modules is considered to be a common mechanism of protein evolution (Patthy 2003), and using domain replacement to produce functional hybrids has become an important approach for studying multidomain proteins in recent years (Bhatt et al. 2004; Sasata et al. 2004; Fan et al. 2005; Lu et al. 2005; Suo 2005). Such studies have been done for the Nt-FDH homologous enzyme, GARFT (Nixon et al. 1997; Nixon and Benkovic 2000; Lee et al. 2003). GARFT is ~200 amino acid residues long and consists of two domains, a folate-binding amino-terminal domain and a GAR-binding carboxy-terminal domain (Greasley et al. 1999). Domain replacement studies have demonstrated that functional hybrids containing GARFT activity can be produced from the GAR-binding domain and the folate-binding domain originated from other 10-formyl-THF utilizing enzymes (Nixon et al. 1997; Nixon and Benkovic 2000; Lee et al. 2003). This implies that individual modules (which can be considered subdomains) of GARFT have at least some functional independence.
The recently solved crystal structure of Nt-FDH revealed that its overall fold and architecture of the catalytic center are remarkably similar to those of FMT or GARFT (Chumanevich et al. 2004). Specifically, the two catalytic residues whose importance has been established for all three enzymes (Inglese et al. 1990; Warren et al. 1996; Krupenko and Wagner 1999; Newton and Mangroo 1999; Shim and Benkovic 1999; Krupenko et al. 2001), aspartate and histidine, overlay very closely in these proteins (Chumanevich et al. 2004). This suggests a similar catalytic mechanism for removing the formyl group from the folate substrate. Nt-FDH, however, is missing an asparagine in position 104, a crucial residue in FMT and GARFT catalysis (Newton and Mangroo 1999; Shim and Benkovic 1999), and instead contains an isoleucine at this position. When compared to GARFT, Nt-FDH and FMT are extended at the carboxyl terminus by ~100 residues (Krupenko and Wagner 1998). This part of their respective sequences represents a separate domain, which in the case of FMT binds methionyl-tRNA (Schmitt et al. 1996). In spite of low sequence identity, these carboxy-terminal regions of FMT and Nt-FDH have surprisingly similar structures (Chumanevich et al. 2004). There is no known function for this region in Nt-FDH, however. We have undertaken the present study to understand the modular nature of FDH through the generation of hybrid enzymes by domain replacement with FMT, and to explore the molecular basis for the distinct catalytic mechanisms of Nt-FDH and related 10-formyl-THF utilizing enzymes.
Results
Construction and activity of FMT/FDH hybrid proteins
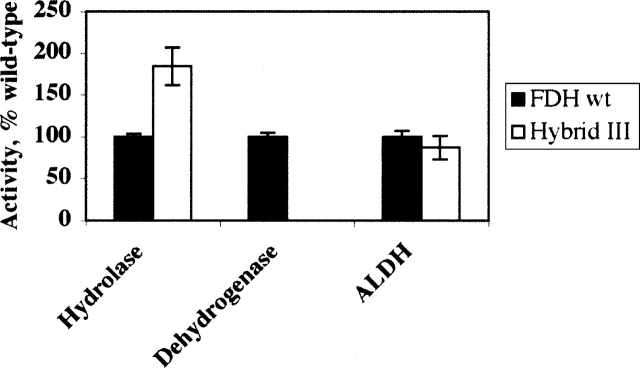
We have expressed and analyzed the catalytic activities of three hybrid FDH enzymes (Fig. 1B). In the first hybrid (hybrid I) the entire amino-terminal hydrolase domain of FDH (Nt-FDH) was replaced with FMT. In the other two hybrids, two subdomains of Nt-FDH, the folate-binding catalytic domain (hybrid II) and the domain equivalent to the methionyl-tRNA-binding domain of FMT (hybrid III), were each substituted with corresponding domains of FMT. The beginning of the linker region (at residue 195) between the amino- and carboxy-terminal domains of FMT was used as the junction point within each hybrid (Fig. 1C). This residue was chosen because it is not a part of ordered secondary structure and, unlike other residues in the linker region, is not conserved among FMT orthologs (Schmitt et al. 1996; Gite et al. 2000). All hybrids were expressed as soluble proteins in insect cells using a baculovirus expression system and were purified similar to wild-type FDH enzymes. The purified hybrid enzymes possessed aldehyde dehydrogenase activity similar to those of the wild-type FDH (data not shown), suggesting that the carboxy-terminal ALDH-homologous domain was properly folded within each hybrid protein. None of the hybrids displayed detectable 10-formyl-THF dehydrogenase activity. Only one of the hybrids (hybrid III) revealed hydrolase activity (Fig. 2) with the Km of 7.5 μM and Vmax of 145 nmol/min per mg (the corresponding parameters for the wild-type FDH are 5.8 μM and 85 nmol/min per mg). These studies demonstrated that the catalytic center of Nt-FDH and the carboxy-terminal domain of FMT (the methionyl-tRNA-binding subdomain) can be assembled into a functional hydrolase.
Figure 2.
Activity of hybrid III as compared to the wild-type FDH. Error bars represent standard error of at least three measurements.
Determination of the size of the functional hydrolase domain
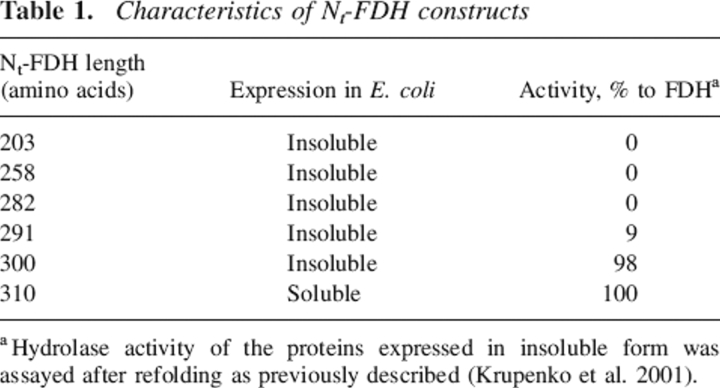
We have earlier reported the expression of two different constructs of the amino-terminal domain of FDH (Krupenko et al. 1997a). Two peptides of different length were expressed: one containing the first 203 amino acids from the amino terminus corresponding to the length of GART, and the other containing the first 310 amino acids corresponding to the length of FMT. The 203–amino acid long protein was insoluble, suggesting that the protein was not folded properly and was not purified and characterized. In contrast, the expressed 310–amino acid N-terminal protein (Nt-FDH) was soluble and displayed 10-formyl-THF hydrolase activity, suggesting proper folding. To determine the minimal length of an amino-terminal peptide (between 203 and 310 residues) that would be able to catalyze the hydrolase reaction, we expressed and evaluated proteins of 258, 282, 291, and 300 amino acid residues in length. The choice of size of the proteins was arbitrary. We observed that all proteins were expressed in Escherichia coli in insoluble form. After dissolving in 6 M urea and refolding by stepwise dialysis, ~90% of each protein was recovered as soluble material. Only the 300-residue-long protein produced full hydrolase activity similar to Nt-FDH and full-length FDH (Table 1). The 291-residue-long protein had ~9% activity of the fully functional hydrolase domain. The two other proteins (258 and 282 amino acids long) were completely inactive. These results demonstrated that ~300 residues of the FDH amino terminus are necessary to produce full hydrolase activity.
Table 1.
Characteristics of Nt-FDH constructs
aHydrolase activity of the proteins expressed in insoluble form was assayed after refolding as previously described (Krupenko et al. 2001).
FMT does not possess hydrolase activity toward the folate substrate
Because the FMT/FDH hybrid protein (hybrid I; Fig. 1) did not reveal 10-formyl-THF hydrolase activity, we expressed FMT separately to test whether the enzyme itself possesses this activity. FMT having a 6xHis tag in the amino terminus was expressed in E. coli as a soluble protein and was purified using metal-affinity chromatography as described in Materials and Methods. We observed that the purified recombinant FMT did not produce a detectable level of hydrolase activity.
Site-directed mutagenesis of residues in the FDH hydrolase catalytic center
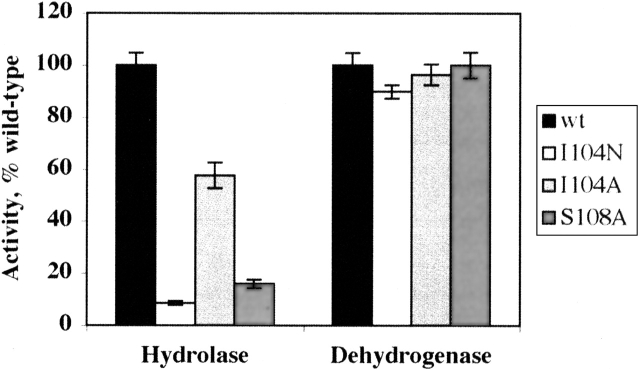
We studied the role of two residues located in the hydrolase catalytic center of FDH, Ile104 and Ser108 (Fig. 3A). The first residue, I104, was selected because it substitutes for the catalytically essential asparagine found in other 10-formyl-THF utilizing enzymes. We therefore suggested that this could differentiate the Nt-FDH hydrolase mechanism from the transferase mechanism of the homologous enzymes. Replacement of Ile104 with alanine resulted in significantly decreased hydrolase activity of the full-length FDH (by nearly 50%) (Fig. 4). Replacement of this residue by asparagine, the catalytic residue found in GARFT and FMT, resulted in even stronger suppression of hydrolase activity, to <10% of the wild-type enzyme. For both mutants, dehydrogenase and aldehyde dehydrogenase activities were similar to those of the wild-type enzyme. Similar results for hydrolase activity were obtained for corresponding mutants of Nt-FDH expressed separately (Table 2).
Figure 3.
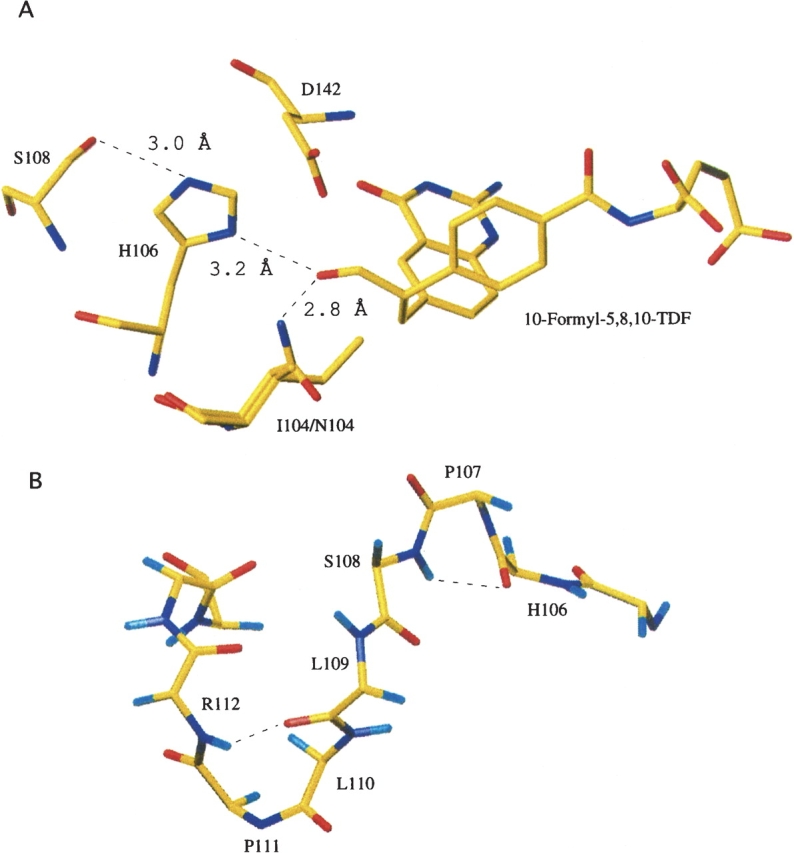
(A) Spatial arrangement of essential catalytic residues and folate substrate within the Nt-FDH active site. This figure was generated by superimposing the structure of Nt-FDH (PDB ID 1S3I) with that of GARFT complexed with the inhibitor 10-formyl-5,8,10-trideazafolate (PDB ID 1C2T). Superimposed isoleucine (wild-type enzyme) and asparagine (I104N mutant) are shown at position 104. (B) Backbone of Nt-FDH showing the SLLP motif. Hydrogen bonds are shown as dashed lines.
Figure 4.
Activities of full-length FDH mutants expressed as a percentage of wild type. Error bars represent standard error of at least three measurements.
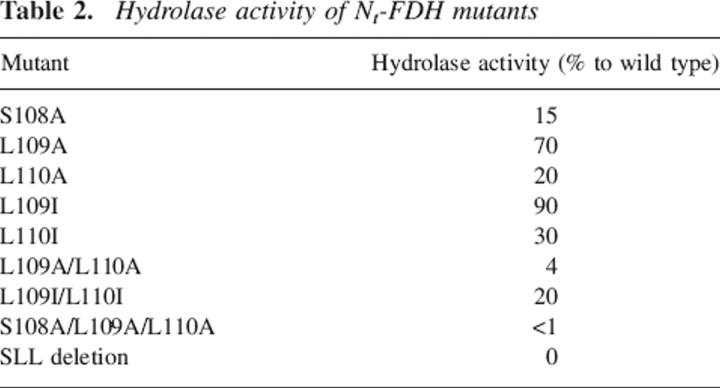
Table 2.
Hydrolase activity of Nt-FDH mutants
The second residue, Ser108, is a part of an SLLP motif that is proposed to be a 10-formyl-THF binding element (Cook et al. 1991; Schmitt et al. 1996). The region between H106 and G115 forms a loop containing two tight turns between two β-strands (β5 and β6) (see Fig. 3B). Pro107 is the i + 1 residue in a γ turn between the catalytic H106 and S108, with the carbonyl oxygen of H106 hydrogen bonding to the amide proton of S108 (Fig. 3B). The crystal structure of Nt-FDH (Chumanevich et al. 2004) revealed that the side chain of S108 is positioned close to the catalytic histidine and can assist in the rearrangement of positions of its electrons during catalysis (see Fig. 3A). Mutation of S108 to alanine caused a sharp reduction in hydrolase activity of both the full-length FDH and the hydrolase domain expressed as a separate protein (Fig. 4), suggesting involvement of this residue in hydrolase catalysis. However, the mutation had no effect on the 10-formyl-THF dehydrogenase activity of FDH (Fig. 4), implying a dehydrogenase mechanism independent from the hydrolase mechanism. As would be expected, there was no effect of this substitution on aldehyde dehydrogenase activity (data not shown). In contrast to the serine, the two downstream leucine residues of the putative folate-binding motif are less crucial for hydrolase catalysis (Table 2). Although the replacement of either of these residues decreases activity, with additive effects for double mutants, mutations to hydrophobic isoleucine had less of an effect than mutations to alanine. This suggests the importance of these residues for supporting an overall hydrophobic environment within the catalytic site pocket.
Discussion
Since the 10-formyl-THF utilizing enzymes, GARFT, FMT, and FDH, have homologous folate binding sites and very similar folds, we have suggested that some steps in the catalytic mechanisms of all three enzymes should be similar. Specifically, we expected that the initial step in catalysis, which is apparently the breaking of the bond between the folate coenzyme and the formyl group, should proceed through similar mechanisms. In the case of GARFT and FMT, the formyl is transferred to a second substrate, while in the case of FDH, it is transferred to another catalytic domain to be further converted to CO2. In the absence of the second catalytic domain, the formyl group is released as formate, which is the end-product of a hydrolase reaction catalyzed by the amino-terminal domain of FDH. The reaction can be thought of as a transferase reaction in which the formyl group is transferred to a water molecule rather than a separately bound substrate. The presence of conserved catalytic residues in the active sites of all three enzymes (Inglese et al. 1990; Warren et al. 1996; Krupenko and Wagner 1999; Newton and Mangroo 1999; Shim and Benkovic 1999; Krupenko et al. 2001) further supports the hypothesis of a similar catalytic mechanism.
Interestingly, substituting the amino-terminal domain of FDH with FMT resulted in an enzyme with no 10-formyl-THF dehydrogenase activity, suggesting that FMT cannot functionally substitute for the amino-terminal domain of FDH. In contrast, studies on GARFT have demonstrated that substitution of the catalytic domain of GARFT with the corresponding domains of 10-formyl-THF utilizing enzymes results in functional hybrids possessing GAR-formyltransferase activity (Nixon et al. 1997; Lee et al. 2003). Both FMT and Nt-FDH consist of two domains: a catalytic folate-binding domain and a carboxy-terminal domain (Chumanevich et al. 2004). While in FMT this latter domain binds the methionyl-tRNA substrate, which is the acceptor of the formyl group (Gite et al. 2000), the function of this domain in Nt-FDH is unclear. Generally, it would be expected that Nt-FDH of a size similar to GARFT (~200 amino acid residues) could produce the 10-formyl-THF hydrolase activity, since GARFT itself can catalyze a more chemically complex reaction. Our studies, however, have demonstrated that the extension of the folate-binding domain by ~100 residues is required for the hydrolase catalysis, suggesting the functional involvement of this part of the molecule in the hydrolase mechanism.
Taking into account the modular organization of FDH, there are several possibilities why the FMT/FDH hybrid was inactive. One is that the catalytic mechanisms of Nt-FDH and FMT related to the removal of the formyl group from the folate substrate are different, and therefore the overall FDH mechanism is not functional with FMT substitution. Another possibility is that, although the FMT catalytic center is functionally capable of substituting for the Nt-FDH hydrolase mechanism in the overall FDH catalysis, the interface between the folate binding site and the aldehyde dehydrogenase catalytic domain was distorted due to misorientation of the two catalytic domains. Structural comparisons of the C-terminal domains of FMT and Nt-FDH (Chumanevich et al. 2004) indicate that the carboxyl termini of each enzyme are oriented differently. FMT contains a carboxy-terminal β-strand that directs the remaining residues back in the general direction of the amino-terminal domain (Schmitt et al. 1996). Nt-FDH, however, is topologically different. The structurally equivalent β-strand appears earlier in the domain, leaving a helix as the last secondary structural element (Chumanevich et al. 2004). This results in the remaining carboxy-terminal residues pointing away from the amino-terminal region rather than toward it (Fig. 1D). Unless the movement of the carboxyl termini in both proteins is relatively unrestricted, this difference could result in a hybrid protein in which the FMT catalytic domain is oriented incorrectly with respect to the aldehyde dehydrogenase domain of FDH.
Apparently, both the inability of FMT to catalyze the hydrolysis of 10-formyl-THF in the absence of a second substrate (methionyl-tRNA), and the mispositioning of the catalytic domain relative to the ALDH-like domain of FDH resulted in a nonactive FMT/FDH hybrid. Indeed, assays of recombinant E. coli FMT revealed that this enzyme does not possess hydrolase activity toward 10-formyl-THF substrate. Likewise, the FMT/FDH hybrid bearing the FMT folate binding site and catalytic center (the first 194 residues from the amino terminus; see hybrid II in Fig. 1) was not capable of the hydrolase catalysis. Interestingly, the other hybrid, containing the carboxy-terminal portion of FMT (residues 196–311; hybrid III in Fig. 1), revealed strong hydrolase activity, demonstrating that the carboxy-terminal domain of FMT is capable of substituting for the corresponding domain of Nt-FDH to generate the hydrolase activity. This hybrid, however, does not catalyze the dehydrogenase reaction, most likely due to incorrect positioning of this hybrid domain relative to the ALDH domain of FDH.
In general, these results showed an intrinsic inability of FMT to catalyze the 10-formyl-THF hydrolase reaction. This in turn implies a fundamental difference between the catalytic centers of Nt-FDH and FMT, but not between their carboxy-terminal subdomains (with regard to hydrolase catalysis). Two critical catalytic residues of Nt-FDH (His106 and Asp142) are homologous to catalytic residues in FMT. However, FMT, as well as GARFT, has a third catalytic residue, an asparagine located two positions upstream of the catalytic histidine. This residue was proposed to act by stabilizing the formyl oxygen of 10-formyl-THF along with the protonated active site histidine, thus enabling their respective substrates to conduct a nucleophilic attack on the formyl carbon (Newton and Mangroo 1999; Shim and Benkovic 1999). FDH contains an isoleucine at this position, which would be incapable of performing the same role (Fig. 3A). Interestingly, the hydrolase activity is also lacking in GARFT (Lee et al. 2003). Since asparagine should help stabilize an oxy-anion intermediate, one might assume that replacing isoleucine with asparagine could enhance FDH hydrolase activity. Surprisingly, our studies have shown that this is not the case: Asparagine is particularly unsuited for hydrolase activity, suggesting instead that the presence of this residue in GARFT and FMT could prevent unwanted hydrolysis of the folate substrate.
Despite the inability of FMT to catalyze the hydrolase reaction, Nt-FDH and FMT catalytic centers have rather more similarities than differences. For example, immediately downstream of the catalytic histidine is an SLLP motif, which is strictly conserved among 10-formyl-THF utilizing enzymes (Schmitt et al. 1996). Although this motif is rather an evolutionary conserved element of the overall active site architecture, the serine residue is essential for the hydrolase catalysis. It is likely that this serine forms a hydrogen bond to the catalytic H106 resulting in rearrangement of the electron density of the imidazole ring. Thus, the combination of His-Ser-Asp in the Nt-FDH active site could be a kind of modified catalytic triad found in serine proteases and other families of enzymes (Dodson and Wlodawer 1998; Polgar 2005). Interestingly, similar to I104, this serine is required for full hydrolase activity but not for full dehydrogenase activity. We have previously suggested that FDH hydrolase catalysis is a part of the overall dehydrogenase mechanism and does not have an independent function. Although in general this hypothesis is likely to be correct, our present studies also imply that there could be a part of the hydrolase mechanism that is not a component of dehydrogenase catalysis. We suggest that residues in the hydrolase catalytic center, which do not participate in dehydrogenase catalysis, are required for the final step of the hydrolase reaction, which releases the product in the form of formate. This function is apparently inhibited by the presence of the ALDH domain of FDH that diverts catalysis toward the transfer of an intermediate to the second catalytic domain rather than releases it as a formate. This also suggests that the hydrolase reaction might proceed when the two functional domains are uncoupled. It is not clear at present whether such a hydrolysis occurs in the cell and what could be the physiological role of this reaction.
Materials and methods
Reagents
10-Formyl-5,8-dideazafolate (10-formyl-DDF) was obtained from Dr. John B. Hynes (Department of Pharmaceutical Chemistry, Medical University of South Carolina). Restriction enzymes were purchased from New England BioLabs. Agarose for preparative and analytical electrophoresis was obtained from Bio-Rad. Grace's Insect cell media was purchased from Invitrogen. Fetal bovine serum was purchased from Atlanta Biologicals. All other chemicals were obtained from Sigma.
Generation of expression constructs
E. coli FMT cDNA from pQE-16 FMTp vector (Ramesh et al. 1997) (a gift of Dr. Uttam RajBhandary, Department of Biology, Massachusetts Institute of Technology) was amplified by PCR, inserted into pCR2.1 vector using TA Cloning Kit (Invitrogen), and then recloned into pRSET expression vector through NdeI/EcoRI restriction sites. The primers for amplification were 5′-GAGGAGAAATTACATATGAGAGGATCCTC-3′ and 5′-GTCCAAGCTCAGCGAATTCAGCTTAGTG-3′ (restriction sites are shown in boldface). The resulting cDNA had an additional sequence on the 3′ end coding for a six-residue His tag.
To generate hybrids between FDH and FMT, the sequences coding for the corresponding domain of FMT or for the full-length FMT were amplified by PCR using pQE-16 FMTp plasmid as a template and were cloned into pCR2.1 vector. Each PCR primer was designed to have a restriction site to allow excision of the cloned fragment from pCR2.1 vector (Table 3). The following combinations of primers were used: primers 1 and 2 (for amplification of the sequence coding for the full-length FMT), primers 1 and 3 (for amplification of the sequence coding for the amino-terminal domain of FMT, residues 1–194), and primers 2 and 4 (for amplification of the sequence coding for the carboxy-terminal domain of FMT, residues 196–311). AflII and SpeI restriction sites were introduced to the FDH coding sequence, cloned into the pVL1393 expression vector (Krupenko et al. 1995a) by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene). These two sites, together with the originally present XbaI site, were unique in the pVL1393/FDH vector. Sequences encoding for Nt-FDH or its amino- or carboxy-terminal domains were excised using respective restriction sites and were replaced, using the same sites, with corresponding sequences encoding for FMT or its domains excised from the pCR2.1 vector. Each of the resulting constructs was confirmed by sequencing the entire vector.
Table 3.
Primers used for the construction of hybrid proteins
aRestriction sites are shown in boldface.
Expression and purification of full-length recombinant proteins
The expression of full-length FDH protein, including FDH/FMT hybrids, was carried out in High Five insect cells using recombinant baculovirus as described previously (Krupenko et al. 1995). The pVL1393 vector containing FDH cDNA was cotransfected with linearized AcNPV baculovirus into Sf9 cells using the BaculoGold transfection kit (BD Pharmingen) according to the manufacturer's protocol. Recombinant virus was amplified to high titer by successive rounds of infection of Sf9 cells. For protein expression, High Five cells were seeded as a monolayer in 185 cm2 flasks and allowed to grow to ~80% confluence. Each flask was infected with 1.0 mL of high titer virus. Cells and media were collected ~5 d post-infection and spun down. FDH was purified from protein secreted into the media. The media were dialyzed overnight against buffer (20 mM Tris-HCl at pH 7.4) containing 0.02% NaN3. The dialyzed media were subjected to a two-step purification process using a Sepharose 5-formyl-THF affinity column and a Sephacryl S-300 size-exclusion column as described previously (Krupenko et al. 1995; Reuland et al. 2003).
Expression and purification of FMT and Nt-FDH mutants
E. coli BL21(DE3) cells (Strategene) were transformed, according to the manufacturer's protocol, with pRSET vector containing cDNA representing the first 310 amino acids of FDH or with the FMT construct described above. The cells were grown in 4 mL of NZCYM medium containing ampicillin (50 μg/mL) overnight at 37°C with shaking. One hundred milliliters of NZCYM medium containing ampicillin were inoculated with the overnight culture and incubated at 37°C with shaking until A600 = 0.6 (~6 h) followed by induction with isopropyl-β-D-thiogalactopyranoside (1 mM final concentration). Three hours after induction, the cells were pelleted by centrifugation (5000g, 10 min), resuspended in 2 mL of buffer (50 mM Tris-HCl at pH 8.0, 0.2 mg/mL lysozyme, and 0.1% Triton X-100), and sonicated (three times for 45 s). Recombinant proteins were purified from the soluble portion of the cell lysate after removal of insoluble material by centrifugation (18,000g, 15 min). Nt-FDH was purified in one step using DEAE ion-exchange chromatography as described previously (Krupenko and Wagner 1998). FMT was purified by one-step metal-affinity chromatography using the HisTrap system (Pharmacia).
Assays of enzyme activity
All assays were performed at 30°C in a Shimadzu 2401PC double-beam spectrophotometer. For measurement of hydrolase activity the reaction mixture contained 0.05 M Tris-HCl (pH 7.8), 100 mM 2-mercaptoethanol, and 5 μM of substrate, 10-formyl-DDF. 10-Formyl-DDF is an alternative, stable substrate for the enzyme (Krupenko et al. 1995b). The reaction was started by the addition of enzyme (1–20 μg) in a final volume of 1.0 mL and read against a blank cuvette containing all components except enzyme. Appearance of product 5,8-dideazafolate was measured at 295 nm using a molar extinction coefficient of 18.9 × 103 (Smith et al. 1981). Addition of NADP+ to the reaction mixture provided a measure of both dehydrogenase and hydrolase activities, i.e., total activity of the enzyme. Hydrolase activity measured in the absence of NADP+ was subtracted from the total activity to give the dehydrogenase activity. Dehydrogenase activity was also measured independently using the increase in absorbance at 340 nm due to production of NADPH and the molar extinction coefficient of 6.2 × 103. Aldehyde dehydrogenase activity was assayed using propanal as described previously (Krupenko et al. 1997b). The reaction mixture contained 50 mM CHES buffer (pH 9.4), 5 mM propanal, 1 mM NADP+, and enzyme in a total volume of 1 mL. Activity was estimated from the increase in absorbance at 340 nm.
Acknowledgments
We thank Drs. Christopher Davies and Natalia Krupenko for carefully reading the manuscript and for their helpful suggestions, and Dr. Uttam RajBhandary for providing pQE-16 FMTp vector. This work was supported by NIH grant DK54388 (S.A.K.) and by a fellowship from the Abney Foundation (S.N.R.).
Footnotes
Reprint requests to: Sergey A. Krupenko, Department of Biochemistry and Molecular Biology, Medical University of South Carolina, 173 Ashley Avenue, Room 512-B BSB, Charleston, SC 29425, USA; e-mail: krupenko@musc.edu; fax: (843) 792-8565.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.052062806.
Abbreviations: ALDH, aldehyde dehydrogenase; FDH, 10-formyltetrahydrofolate dehydrogenase; FMT, L-methionyl-tRNA formyltransferase; GARFT, glycinamide ribonucleotide formyltransferase; THF, tetrahydrofolate.
References
- Bhatt A.N., Khan M.Y., Bhakuni V. 2004. The C-terminal domain of dimeric serine hydroxymethyltransferase plays a key role in stabilization of the quaternary structure and cooperative unfolding of protein: Domain swapping studies with enzymes having high sequence identity Protein Sci. 13: 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E., Beaussart F., Kummerfeld S.K., Teichmann S.A., Weiner J. 3rd. 2005. The evolution of domain arrangements in proteins and interaction networks Cell. Mol. Life Sci. 62: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumanevich A.A., Krupenko S.A., Davies C. 2004. The crystal structure of the hydrolase domain of 10-formyltetrahydrofolate dehydrogenase: Mechanism of hydrolysis and its interplay with the dehydrogenase domain J. Biol. Chem. 279: 14355–14364. [DOI] [PubMed] [Google Scholar]
- Cook R.J. and Wagner C. 1995. Enzymatic activities of rat liver cytosol 10-formyltetrahydrofolate dehydrogenase Arch. Biochem. Biophys. 321: 336–344. [DOI] [PubMed] [Google Scholar]
- Cook R.J., Lloyd R.S., Wagner C. 1991. Isolation and characterization of cDNA clones for rat liver 10-formyltetrahydrofolate dehydrogenase J. Biol. Chem. 266: 4965–4973. [PubMed] [Google Scholar]
- Dodson G. and Wlodawer A. 1998. Catalytic triads and their relatives Trends Biochem. Sci. 23: 347–352. [DOI] [PubMed] [Google Scholar]
- Fan H.Y., Trotter K.W., Archer T.K., Kingston R.E. 2005. Swapping function of two chromatin remodeling complexes Mol. Cell 17: 805–815. [DOI] [PubMed] [Google Scholar]
- Gite S., Li Y., Ramesh V., Rajbhandary U.L. 2000. Escherichia coli methionyl-tRNA formyltransferase: Role of amino acids conserved in the linker region and in the C-terminal domain on the specific recognition of the initiator tRNA Biochemistry 39: 2218–2226. [DOI] [PubMed] [Google Scholar]
- Greasley S.E., Yamashita M.M., Cai H., Benkovic S.J., Boger D.L., Wilson I.A. 1999. New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with β-GAR and 10-formyl-5,8,10-trideazafolic acid Biochemistry 38: 16783–16793. [DOI] [PubMed] [Google Scholar]
- Inglese J., Smith J.M., Benkovic S.J. 1990. Active-site mapping and site-specific mutagenesis of glycinamide ribonucleotide transformylase from Escherichia coli Biochemistry 29: 6678–6687. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures J. Appl. Crystallogr. 24: 946–950. [Google Scholar]
- Krupenko S.A. and Oleinik N.V. 2002. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells Cell Growth Differ. 13: 227–236. [PubMed] [Google Scholar]
- Krupenko S.A. and Wagner C. 1998. Overexpression of functional hydrolase domain of rat liver 10-formyltetrahydrofolate dehydrogenase in Escherichia coli Protein Expr. Purif. 14: 146–152. [DOI] [PubMed] [Google Scholar]
- Krupenko S.A. and Wagner C. 1999. Aspartate 142 is involved in both hydrolase and dehydrogenase catalytic centers of 10-formyltetrahydrofolate dehydrogenase J. Biol. Chem. 274: 35777–35784. [DOI] [PubMed] [Google Scholar]
- Krupenko S.A., Horstman D.A., Wagner C., Cook R.J. 1995a. Baculovirus expression and purification of rat 10-formyltetrahydrofolate dehydrogenase Protein Expr. Purif. 6: 457–464. [DOI] [PubMed] [Google Scholar]
- Krupenko S.A., Wagner C., Cook R.J. 1995b. Recombinant 10-formyltetrahydrofolate dehydrogenase catalyses both dehydrogenase and hydrolase reactions utilizing the synthetic substrate 10-formyl-5,8-dideazafolate Biochem. J. 306: 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupenko S.A., Wagner C., Cook R.J. 1997a. Domain structure of rat 10-formyltetrahydrofolate dehydrogenase. Resolution of the amino-terminal domain as 10-formyltetrahydrofolate hydrolase J. Biol. Chem. 272: 10273–10278. [DOI] [PubMed] [Google Scholar]
- Krupenko S.A., Wagner C., Cook R.J. 1997b. Expression, purification, and properties of the aldehyde dehydrogenase homologous carboxy-terminal domain of rat 10-formyltetrahydrofolate dehydrogenase J. Biol. Chem. 272: 10266–10272. [DOI] [PubMed] [Google Scholar]
- Krupenko S.A., Vlasov A.P., Wagner C. 2001. On the role of conserved histidine 106 in 10-formyltetrahydrofolate dehydrogenase catalysis: Connection between hydrolase and dehydrogenase mechanisms J. Biol. Chem. 276: 24030–24037. [DOI] [PubMed] [Google Scholar]
- Lee S.G., Lutz S., Benkovic S.J. 2003. On the structural and functional modularity of glycinamide ribonucleotide formyltransferases Protein Sci. 12: 2206–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.J., White S.W., Rock C.O. 2005. Domain swapping between Enterococcus faecalis FabN and FabZ proteins localizes the structural determinants for isomerase activity J. Biol. Chem. 280: 30342–30348. [DOI] [PubMed] [Google Scholar]
- Min H., Shane B., Stokstad E.L. 1988. Identification of 10-formyltetrahydrofolate dehydrogenase-hydrolase as a major folate binding protein in liver cytosol Biochim. Biophys. Acta 967: 348–353. [DOI] [PubMed] [Google Scholar]
- Newton D.T. and Mangroo D. 1999. Mapping the active site of the Haemophilus influenzae methionyl-tRNA formyltransferase: Residues important for catalysis and tRNA binding Biochem. J. 339: 63–69. [PMC free article] [PubMed] [Google Scholar]
- Nixon A.E. and Benkovic S.J. 2000. Improvement in the efficiency of formyl transfer of a GAR transformylase hybrid enzyme Protein Eng. 13: 323–327. [DOI] [PubMed] [Google Scholar]
- Nixon A.E., Warren M.S., Benkovic S.J. 1997. Assembly of an active enzyme by the linkage of two protein modules Proc. Natl. Acad. Sci. 94: 1069–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleinik N.V. and Krupenko S.A. 2003. Ectopic expression of 10-formyltetrahydrofolate dehydrogenase in a549 cells induces g(1) cell cycle arrest and apoptosis Mol. Cancer Res. 1: 577–588. [PubMed] [Google Scholar]
- Patthy L. 2003. Modular assembly of genes and the evolution of new functions Genetica 118: 217–231. [PubMed] [Google Scholar]
- Polgar L. 2005. The catalytic triad of serine peptidases Cell. Mol. Life Sci. 62: 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh V., Gite S., Li Y., Rajbhandary U.L. 1997. Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: Role of a 16-amino acid insertion module in initiator tRNA recognition Proc. Natl. Acad. Sci. 94: 13524–13529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuland S.N., Vlasov A.P., Krupenko S.A. 2003. Disruption of a calmodulin central helix-like region of 10-formyltetrahydrofolate dehydrogenase impairs its dehydrogenase activity by uncoupling the functional domains J. Biol. Chem. 278: 22894–22900. [DOI] [PubMed] [Google Scholar]
- Sasata R.J., Reed D.W., Loewen M.C., Covello P.S. 2004. Domain swapping localizes the structural determinants of regioselectivity in membrane-bound fatty acid desaturases of Caenorhabditis elegans J. Biol. Chem. 279: 39296–39302. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Blanquet S., Mechulam Y. 1996. Structure of crystalline Escherichia coli methionyl-tRNA(f)Met formyltransferase: Comparison with glycinamide ribonucleotide formyltransferase EMBO J. 15: 4749–4758. [PMC free article] [PubMed] [Google Scholar]
- Shim J.H. and Benkovic S.J. 1999. Catalytic mechanism of Escherichia coli glycinamide ribonucleotide transformylase probed by site-directed mutagenesis and pH-dependent studies Biochemistry 38: 10024–10031. [DOI] [PubMed] [Google Scholar]
- Smith G.K., Mueller W.T., Benkovic P.A., Benkovic S.J. 1981. On the cofactor specificity of glycinamide ribonucleotide and 5-aminoimidazole-4-carboxamide ribonucleotide transformylase from chicken liver Biochemistry 20: 1241–1245. [DOI] [PubMed] [Google Scholar]
- Suo Z. 2005. Thioesterase portability and peptidyl carrier protein swapping in yersiniabactin synthetase from Yersinia pestis Biochemistry 44: 4926–4938. [DOI] [PubMed] [Google Scholar]
- Vogel C., Bashton M., Kerrison N.D., Chothia C., Teichmann S.A., Gokhale R.S., Khosla C. 2004. Structure, function and evolution of multidomain proteins Curr. Opin. Struct. Biol. 14: 208–216. [DOI] [PubMed] [Google Scholar]
- Warren M.S., Marolewski A.E., Benkovic S.J. 1996. A rapid screen of active site mutants in glycinamide ribonucleotide transformylase Biochemistry 35: 8855–8862. [DOI] [PubMed] [Google Scholar]