Abstract
Cyanobacteria, such as Anabaena, produce a variety of bioactive natural products via polyketide synthases (PKS), nonribosomal peptide synthetases (NRPS), and hybrid peptide/polyketide pathways. The protein Asl1650, which is a member of the acyl carrier protein family from the cyanobacterium Anabaena sp. PCC 7120, is encoded in a region of the Anabaena genome that is rich in PKS and NRPS genes. To gain new insight into the physiological role of acyl carriers in Anabaena, the solution structure of Asl1650 has been solved by NMR spectroscopy. The protein adopts a twisted antiparallel four-helix bundle fold, with a variant phosphopantetheine-attachment motif positioned at the start of the second helix. Structure comparisons with proteins from other organisms suggest a likely physiological function as a discrete peptidyl carrier protein.
Keywords: NMR structure determination, acyl carrier protein, peptidyl carrier protein, polyketide synthases, nonribosomal peptide synthetases, cyanobacteria
Polyketides and nonribosomally synthesized peptides form two large groups of natural products synthesized by microbes as secondary metabolites. These natural products include compounds with antibiotic, antifungal, immunosuppressant, and anticancer activities (O'Hagan 1991; Carreras et al. 1997; Cane and Walsh 1999; Sankawa 1999; Finking and Marahiel 2004). The actinomycetes (Omura et al. 2001; Bentley et al. 2002) and the filamentous cyanobacteria (Dittmann et al. 2001; Hoffmann et al. 2003) are particularly rich producers of these compounds, and the genome of the filamentous, heterocyst-forming cyanobacterium Anabaena sp. PCC 7120 contains multiple polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) genes (Nakamura et al. 1998; Kaneko et al. 2001).
Asl1650 is an acyl carrier protein encoded in a region of the Anabaena genome that is particularly rich in PKS and NRPS genes, and the surrounding genes encode enzymes with sequence homology with both PKS and NRPS enzymes. Modular type I PKS systems are large, multifunctional proteins with multiple enzyme activities present on a single polypeptide chain, whereas iterative type II PKS systems are assemblies of discrete polypeptide chains, where each chain has one enzyme activity, and noncovalent protein–protein associations presumably are important for the system to function. In both types of systems, the acyl carrier protein (ACP) plays a central role, acting as the anchor point for the growing acyl chain, which is attached by a thioester linkage to the 4′-phosphopantetheine prosthetic group of the ACP. This prosthetic group is covalently attached to a serine residue found in a conserved three-residue sequence, most commonly −Asp−Ser−Leu− (DSL). Polyketide biosynthesis begins with the attachment of an acyl- (or aryl)-coenzyme A “starter unit” to the ACP. This starter unit remains attached while “extender” acyl-coenzyme A units are added on in a series of decarboxylative condensation reactions, and are then further modified by tailoring enzymes. PKSs share their basic organization with fatty acid synthases (FASs) (Hopwood 1997), in which an ACP domain carries growing fatty acid chains; the FASs of yeasts, animals, and plants are large, modular type I enzymes, while those of bacteria and mitochondria (Zhang et al. 2003) have the type II organization.
Nonribosomal peptide synthetases use activated aminoacyl units as building blocks for the synthesis of bioactive peptides, which often contain unusual amino acids (Marahiel et al. 1997; Cane and Walsh 1999; Finking and Marahiel 2004; Walsh 2004). Here, the carrier function is performed by peptidyl carrier protein (PCP) domains homologous to ACPs, and the peptide substrate is bound to the 4′-phosphopantetheinyl group of the PCP domain as a peptidyl thioester. Hybrid PKS/NRPS systems also exist. In contrast to the PKSs and FASs, almost all nonribosomal peptide synthetases characterized to date are of the modular type I organization, with the PCP function represented by one domain of the multifunctional polypeptide chain. Nonetheless, the existence of discrete PCPs has been reported in two hybrid PKS/NRPS systems, i.e., the gene clusters for the biosynthesis of bleomycin in Streptomyces verticillus (Du and Shen 1999) and for the biosynthesis of leinamycin in Streptomyces atroolivaceus (Tang et al. 2004).
Cyanobacterial PKSs and NRPSs are rich sources of potentially useful therapeutic compounds, and to date only a few of these gene clusters have been characterized (Dittmann et al. 2001; Hoffmann et al. 2003). The genomic context of Asl1650 is interesting in that adjacent genes show homology with both PKS and NRPS enzymes. Furthermore, these genes show only low sequence homology with the previously characterized PKS and NRPS systems. To investigate whether Asl1650 functions as part of a PKS, NRPS, or hybrid pathway, and as a step toward better characterization of these pathways, we have determined the solution structure of Asl1650 by NMR spectroscopy.
As a complement to these considerations on its functional role in essential physiological processes in Anabaena, Asl1650 was identified as an ortholog of a mouse fatty acid synthase ACP domain (SwissProt P19096), with which it shares ∼18% sequence identity. This finding resulted from a bioinformatics strategy aimed at extending the coverage of protein fold space of eukaryotic proteins. Structural studies of remote orthologs (distantly related proteins with low levels of sequence identity) are of interest in expanding the characterization of large protein fold families that are represented in all kingdoms of life, and in delineating functional relationships within these families. The evolutionary/organizational similarities between FAS, PKS, and NRPS have previously been intensively studied, and similarity between ACPs from different species has been implicated from multiple three-dimensional structure determinations (Kim and Prestegard 1990; Crump et al. 1997; Parris et al. 2000; Weber et al. 2000; Volkman et al. 2001; Xu et al. 2001; Roujeinikova et al. 2002; Wong et al. 2002; Findlow et al. 2003; Li et al. 2003; Reed et al. 2003; Park et al. 2004; Qiu and Janson 2004; http://www.jcsg.org/). These include ACPs involved in fatty acid synthesis (Bacillus subtilis, Escherichia coli, Helicobacter pylori, Thermotoga maritima, Mycobacterium tuberculosis, rat), polyketide synthesis (act, fren, otc), as well as in nonribosomal peptide biosynthesis (Bacillus brevis). In view of the high diversity of bacterial PKS and NRPS systems, this study of Asl1650 will be an important addition to the structural database that can be used for continued attempts at complete characterization of these systems.
Results
The NMR experiments were performed with recombinant Asl1650 expressed in E. coli. The construct used is a variant of the wild-type sequence, with a designed replacement of Cys 7 by Ala, to prevent intermolecular disulfide formation, and an extra N-terminal tripeptide segment, Gly–Ser–His. The protein was derived from the thrombin-cleavage site of the expression and purification tag prepared with natural isotope distribution, and uniformly labeled either with 15N or with 15N and 13C (for details, see Materials and Methods).
NMR structure determination of Asl1650
The structure of Asl1650 has been determined using multidimensional heteronuclear NMR spectroscopy. Because of a tendency for aggregation at higher protein concentrations, the Asl1650 concentration was adjusted to 2 mM, and 250 mM NaCl was added. Under these conditions, a stable protein solution was obtained, with no changes in the [1H,15N]-HSQC spectrum (Fig. 1) observed over a period of several months.
Figure 1.
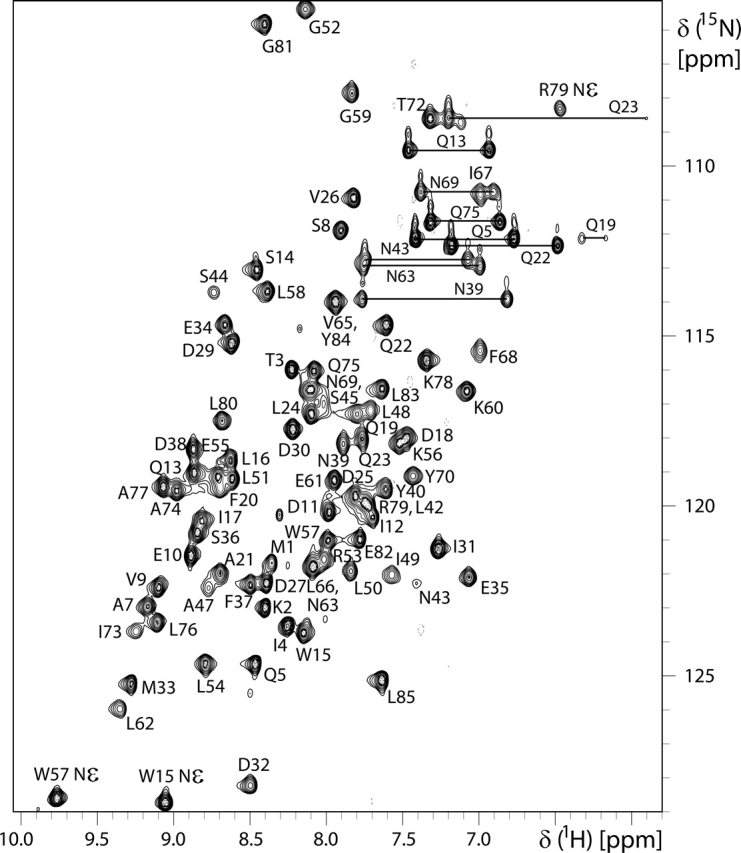
2D [1H,15N]-HSQC spectrum of uniformly 15N-enriched Asl1650 (600 MHz, 303 K, protein concentration 2 mM in 20 mM sodium phosphate buffer at pH 6.0, 250 mM NaCl). Backbone 15N−1H cross peaks are identified with the one-letter amino acid symbol and the sequence number; for side chain 15N−1H correlations, the atom positions for Arg and Trp and horizontal lines for the −15N1H2 moieties of Asn and Gln are also indicated.
All backbone 15N and 1HN resonances were assigned except for those of Ser (−2), His (−1), and Lys 46 (negative sequence numbers are used for the residues in the N-terminal tripeptide segment preceding the Asl1650 sequence). All aliphatic and aromatic side chain resonances were assigned, except for all of the resonances of Ser 44, ɛCH3 of Met 1 and Met 33, δCH2 of Lys 46, and ζCH of Phe 68. All labile side chain protons of Asn, Gln, and Arg were assigned, with the sole exception of Arg 53 ηH.
The structure was determined with fully automated analysis of the [1H,1H]-NOESY spectra, using the standard protocol for the ATNOS/CANDID/DYANA suite of programs (Güntert et al. 1997; Herrmann et al. 2002a, b; for details, see Materials and Methods). The correct fold was obtained in the first cycle of calculation, with no major changes to the structure occurring during the subsequent six cycles. The statistics for the structure determination and for the bundle of 20 conformers with the lowest residual DYANA target function values, which is used to represent the solution structure of Asl1650, are summarized in Table 1. The structure is well defined with the exception of the N-terminal polypeptide segment −3 to 2 (Fig. 2A).
Table 1.
Input for the structure calculation of the 88-residue protein construct Asl1650(−3–85) and statistics of the bundle of 20 energy-minimized DYANA conformers used to represent the NMR structure
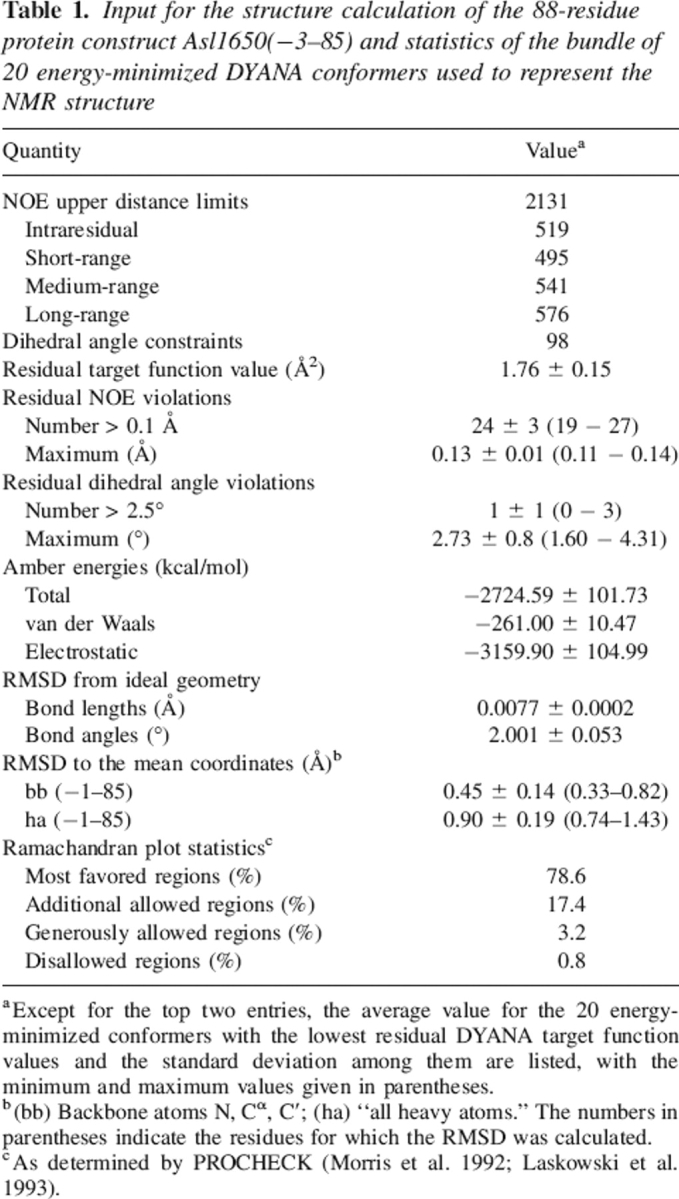
aExcept for the top two entries, the average value for the 20 energy-minimized conformers with the lowest residual DYANA target function values and the standard deviation among them are listed, with the minimum and maximum values given in parentheses.
b(bb) Backbone atoms N, Cα, C′; (ha) “all heavy atoms.” The numbers in parentheses indicate the residues for which the RMSD was calculated.
cAs determined by PROCHECK (Morris et al. 1992; Laskowski et al. 1993).
Figure 2.
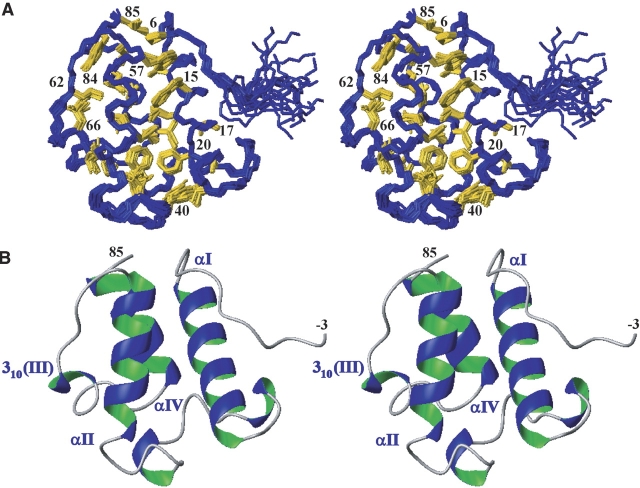
Wall-eye stereo views of the NMR structure of the protein Asl1650. (A) Bundle of 20 energy-minimized DYANA conformers. (Blue) Polypeptide backbone, (gold) hydrophobic side chains of the protein core. The positions of selected hydrophobic core residues are identified with the sequence numbers. (B) Ribbon diagram of the Asl1650 conformer with the lowest RMSD to the mean coordinates of the bundle of 20 conformers in panel A. The four helices forming a helix bundle (see text) are labeled αI, αII, 310(III), and αIV at their N termini. The chain-terminal residues −3 and 85 are indicated.
The NMR structure of Asl1650
The protein forms a twisted, antiparallel four-helix bundle with an up-down-up-down topology (Fig. 2B). Three α-helices, αI, αII, and αIV, span residues 9–24, 45–58, and 73–83. A long, well-ordered loop connects αI and αII. The remaining helix consisting of residues 64–66 is a 310-helix, within the loop connecting helices αII and αIV.
The N-terminal tripeptide segment of residues −3 to −1, which was added by cloning, does not associate with the protein. Together with the first two residues of the Asl1650 sequence, Met 1 and Lys 2, it forms a disordered “tail.” The three α-helices have amphipathic character, each donating hydrophobic residues to the protein core on the inside of the helix bundle, while charged residues are exposed on the protein surface. The well-defined hydrophobic core is a striking feature of the structure (Fig. 2A) and includes the side chains of Ile 12, Trp 15, Leu 16, and Phe 20 of helix αI; Leu 50, Leu 54, Trp 57, and Leu 58 of helix αII; Leu 76 and Leu 80 of helix αIV; and Tyr 84 and Leu 85. At one end (Fig. 2A), Pro 6 stacks with Trp 15 (from helix αI) and Trp 57 (from helix αII). Well-defined structure persists all the way to the C terminus, since the side chains of Tyr 84 and Leu 85 also form part of the hydrophobic core (Fig. 2A).
The long loop of residues 25–44, which connects the helices αI and αII, is well defined in spite of the fact that it contains only two recognizable tight turns and no regular secondary structure. This is likely due to the presence of several hydrophobic residues that form part of the protein interior, where they associate with side chains from the helices αI, αII, and αIV. Thus, the hydrophobic residues Ile 31, Phe 37, Tyr 40, and Leu 42 are all in close contact with Phe 20 and Leu 24 of helix αI. The tight turns in this loop are also stabilized by hydrogen bonds. In particular, in the 310-turn of residues 28–30, the carbonyl oxygens of Asp 27 and Pro 28 are linked to the amide protons of Asp 30 and Ile 31, and similar (i, i + 3) hydrogen bonds from the carbonyl oxygens of Ser 36 and Phe 37 to the amide protons of Asn 39 and Tyr 40 are identified in a 310-turn of residues 37–39. In addition, the side chain hydroxyl group of Tyr 40 forms a hydrogen bond to the amide proton of Asp 32.
Discussion
A detailed analysis of the solution structure of Asl1650 and comparison with the structural and functional data available for other proteins of the acyl carrier protein (ACP) family lead us to propose that this protein functions as a discrete peptidyl carrier protein in either a nonribosomal peptide synthetase (NRPS) or a hybrid polyketide synthase (PKS)/NRPS biosynthetic system. This prediction is based on the following pieces of evidence: (1) Asl1650 adopts a twisted, antiparallel, four-helix bundle fold characteristic of the ACP family; (2) the fold of Asl1650 shows the closest similarity to the peptidyl carrier protein (PCP) and PKS ACP proteins but significantly larger differences to fatty acid synthase (FAS) ACP proteins; (3) a close structural resemblance to B. brevis PCP in the molecular recognition region of helix III, including the hydrophobic character of its molecular surface; (4) apart from helix III, the molecular surface is mainly negatively charged, which would be consistent with Asl1650 being a discrete soluble protein as predicted by genome annotation (Nakamura et al. 1998; Kaneko et al. 2001); (5) Asl1650 is encoded in a region of the Anabaena genome that is rich in both PKS and NRPS genes; (6) Asl1650 contains a variant active-site sequence which differs from those in all presently known ACP structures but has occasionally been found in NRPS modules; (7) Asl1650 lacks conserved Arg and Asn residues found in all PKS ACPs characterized to date, but retains a conserved Lys residue known to be important in PCP function. In the following, we evaluate these experimental data in more detail.
Global fold comparison of Asl1650 with related proteins
The energy-minimized Asl1650 conformer with the lowest root-mean-square deviation (RMSD) to the mean coordinates of the bundle of conformers in Figure 2A was superimposed on the other known ACP structures for which coordinates are available (Kim and Prestegard 1990; Crump et al. 1997; Parris et al. 2000; Weber et al. 2000; Volkman et al. 2001; Xu et al. 2001; Roujeinikova et al. 2002; Wong et al. 2002; Findlow et al. 2003; Li et al. 2003; Reed et al. 2003; Qiu and Janson 2004; JCSG, http://www.jcsg.org/). This comparison revealed that Asl1650 generally resembles PKS ACPs more closely than FAS ACPs. An analysis by Li et al. (2003) has previously noted structural differences between PKS ACPs and FAS ACPs. The highest overall structural similarities are with otc ACP, a type II ACP involved in oxytetracycline biosynthesis in Streptomyces rimosus (Findlow et al. 2003); fren ACP, which is involved in frenolicin biosynthesis in Streptomyces roseofulvus (Li et al. 2003); Lactobacillus rhamnosus Dcp, a D-alanyl carrier protein involved in lipoteichoic acid biosynthesis (Volkman et al. 2001); and TycC3 PCP, a peptidyl carrier protein domain of the NRPS responsible for tyrocidine biosynthesis in B. brevis (Weber et al. 2000). Comparison between Asl1650 and each of these four proteins showed that the backbone atoms of the corresponding regions of the helices αI, αII, and αIV can be superimposed with an RMSD <1.70 Å (see Fig. 3 for the identification of the polypeptide segments used for the RMSD calculations). In spite of these close fits between the three-dimensional structures, the sequence identity between Asl1650 and the other four proteins ranges from only 14% to 22% (Fig. 3). Some key residues are also found in similar positions. For example, Phe 37, which is highly conserved throughout the ACP family, adopts nearly identical positions in the three-dimensional structures, and the orientation of several leucine side chains within the core is also conserved. Significant differences arise only in the lengths of some of the helices (Fig. 3). Thus, helix αI of Asl1650 is extended at the N terminus by a full turn relative to αI in otc and fren ACPs, and Asl1650 lacks the flexible C-terminal extension of otc ACP. In terms of relative helix disposition, Asl1650 is also very similar to B. brevis TycC3 PCP and to L. rhamnosus Dcp.
Figure 3.
Sequence alignment of Asl1650 with other structurally characterized proteins in the ACP family. The PDB codes for the comparison proteins are included in parentheses in the top part of the figure. The alignment was obtained with the software FFAS (Rychlewski et al. 2000). Residue numbers for Asl1650 are indicated above the sequence. (Box) The three-residue phosphopantetheinylation-site sequence; (periods) residues not used by FFAS for the alignment; (stars) sequence positions 46, 74, and 81, which are discussed in the text as having possible functional importance. The sequences of the otc, fren, M. tub., TM0175, act, and rat ACPs were truncated so as not to extend beyond the C terminus of Asl1650. The insertion in L. rhamnosus Dcp at the positions corresponding to 64–65 of Asl1650 was deleted by the “master-slave” alignment format of FFAS and was added interactively based on inspection of the three-dimensional structure. The positions of the helices that form the four-helix bundle in Asl1650 are indicated above the Asl1650 sequence. For all the proteins, helical residues, as identified by MOLMOL (Koradi et al. 1996), are shown in bold lettering. The residues used for the RMSD calculations in the three-dimensional structure superpositions with Asl1650 (see text) are underlined.
In contrast, a comparison with FAS ACPs shows that although the global fold is maintained, the helix orientations diverge and the conformations of the connecting loops also show larger differences. Thus, if αII and αIV are superimposed for best fit with the corresponding helices in the FAS ACPs of M. tuberculosis (Wong et al. 2002), E. coli (Kim and Prestegard 1990; Roujeinikova et al. 2002; Qiu and Janson 2004), B. subtilis (Parris et al. 2000; Xu et al. 2001), and T. maritima (JCSG, http://www.jcsg.org/; PDB code 1VKU), αI then has a noticeably different orientation, and the arrangements of the third helix [310(III) or αIII] are also quite widely different. Similar differences are also observed when comparing Asl1650 with act ACP, which is involved in actinorhodin biosynthesis in Streptomyces coelicolor (Crump et al. 1997). Finally, rat FAS ACP (Reed et al. 2003) has a much shorter helix αI; αII is also shorter by a full turn (Fig. 3); and the spatial arrangement of the loops linking the helices is substantially different from Asl1650. Superposition of the coordinates of Asl1650 with those of the proteins described in this paragraph, using the backbone atoms of helices αI, αII, and αIV (Fig. 3), yielded RMSD values ranging from 1.8 to 4.45 Å.
Structure and dynamics of the loop linking the helices αI and αII
Several studies of ACPs and related proteins have revealed conformational disorder in the loop linking the first two helices in the four-helix bundle. Thus, roughly the first half of this loop was disordered in act ACP (Crump et al. 1997) and in rat FAS ACP (Reed et al. 2003), as shown by high displacement values among the bundle of conformers representing the NMR structure and by rapid amide proton exchange with the solvent, and in the fren (Li et al. 2003) and otc (Findlow et al. 2003) ACPs, as shown by corresponding observations and by 15N relaxation data indicating high-frequency mobility. Two different local conformations were apparent in the second half of this loop in TycC3 PCP, and 15N relaxation data also indicated high flexibility (Weber et al. 2000); in the B. subtilis FAS ACP, structural disorder and flexibility were apparent throughout much of the loop (Xu et al. 2001). In stark contrast, no evidence for loop disorder (Fig. 2A) or flexibility was obtained in Asl1650. 15N{1H}-NOE values were between 0.7 and 0.8 for almost all residues within the loop, similar to the values for the residues in the helices. The well-structured loop in Asl1650 can be rationalized by the fact that the side chains of Ile 31, Phe 37, Tyr 40, and Leu 42 all contribute to the protein core and thus anchor the loop. In this respect, Asl1650 resembles the FAS ACP from M. tuberculosis (Wong et al. 2002) and the D-alanyl carrier protein (Dcp) from L. rhamnosus (Volkman et al. 2001), in which the loop is also as well ordered as the rest of the protein.
Helix 310(III)
The short helix 310(III) in the loop connecting the helices αII and αIV varies considerably in different ACPs. Although a corresponding helix is present in almost all known ACP structures (Fig. 3), it is structurally disordered in act ACP and B. subtilis FAS ACP, and two different conformations in slow exchange were found in fren ACP. In Asl1650, this 310-helix of residues 64–66 is well defined, with no evidence for increased dynamics.
Conservation of putative functional residues
The active site of ACPs comprises a tripeptide segment that includes an invariant serine, which is the site of covalent modification by a phosphopantetheinyl group donated by coenzyme A in a reaction catalyzed by phosphopantetheinyltransferases (PPTases). In Asl1650, the active site has the sequence NSS (residues 43–45). This is at variance with most of the other characterized ACPs, which contain the sequence DSX, where X is either leucine or another hydrophobic residue (Fig. 3). After spatial alignment of helices αI, αII, and αIV, the position and orientation of the NSS active-site segment in Asl1650 were essentially identical to those of the DSX sequences in the other ACPs. In all cases, the active-site sequence is found at the beginning of helix αII (Fig. 3), and all three residues are exposed on the protein surface and available for recognition and covalent modification by PPTases. The presence of a second serine in the Asl1650 recognition sequence raises the possibility of two alternative sites for the covalent modification, although structural similarity would suggest the central Ser as the primary modification site.
The Asp to Asn substitution in the first position of the active site tripeptide segment is not common, but occurs in several other CP domains, including domains of hybrid PKS/NRPS pathways such as those of microcystin biosynthesis (Rouhiainen et al. 2004), rapamycin biosynthesis (Aparicio et al. 1996), and bleomycin biosynthesis (Du and Shen 1999), as well as various predicted CP domains in Mycobacteria, Bacillus, Streptomyces, and other species. By analogy to type II FAS systems, this modification is likely to be relevant for the activation by PPTases. The structure of the B. subtilis FAS ACP–ACP synthase complex (Parris et al. 2000) showed that the Asp residue is directly involved in intermolecular recognition. Although it is possible that PKS and NRPS systems would function differently, it is a likely hypothesis that in these systems also, the residue preceding the central Ser might be involved in recognition. The residue at this position has been shown to affect interactions between the PCP and epimerization domains in NRPS modules (Linne et al. 2001). Different residues are conserved at this position between PCP domains located N-terminally to epimerization domains versus those located N-terminally to condensation domains (D and H, respectively) (Linne et al. 2001).
Two conserved residues corresponding to Arg 72 and Asn 79 in act ACP have been identified as being characteristic of PKS ACPs (Crump et al. 1997), and might be involved in intermolecular interactions within PKS systems. Asl1650 lacks these conserved Arg and Asn residues, and Ala 74 and Gly 81 occupy the corresponding positions (Fig. 3).
Crystallography (Parris et al. 2000), NMR spectroscopy (Jain et al. 2004), molecular modeling (Zhang et al. 2001; Keatinge-Clay et al. 2003), and mutational studies (Flaman et al. 2001; Zhang et al. 2001; Mofid et al. 2002; Worsham et al. 2003) have implicated the helices αII and αIII as important sites for recognition by PPTases and other biosynthetic enzymes. A conserved trait of peptidyl carrier proteins (PCPs) is a Lys or Arg immediately following the active-site sequence at the beginning of helix αII, as distinct from ACPs, which often have an acidic or small polar residue at this position (Finking et al. 2004). This residue is thought to be responsible for allowing recognition of the PCP by promiscuous PPTases of secondary metabolism (Sfp-type), which contain a key acidic residue in their active sites, while preventing recognition by PPTases of primary metabolism (AcpS-type), where the relevant residues are basic (Finking et al. 2004). In common with PCP domains, this residue is Lys 46 in Asl1650 (Fig. 3).
At positions 49 and 56, Asl1650 shows closer similarity to PCP/PKS ACP domains; for example, position 49 is most commonly Glu in FAS and PKS ACPs, while in Asl1650 it is Ile and in PCP it is Ala. These residues are all exposed on the protein surface in the Asl1650 structure.
Other molecular surface features
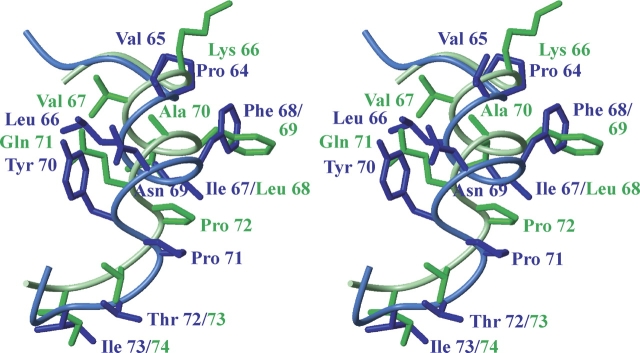
An unusual feature of the short helix 310(III) of Asl1650 is that Phe 68 forms a hydrophobic patch with Pro 64, Val 65, Tyr 70, and Pro 71 on the protein surface. This contrasts with the hydrophilic nature of the third helix in most of the other known ACPs, but strongly resembles the corresponding region of TycC3 PCP (Weber et al. 2000), which forms one domain of a large modular, multifunctional NRPS. As shown in Figure 4, significant structural similarity exists between the two proteins throughout the helix 310(III), with the surface-exposed residues Phe 68/69 and Pro 71/72 in nearly identical positions. The similarity extends beyond the end of 310(III) toward the start of helix αIV at residue 73. However, in PCP, the hydrophobic side chains from helix 310(III) form part of a large hydrophobic region, which extends across nearly half of the protein surface and includes the phosphopantetheinylation site, whereas in Asl1650 the side chains from 310(III) form an isolated hydrophobic patch on the protein surface (Fig. 5). This characteristic of helix III contrasts with that in ACPs of both PKS and FAS systems, where most of the solvent-exposed residues of helix III are hydrophilic. For example, in L. rhamnosus Dcp, Arg and Lys residues are located on the surface of helix III (Fig. 5), while in the B. subtilis holo-ACP–ACP synthase complex (Parris et al. 2000), the acidic side chains of Asp 56 and Glu 60 are surface-exposed. The crystal structure of this complex revealed that these side chains form part of the molecular surface recognized by the ACP synthase. This area is near the active-site Ser on the molecular surface. In both PKS and FAS ACPs, acidic residues are usually found in the positions corresponding to Pro 64 and Val 65 in Asl1650 (Fig. 3). Overall, the proximity to the active site and the surprisingly close three-dimensional structure similarity to PCP in this region implicate the helix III region as an important area for molecular recognition.
Figure 4.
Wall-eye stereo view illustrating structural similarity in the region of the helix 310(III) between Asl1650 (blue) and TycC3 PCP (green). The backbone is represented by a spline function through the Cα positions, and the side chain heavy atoms are shown as stick diagrams. The orientation is such that side chains in the front right of the figure are exposed on the protein surface.
Figure 5.
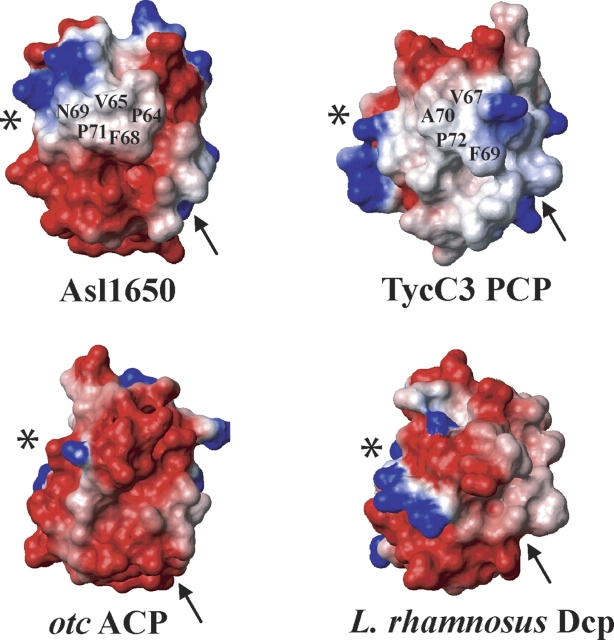
Molecular surface presentations of Asl1650 and TycC3 PCP (see Fig. 3), and two other ACPs, colored according to electrostatic potential (positive, blue; negative, red; neutral, white). The Asl1650 structure has been rotated by 90° about a vertical axis relative to the views in Figure 2. The other proteins have been oriented after a best-fit superposition of the corresponding residues in the helices αI, αII, and αIV (see Fig. 3). For each protein, the C terminus of the helix III is marked with a star, from where this helix extends horizontally to the right across each protein surface. For Asl1650 and TycC3 PCP, the surface residues introduced in Figure 4 are also identified. Furthermore, the location of the active-site tripeptide segment is indicated with an arrow. The surfaces were calculated with MOLMOL (Koradi et al. 1996), using an all-heavy-atom presentation and “simple” charges.
It is apparent in Figure 5 that Asl1650, with the helix III hydrophobic surface surrounded by charged residues, presents neither the uniformly negatively charged surface of ACPs, as represented by otc ACP, nor the crescent of positive charge of the corresponding region in Dcp. This would be consistent with a function as a discrete soluble protein, in contrast to PCP, which functions as a domain within a larger type I NRPS protein. These observations lead us to hypothesize that the hydrophobic helix III region may be important for intermolecular interactions within a NRPS system and that Asl1650 may function as part of a NRPS or hybrid NRPS/PKS system, rather than a PKS system.
Genomic context
Within the Anabaena genome (Nakamura et al. 1998; Kaneko et al. 2001), the asl1650 gene lies directly downstream of a putative transcriptional regulator (all1651) and upstream of several other NRPS and PKS genes (all1646–1649). The open reading frame all1647 has sequence similarity to core NRPS adenylation and condensation domains but contains no PCP region. It is possible that Asl1650 might function within the same biosynthetic pathway as the protein encoded by all1647, and might provide peptidyl carrier protein activity through an intermolecular interaction. Such an arrangement would not conform to the classical type I/type II organization, with either all enzyme activities on one polypeptide chain (modular, type I), or separate proteins for each enzyme activity (iterative, type II) (see the introduction). In this context, we note further that Asl1650 shares more sequence characteristics with PCPs located N-terminally to epimerization domains rather than with those of PCPs located N-terminally to condensation domains (Marahiel et al. 1997; Finking and Marahiel 2004; Finking et al. 2004).
The ORFs all1646, all1648, and all1649 encode proteins with similarity to type I PKSs. These are followed by several proteins of unknown function. We note that a similar gene cluster with a nearly identical arrangement is found in the genome of the closely related strain Anabaena variabilis. The protein encoded by all1646 is also known as HglE2 and shares 51% sequence identity with HglE, a protein essential for heterocyst glycolipid formation in Nostoc punctiforme (Black and Wolk 1994; Campbell et al. 1997). Anabaena sp. PCC 7120 contains two orthologs of HglE, with HglE1 being more similar than HglE2. HglE1 is located in a cluster of genes known to be involved in heterocyst glycolipid biosynthesis (Black and Wolk 1994; Campbell et al. 1997). Expression of both HglE1 and HglE2 was absent in a mutant with an inactivated copy of devH, which encodes a transcriptional regulator involved in heterocyst differentiation. The mutant strain was deficient in heterocyst glycolipid biosynthesis (Ramírez et al. 2005). This process may therefore represent a possible cellular role for Asl1650 and/or the neighboring genes. Alternatively, cyanobacteria produce a variety of other hybrid peptide–polyketide metabolites; for example, Nostoc strains produce the potent cytotoxic cryptophycins via hybrid pathways (Golakoti et al. 1995). The greatest diversity of PKS/NRPS genes occurs in the filamentous cyanobacteria as opposed to the unicellular cyanobacteria (Dittmann et al. 2001). Consistent with this general trend, many proteins or protein domains with similarity to Asl1650 (at a cutoff E-value ≤ 5) are detectable by a BLAST search in the genome sequences of the filamentous cyanobacteria Anabaena sp. PCC 7120, Anabaena variabilis, and Nostoc punctiforme (15, 19, and 38 orthologs, respectively), whereas fewer orthologs are detectable in the unicellular cyanobacteria Synechocystis sp. PCC 6803, Prochlorococcus marinus str. MIT 9313, and Synechococcus sp. WH8102, which do not have such a wide variety of PKS/NRPS gene clusters (6, 2, and 6 orthologs, respectively).
Classification of acyl carrier proteins
The structural observations described above lead us to some guidelines for the classification of proteins in the ACP family. We suggest that the helix III region should be primarily considered in addition to helix II when attempting to classify these proteins; in particular, the residues at positions 64, 68, 69, and 71, which are surface-exposed and uncharged in Asl1650 and B. brevis PCP (and also in type I PKS ACP domains where, however, a histidine residue at position 70 is more common). Furthermore, positions 64 and 65 in helix III are usually acidic in FAS and type II PKS ACPs.
Functionally important surface-exposed residues of helix II can provide a basis for further discrimination among these proteins. The active-site tripeptide sequence is at the start of helix II, and the residue immediately following it (position 46 in Asl1650) is often a lysine in PCPs (Finking and Marahiel 2004), aspartate or a small polar residue in FAS ACPs, and alanine in PKS ACPs. In type I PKS ACPs, this residue is most often an alanine, but may also be serine, threonine, valine, or methionine. Position 49 is a highly conserved glutamate residue in FAS ACPs and in PKS ACPs of both types. In PCPs, this position is variable, and may be neutral or basic. In contrast to the helices II and III, there are no identifying residues located in the first half of ACP/PCP sequences, consistent with our structural analysis, which shows that the position, length, and orientation of helix αI and of the long loop following it vary more extensively than the helices II, III, and IV (Fig. 3).
Since the ACP family is diverse and exceptions to many of the sequence conservation trends exist, the use of a profile/profile alignment method, such as FFAS (Rychlewski et al. 2000), is recommended as an additional tool when comparing new candidate proteins to known ACPs and PCPs. The FFAS server (http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) calculates an overall score based on the match to sequence profiles generated from an existing database. The score takes into account weak sequence similarity over the entire protein, which is otherwise difficult to detect. Theoretical considerations show that a high score implies overall structural similarity, and practical experience indicates that this method is able to help in differentiating subclasses of proteins, even within protein families such as the ACPs that share only low sequence identity. Overall, we recommend that classification of ACPs be based on matching of the structural features described in the first two paragraphs of this section, supplemented by results obtained using such profile alignment methods.
Materials and methods
Identification of Asl1650 as an acyl carrier protein ortholog
Bacterial orthologs of predicted proteins from the mouse genome that are represented in the Protein Family database (Pfam) were selected. Pfam domain PF00550 represents the acyl carrier protein family. Asl1650 was identified as an ortholog of the acyl carrier protein domain of the mouse fatty acid synthase (residues 2108–2185; SwissProt accession no. P19096). Although the overall level of sequence identity is low (18%), the relationship could be substantiated by PSI-Blast (Altschul et al. 1997), an iterative, profile/sequence alignment method that is sensitive to weak sequence similarity. In the rest of the ACP family, similarity is most evident near the Ser residue in the consensus motif DSX (X = hydrophobic), which is the site of covalent modification by a phosphopantetheinyl group.
Expression and purification
Asl1650 (previously amplified from genomic DNA, as described in Lesley et al. 2002), was subcloned into the NdeI and EcoRI sites of plasmid pET-28b(+) (Novagen), yielding a construct encoding residues 1–85 and a thrombin-cleavable N-terminal His6-tag (MGSSHHHHHHSSGLVPRGSH). Restriction enzymes were obtained from New England BioLabs. Replacement of the single Cys residue in position 7 of the wild-type sequence by Ala was achieved with the QuikChange site-directed mutagenesis system (Stratagene). A comparison of the 2D [1H,15N]-HSQC spectra of the wild-type Asl1650 and Asl1650 [C7A] showed essentially no differences outside of the peptide segment of residues 6–8. BL21-CodonPlus(DE3)-RIL cells (Stratagene) were used for protein expression. Cultures were grown at 37°C to an OD600 value of ∼0.7, induced with 1 mM IPTG, and grown for another 3 h. Cells were lysed by sonication in 50 mM Tris (pH 8.0) with 500 mM NaCl, 5 mM imidazole, and protease inhibitors (Complete EDTA-free, Roche). The lysate was clarified by centrifugation, filtered, and applied to a HiTrap chelating HP (iminodiacetic acid) column (Amersham) charged with Ni2+, and eluted with a 5–250 mM imidazole gradient. The sample was concentrated, diluted into 50 mM Tris buffer (pH 8.0) containing 50 mM NaCl and 10 mM CaCl2, and 250 μL thrombin-agarose resin (Sigma) was added; cleavage was complete after gentle shaking (90 rpm) for 42 h at 37°C. The cleaved His6-tag was removed by a second Ni2+-affinity chromatographic step. A final anion-exchange step (HiTrap Q FF, 20 mM Tris at pH 8.0, 0–1.5 M NaCl gradient) then yielded pure Asl1650, as assessed by SDS-PAGE and MALDI-TOF mass spectrometry. An N-terminal tripeptide segment not present in the wild-type sequence (GSH) remained after thrombin cleavage. Mass spectrometry also showed that the recombinant protein had not been covalently modified with a phosphopantetheine group by the E. coli holo-(acyl carrier protein) synthase (AcpS).
NMR sample preparation
Samples with natural isotope abundance were produced from cultures grown in LB broth, while isotope labeling was accomplished by growing cultures in minimal medium containing either 1 g/L 15NH4Cl for a uniformly 15N-labeled sample, or 1 g/L 15NH4Cl and 4 g/L 13C6-D-glucose for a uniformly 15N,13C-labeled sample. Both procedures yielded 50 mg of pure protein from 1 L of culture. NMR samples of volume 550 μL were produced by exchanging the pure protein into 20 mM sodium phosphate buffer (pH 6.0) containing 250 mM NaCl and 2 mM NaN3, using Millipore Ultrafree centrifugal concentrators (Biomax-5 membrane) to obtain a final protein concentration of 2 mM.
NMR spectroscopy
NMR spectra were recorded at 303 K on Bruker Avance 600 and Avance 900 MHz spectrometers equipped with TXI HCN z- or xyz-gradient probes, and on a Bruker Avance 500 MHz spectrometer equipped with a TXI HCN z-gradient cryoprobe. For the backbone and side chain resonance assignment, the following experiments were recorded with a solution of the uniformly 15N,13C-labeled protein in 90% H2O/10% D2O (v/v): 3D HNCACB (Wittekind and Mueller 1993), 3D CBCA(CO)NH, 3D HBHA(CO)NH (Grzesiek and Bax 1992), 3D HNCO (Ikura et al. 1990), (H)CC(CO)NH-TOCSY (Logan et al. 1993) (DIPSI-2 sequence [Shaka et al. 1988] for the 13C, 13C mixing, 10 kHz field, τm = 12 ms), HC(C)H-TOCSY (DIPSI-3 sequence [Shaka et al. 1988] for 13C, 13C mixing, 10 kHz field, τm = 12 ms, z-filter version [Peti et al. 2000]), and 3D 13C-resolved [1H,1H]-NOESY (τm = 80 ms) (Muhandiram et al. 1993). The following additional experiments were recorded with a uniformly 15N-labeled sample in 90% H2O/10% D2O (v/v): 2D [1H,15N]-HSQC, 3D 15N-resolved [1H,1H]-TOCSY (τm = 65 ms; DIPSI-2 mixing sequence [Shaka et al. 1988], field strength 8 kHz for 1H, 1H mixing), and 3D 15N-resolved [1H,1H]-NOESY (τm = 80 ms). Assignment of aromatic side chain resonances was based on a 2D [1H,1H]-NOESY spectrum (900 MHz, τm = 80 ms) obtained after exchanging the sample into D2O (Cambridge Isotope Laboratories), in combination with 2D [1H,13C]-HSQC and 3D 13C-resolved [1H,1H]-NOESY (τm = 80 ms) spectra. 15N{1H}-NOE values were measured from 2D [1H,15N]-HSQC spectra recorded in an interleaved manner (Farrow et al. 1994) with and without 1H saturation by a series of 120° high-power 1H pulses separated by 5-msec delays. Internal 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) was used as a chemical shift reference for 1H, 15N, and 13C (Wishart et al. 1995). Data processing and analysis were carried out using the programs XWINNMR 3.5 (Bruker) and XEASY (Bartels et al. 1995).
Three-dimensional structure determination
Structure determination was based on experimental NOE data obtained from 3D 15N-resolved [1H,1H]-NOESY, 3D 13C-resolved [1H,1H]-NOESY, and 2D [1H,1H]-NOESY spectra recorded at 900 MHz with τm = 80 msec, whereby the 2D [1H,1H]-NOESY spectrum was obtained in D2O solution. Structure calculations employed the programs ATNOS (Herrmann et al. 2002a) for automated NOE peak picking, CANDID (Herrmann et al. 2002b) for automated NOE assignment, and DYANA (Güntert et al. 1997) for torsion angle dynamics structure calculation. The chemical shift lists from the resonance assignment and the three aforementioned NOESY spectra were used as input for ATNOS. The standard ATNOS/CANDID/DYANA protocol (Herrmann et al. 2002a, b) consisting of seven cycles of NOE peak picking, NOE assignment, distance restraint generation, and structure calculation was employed. Each cycle of calculation produced an intermediate ensemble of three-dimensional structures, which was used as additional input for the subsequent cycle to re-evaluate the experimental NOESY data. Supplemental dihedral angle restraints were also employed in each cycle, as obtained from secondary structure identification using Cα chemical shifts (Spera and Bax 1991; Luginbühl et al. 1995). The final set of unambiguous NOE assignments obtained in the last cycle led to 2131 meaningful distance restraints, i.e., on average, 24 restraints/residue. The 20 structures with the lowest residual DYANA target function values obtained from the final cycle 7 were subjected to energy minimization in a shell of water molecules, using the program OPALp (Luginbühl et al. 1996; Koradi et al. 2000) with the Amber force field (Cornell et al. 1995). The quality of the final structures was assessed with PROCHECK (Morris et al. 1992; Laskowski et al. 1993).
The atomic coordinates of the bundle of 20 Asl1650 conformers of Figure 2A (accession no. 2AFD) and of the conformer closest to their mean coordinates (accession no. 2AFE; Fig. 2B) have been deposited in the Brookhaven Protein Data Bank (http://www.rcsb.org/pdb/). The sequence-specific resonance assignments have been deposited in the BioMagResBank (http://www.bmrb.wisc.edu) with accession number 6751.
Structural analysis and comparison with other ACP family proteins
Superposition of Asl1650 with other ACP family proteins was performed using the program MOLMOL. Best fit of the backbone N, Cα, and C′ atoms of sets of 10, 10, and 9 consecutive residues in the three major helices I, II, and IV of Asl1650 with counterparts in the other proteins was considered, since the shorter helix III and the connecting loops are quite variable. For Asl1650, the residues used were 14–23, 48–57, and 73–81, and for the other proteins, corresponding polypeptide segments were identified as those that produced the lowest RMSD value for superposition (Fig. 3). For the rat FAS ACP (Reed et al. 2003), the region for superposition thus included nonhelical residues adjoining the helices I, II, and IV, since this protein contains shorter helices (Fig. 3). Nonhelical residues at the ends of helices were also included for the helices II and IV in fren ACP, and for helix II of L. rhamnosus Dcp (Fig. 3).
The coordinates of the ACP structures used for these comparisons were obtained from the Protein Data Bank, and in the case of ensembles of NMR structures, the conformer with the lowest RMSD to the mean coordinates (RMSD calculated over the ordered regions of the protein as identified by the investigators in the publications) was used for analysis. The conformer with the lowest RMSD to the mean coordinates (i.e., 2AFE) was also used to represent the Asl1650 solution structure in these comparisons.
Acknowledgments
We thank Dr. Rebecca Page and Jeffrey Velasquez for providing the plasmid encoding Asl1650. Funding from the Joint Center for Structural Genomics (NIH NIGMS Protein Structure Initiative, Grant no. P50 GM62411), the Skaggs Institute for Chemical Biology, the Canadian Institutes of Health Research (fellowship to M.A.J.), and the FWF Austrian Science Foundation (Erwin Schrödinger Fellowship to W.P.) is gratefully acknowledged. K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at TSRI. We also acknowledge the use of the high-performance computing facility of TSRI.
Footnotes
Reprint requests to: Kurt Wüthrich, The Scripps Research Institute, Department of Molecular Biology and Joint Center for Structural Genomics, 10550 North Torrey Pines Rd., MB-44, La Jolla, CA 92037, USA; e-mail: wuthrich@scripps.edu; fax: (858) 784-8014.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051964606.
Abbreviations:ACP, acyl carrier protein; act, actinorhodin; Dcp, D-alanyl carrier protein; DSS, 2,2-dimethyl-2-silapentane-5-sulfonate; FAS, fatty acid synthase; fren, frenolicin; HSQC, heteronuclear single quantum correlation; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; NMR, nuclear magnetic resonance; NRPS, nonribosomal peptide synthetase; otc, oxytetracycline; PCP, peptidyl carrier protein; PKS, polyketide synthase; PPTase, phosphopantetheinyltransferase; RMSD, root-mean-square deviation; TOCSY, total correlation spectroscopy.
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio J.F., Molnár I., Schwecke T., König A., Haydock S.F., Khaw L.E., Staunton J., Leadlay P.F. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase Gene 169 9–16. [DOI] [PubMed] [Google Scholar]
- Bartels C., Xia T., Billeter M., Güntert P., Wüthrich K. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- Bentley S.D., Chater K.F., Cerdeño-Tárraga A.-M., Challis G.L., Thomson N.R., James K.D., Harris D.E., Quail M.A., Kieser H., Harper D.et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature 417 141–147. [DOI] [PubMed] [Google Scholar]
- Black T.A. and Wolk C.P. 1994. Analysis of a Het- mutation in Anabaena sp. strain PCC 7120 implicates a secondary metabolite in the regulation of heterocyst spacing J. Bacteriol. 176 2282–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell E.L., Cohen M.F., Meeks J.C. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133 Arch. Microbiol. 167 251–258. [DOI] [PubMed] [Google Scholar]
- Cane D.E. and Walsh C.T. 1999. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases Chem. Biol. 6 R319–R325. [DOI] [PubMed] [Google Scholar]
- Carreras C.W., Pieper R., Khosla C. 1997. The chemistry and biology of fatty acid, polyketide, and nonribosomal peptide biosynthesis In Deoxysugars, polyketides and related classes: Synthesis, biosynthesis, enzymes (ed. Rohr J.) . pp. 85–126. Springer-Verlag, Berlin vol. 188. [Google Scholar]
- Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz Jr. K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules J. Am. Chem. Soc. 117 5179–5197. [Google Scholar]
- Crump M.P., Crosby J., Dempsey C.E., Parkinson J.A., Murray M., Hopwood D.A., Simpson T.J. 1997. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry 36 6000–6008. [DOI] [PubMed] [Google Scholar]
- Dittmann E., Neilan B.A., Börner T. 2001. Molecular biology of peptide and polyketide biosynthesis in cyanobacteria Appl. Microbiol. Biotechnol. 57 467–473. [DOI] [PubMed] [Google Scholar]
- Du L. and Shen B. 1999. Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003 Chem. Biol. 6 507–517. [DOI] [PubMed] [Google Scholar]
- Farrow N.A., Muhandiram R., Singer A.U., Pascal S.M., Kay C.M., Gish G., Shoelson S.E., Pawson T., Forman-Kay J.D., Kay L.E. 1994. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation Biochemistry 33 5984–6003. [DOI] [PubMed] [Google Scholar]
- Findlow S.C., Winsor C., Simpson T.J., Crosby J., Crump M.P. 2003. Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus Biochemistry 42 8423–8433. [DOI] [PubMed] [Google Scholar]
- Finking R. and Marahiel M.A. 2004. Biosynthesis of nonribosomal peptides Annu. Rev. Microbiol. 58 453–488. [DOI] [PubMed] [Google Scholar]
- Finking R., Mofid M.R., Marahiel M.A. 2004. Mutational analysis of peptidyl carrier protein and acyl carrier protein synthase unveils residues involved in protein–protein recognition Biochemistry 43 8946–8956. [DOI] [PubMed] [Google Scholar]
- Flaman A.S., Chen J.M., Van Iderstine S.C., Byers D.M. 2001. Site-directed mutagenesis of acyl carrier protein (ACP) reveals amino acid residues involved in ACP structure and acyl-ACP synthetase activity J. Biol. Chem. 276 35934–35939. [DOI] [PubMed] [Google Scholar]
- Golakoti T., Ogino J., Heltzel C.E., Husebo T.L., Jensen C.M., Larsen L.K., Patterson G.M.L., Moore R.E., Mooberry S.L., Corbett T.H.et al. 1995. Structure determination, conformational analysis, chemical stability studies, and antitumor evaluation of the cryptophycins. Isolation of 18 new analogs from Nostoc sp. strain GSV 224 J. Am. Chem. Soc. 117 12030–12049. [Google Scholar]
- Grzesiek S. and Bax A. 1992. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR J. Am. Chem. Soc. 114 6291–6293. [Google Scholar]
- Güntert P., Mumenthaler C., Wüthrich K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert P., Wüthrich K. 2002a. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS J. Biomol. NMR 24 171–189. [DOI] [PubMed] [Google Scholar]
- Herrmann T., Güntert P., Wüthrich K. 2002b. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA J. Mol. Biol. 319 209–227. [DOI] [PubMed] [Google Scholar]
- Hoffmann D., Hevel J.M., Moore R.E., Moore B.S. Sequence analysis and biochemical characterization of the nostopeptolide A biosynthetic gene cluster from Nostoc sp. GSV224. Gene 2003. 311 171–180. [DOI] [PubMed] [Google Scholar]
- Hopwood D.A. 1997. Genetic contributions to understanding polyketide synthases Chem. Rev. 97 2465–2498. [DOI] [PubMed] [Google Scholar]
- Ikura M., Kay L.E., Bax A. 1990. A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: Heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin Biochemistry 29 4659–4667. [DOI] [PubMed] [Google Scholar]
- Jain N.U., Wyckoff T.J.O., Raetz C.R.H., Prestegard J.H. 2004. Rapid analysis of large protein–protein complexes using NMR-derived orientational constraints: The 95 kDa complex of LpxA with acyl carrier protein J. Mol. Biol. 343 1379–1389. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Nakamura Y., Wolk C.P., Kuritz T., Sasamoto S., Watanabe A., Iriguchi M., Ishikawa A., Kawashima K., Kimura T.et al. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120 DNA Res. 8 205–213. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay A.T., Shelat A.A., Savage D.F., Tsai S.-C., Miercke L.J.W., O'Connell J.D., Khosla C., Stroud R.M. 2003. Catalysis, specificity, and ACP docking site of Streptomyces coelicolor malonyl-CoA:ACP transacylase Structure 11 147–154. [DOI] [PubMed] [Google Scholar]
- Kim Y. and Prestegard J.H. 1990. Refinement of the NMR structures for acyl carrier protein with scalar coupling data Proteins 8 377–385. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wüthrich K. 1996. MOLMOL: A program for display and analysis of macromolecular structures J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Güntert P. 2000. Point-centered domain decomposition for parallel molecular dynamics simulation Comput. Phys. Commun. 124 139–147. [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Lesley S.A., Kuhn P., Godzik A., Deacon A.M., Mathews I., Kreusch A., Spraggon G., Klock H.E., McMullan D., Shin T.et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline Proc. Natl. Acad. Sci. 99 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Khosla C., Puglisi J.D., Liu C.W. 2003. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein Biochemistry 42 4648–4657. [DOI] [PubMed] [Google Scholar]
- Linne U., Doekel S., Marahiel M.A. 2001. Portability of epimerization domain and role of peptidyl carrier protein on epimerization activity in nonribosomal peptide synthetases Biochemistry 40 15824–15834. [DOI] [PubMed] [Google Scholar]
- Logan T.M., Olejniczak E.T., Xu R.X., Fesik S.W. 1993. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments J. Biomol. NMR 3 225–231. [DOI] [PubMed] [Google Scholar]
- Luginbühl P., Szyperski T., Wüthrich K. 1995. Statistical basis for the use of 13Cαchemical shifts in protein structure determination J. Magn. Reson. B. 109 229–233. [Google Scholar]
- Luginbühl P., Güntert P., Billeter M., Wüthrich K. 1996. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules J. Biomol. NMR 8 136–146. [DOI] [PubMed] [Google Scholar]
- Marahiel M.A., Stachelhaus T., Mootz H.D. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis Chem. Rev. 97 2651–2673. [DOI] [PubMed] [Google Scholar]
- Mofid M.R., Finking R., Marahiel M.A. 2002. Recognition of hybrid peptidyl carrier proteins/acyl carrier proteins in nonribosomal peptide synthetase modules by the 4′-phosphopantetheinyl transferases AcpS and Sfp J. Biol. Chem. 277 17023–17031. [DOI] [PubMed] [Google Scholar]
- Morris A.L., MacArthur M.W., Hutchinson E.G., Thornton J.M. 1992. Stereochemical quality of protein structure coordinates Proteins 12 345–364. [DOI] [PubMed] [Google Scholar]
- Muhandiram D.R., Farrow N.A., Xu G.-Y., Smallcombe S.H., Kay L.E. 1993. A gradient 13C NOESY-HSQC experiment for recording NOESY spectra of 13C-labeled proteins dissolved in H2O J. Magn. Reson. B. 102 317–321. [Google Scholar]
- Nakamura Y., Kaneko T., Hirosawa M., Miyajima N., Tabata S. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803 Nucleic Acids Res. 26 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan D. The polyketide metabolites. Ellis Horwood, Chichester, UK 1991.
- Omura S., Ikeda H., Ishikawa J., Hanamoto A., Takahashi C., Shinose M., Takahashi Y., Horikawa H., Nakazawa H., Osonoe T.et al. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites Proc. Natl. Acad. Sci 98 12215–12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Kim J.-S., Son W.-S., Lee B.J. 2004. pH-induced conformational transition of H. pylori acyl carrier protein: Insight into the unfolding of local structure J. Biochem. 135 337–346. [DOI] [PubMed] [Google Scholar]
- Parris K.D., Lin L., Tam A., Mathew R., Hixon J., Stahl M., Fritz C.C., Seehra J., Somers W.S. 2000. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites Structure 8 883–895. [DOI] [PubMed] [Google Scholar]
- Peti W., Griesinger C., Bermel W. 2000. Adiabatic TOCSY for C, C and H, H J-transfer J. Biomol. NMR 18 199–205. [DOI] [PubMed] [Google Scholar]
- Qiu X. and Janson C.A. 2004. Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions Acta Crystallogr. D Biol. Crystallogr. 60 1545–1554. [DOI] [PubMed] [Google Scholar]
- Ramírez M.E., Hebbar P.B., Zhou R., Wolk C.P., Curtis S.E. 2005. Anabaena sp. strain PCC 7120 gene devH is required for synthesis of the heterocyst glycolipid layer J. Bacteriol. 187 2326–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M.A.C., Schweizer M., Szafranska A.E., Arthur C., Nicholson T.P., Cox R.J., Crosby J., Crump M.P., Simpson T.J. 2003. The type I rat fatty acid synthase ACP shows structural homology and analogous biochemical properties to type II ACPs Org. Biomol. Chem. 1 463–471. [DOI] [PubMed] [Google Scholar]
- Rouhiainen L., Vakkilainen T., Siemer B.L., Buikema W., Haselkorn R., Sivonen K. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90 Appl. Environ. Microbiol. 70 686–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roujeinikova A., Baldock C., Simon W.J., Gilroy J., Baker P.J., Stuitje A.R., Rice D.W., Slabas A.R., Rafferty J.B. 2002. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site Structure 10 825–835. [DOI] [PubMed] [Google Scholar]
- Rychlewski L., Jaroszewski L., Li W., Godzik A. 2000. Comparison of sequence profiles. Strategies for structural predictions using sequence information Protein Sci. 9 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankawa U. In Comprehensive natural products chemistry: Polyketides and other secondary metabolites including fatty acids and their derivatives (eds. Barton D.H.R.et al.) . . 1999. Elsevier, New York Vol. 1 series.
- Shaka A.J., Lee C.J., Pines A. 1988. Iterative schemes for bilinear operators; Application to spin decoupling J. Magn. Reson. 77 274–293. [Google Scholar]
- Spera S. and Bax A. 1991. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts J. Am. Chem. Soc. 113 5490–5492. [Google Scholar]
- Tang G.-L., Cheng Y.-Q., Shen B. 2004. Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase Chem. Biol. 11 33–45. [DOI] [PubMed] [Google Scholar]
- Volkman B.F., Zhang Q., Debabov D.V., Rivera E., Kresheck G.C., Neuhaus F.C. 2001. Biosynthesis of D-alanyl-lipoteichoic acid: The tertiary structure of apo-D-alanyl carrier protein Biochemistry 40 7964–7972. [PubMed] [Google Scholar]
- Walsh C.T. 2004. Polyketide and nonribosomal peptide antibiotics: Modularity and versatility Science 303 1805–1810. [DOI] [PubMed] [Google Scholar]
- Weber T., Baumgartner R., Renner C., Marahiel M.A., Holak T.A. 2000. Solution structure of PCP, a prototype for the peptidyl carrier domains of modular peptide synthetases Structure 8 407–418. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Bigam C.G., Yao J., Abildgaard F., Dyson H.J., Oldfield E., Markley J.L., Sykes B.D. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR J. Biomol. NMR 6 135–140. [DOI] [PubMed] [Google Scholar]
- Wittekind M. and Mueller L. 1993. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins J. Magn. Reson. B. 101 201–205. [Google Scholar]
- Wong H.C., Liu G., Zhang Y.-M., Rock C.O., Zheng J. 2002. The solution structure of acyl carrier protein from Mycobacterium tuberculosis J. Biol. Chem. 277 15874–15880. [DOI] [PubMed] [Google Scholar]
- Worsham L.M.S., Earls L., Jolly C., Langston K.G., Trent M.S., Ernst-Fonberg M.L. 2003. Amino acid residues of Escherichia coli acyl carrier protein involved in heterologous protein interactions Biochemistry 42 167–176. [DOI] [PubMed] [Google Scholar]
- Xu G.-Y., Tam A., Lin L., Hixon J., Fritz C.C., Powers R. 2001. Solution structure of B. subtilis acyl carrier protein Structure 9 277–287. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-M., Rao M.S., Heath R.J., Price A.C., Olson A.J., Rock C.O., White S.W. 2001. Identification and analysis of the acyl carrier protein (ACP) docking site on β-ketoacyl-ACP synthase III J. Biol. Chem. 276 8231–8238. [DOI] [PubMed] [Google Scholar]
- Zhang L., Joshi A.K., Smith S. 2003. Cloning, expression, characterization, and interaction of two components of a human mitochondrial fatty acid synthase. Malonyltransferase and acyl carrier protein J. Biol. Chem. 278 40067–40074. [DOI] [PubMed] [Google Scholar]