Abstract
(R)- and (S)-dichlorprop/α-ketoglutarate dioxygenases (RdpA and SdpA) catalyze the oxidative cleavage of 2-(2,4-dichlorophenoxy)propanoic acid (dichlorprop) and 2-(4-chloro-2-methyl-phenoxy)propanoic acid (mecoprop) to form pyruvate plus the corresponding phenol concurrent with the conversion of α-ketoglutarate (αKG) to succinate plus CO2. RdpA and SdpA are strictly enantiospecific, converting only the (R) or the (S) enantiomer, respectively. Homology models were generated for both enzymes on the basis of the structure of the related enzyme TauD (PDB code 1OS7). Docking was used to predict the orientation of the appropriate mecoprop enantiomer in each protein, and the predictions were tested by characterizing the activities of site-directed variants of the enzymes. Mutant proteins that changed at residues predicted to interact with (R)- or (S)-mecoprop exhibited significantly reduced activity, often accompanied by increased Km values, consistent with roles for these residues in substrate binding. Four of the designed SdpA variants were (slightly) active with (R)-mecoprop. The results of the kinetic investigations are consistent with the identification of key interactions in the structural models and demonstrate that enantiospecificity is coordinated by the interactions of a number of residues in RdpA and SdpA. Most significantly, residues Phe171 in RdpA and Glu69 in SdpA apparently act by hindering the binding of the wrong enantiomer more than the correct one, as judged by the observed decreases in Km when these side chains are replaced by Ala.
Keywords: dioxygenase, enantiospecificity, mecoprop, site-directed mutagenesis, structural modeling, docking
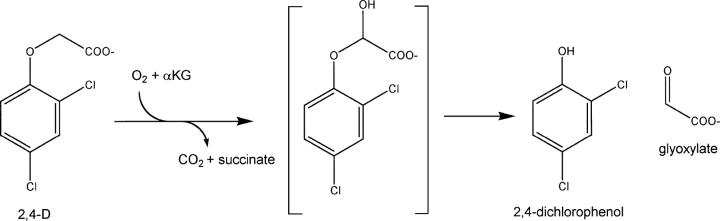
Phenoxyalkanoic acids are systemic and post-emergence inhibitors of broadleaf weeds and are among the most widely applied herbicides in the world (Worthing and Hance 1991; Ahrens 1994; Donaldson et al. 2002). These synthetic auxins (Åberg 1973; Loos 1975; Ahrens 1994) include 2,4-dichlorophenoxyacetic acid (2,4-D) along with the chiral representatives 2-(2,4-dichlorophenoxy)propanoic acid (dichlorprop) and 2-(4-chloro-2-methyl-phenoxy)propanoic acid (mecoprop), of which only the (R)-enantiomers are herbicidally active (Matell 1953). Microorganisms able to degrade these phenoxyalkanoic acid herbicides have been isolated from different environments, and their degradative pathways have been elucidated (Zipper et al. 1996; Hausinger et al. 1997; Tett et al. 1997; Müller et al. 1999, 2001). For example, the first step in 2,4-D metabolism is side-chain hydroxylation to form an unstable intermediate that decomposes by elimination of the phenol derivative (Scheme 1).
Scheme 1.
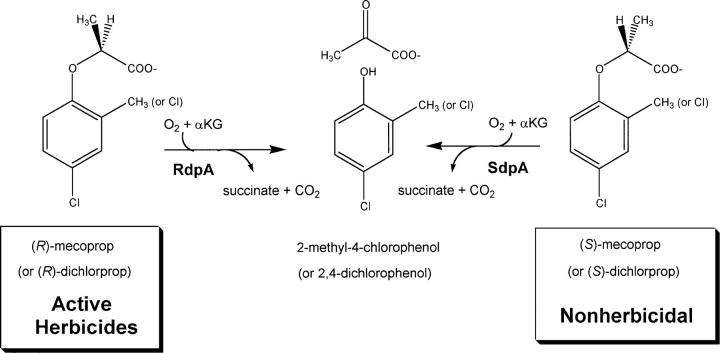
The 2,4-D hydroxylase (TfdA) from Cupriavidus necator (formerly Ralstonia eutropha) JMP134(pJP4) has been intensively studied and shown to require FeII as a cofactor and α-ketoglutarate (αKG) as a cosubstrate (Fukumori and Hausinger 1993a,b; Saari and Hausinger 1998; Hegg et al. 1999; Hogan et al. 2000; Dunning Hotopp and Hausinger 2002). For mecoprop or dichlorprop metabolism, best studied in the soil bacterium Sphingomonas herbicidovorans MH, the enantiomers are separately transported into the cell by distinct uptake systems (Nickel et al. 1997), and enantiomer-specific (R)- and (S)-dichlorprop/αKG dioxygenases (RdpA and SdpA) catalyze the initial degradation steps as illustrated in Scheme 2 (Nickel et al. 1997; Müller 2004; Müller et al. 2004b). RdpA and SdpA share 30% amino acid sequence identity to each other and 30% and 37% identity, respectively, to TfdA, with no significant gaps in alignment quality, indicating that they are all close structural homologs (Sander and Schneider 1991). The substituted phenol products released from these FeII/αKG-dependent dioxygenases are subsequently converted to the corresponding catechols and further metabolized by the modified ortho-cleavage pathway.
Scheme 2.
The herbicide-degrading dioxygenases belong to a large family of mononuclear, nonheme FeII enzymes that catalyze a broad array of reactions (for review, see Hausinger 2004; Clifton et al. 2006) including hydroxylations, epoxidations, desaturations, ring formation, ring expansion, and as only recently discovered, chlorinations (Vaillancourt et al. 2005a,b). Crystal structures have been elucidated for several family members including taurine/αKG dioxygenase (TauD) (Elkins et al. 2002; O'Brien et al. 2003), alkyl sulfatase (AtsK) (Müller et al. 2004a, 2005), clavaminate synthase (CAS) (Zhang et al. 2000), deacetoxycephalosporin C synthase (DAOCS) (Valegård et al. 1998), anthocyanidin synthase (ANS) (Wilmouth et al. 2002), carbapenam synthase (CarC) (Clifton et al. 2003), proline 3-hydroxylase (Clifton et al. 2001), the factor inhibiting hypoxia-inducible factor (FIH) (Dann et al. 2002; Elkins et al. 2003), phytanoyl-coenzyme A 2-hydroxylase (PAHX) (McDonough et al. 2005), and the DNA repair enzyme AlkB (Yu et al. 2006). The structures reveal a common β-jelly roll or double-stranded β-helix fold containing a metal ion-binding motif: His1-X-Asp/Glu-Xn-His2 (where n varies from 40 to 153 residues). Three water molecules occupy the remaining metal ligand positions in the resting enzyme. Two water molecules are displaced upon binding of the cosubstrate, with the αKG C-2 keto group coordinating opposite the carboxylate side chain and the αKG C-1 carboxyl group binding opposite either His1 (TauD, AtsK, CAS, and FIH) or opposite His2 (DAOCS, ANS, CarC, PAHX, and AlkB), with a nearby Arg residue (located 15–22 residues beyond His2 in the sequence) providing additional stabilization to the C-1 carboxylate in the cases of TauD, AtsK, CAS, CarC, and AlkB. Another Arg residue (located in the sequence about 10 residues beyond His2) is positioned to form an ion pair with the C-5 carboxylate of αKG in all structures except FIH, where a Lys located elsewhere in the sequence provides stabilization. Unlike other FeII sites, the αKG-bound metallocenters exhibit a characteristic metal-to-ligand charge-transfer transition (Pavel et al. 1998; Hegg et al. 1999; Ryle et al. 1999; Trewick et al. 2002) conferring a lilac color to this state of the enzymes. The primary substrate (e.g., taurine in the case of TauD) does not bind to the metal center, but the aforementioned crystallographic studies and additional spectroscopic evidence (Ho et al. 2001; Zhou et al. 2001) indicate that substrate binding leads to the loss of the final water molecule, thus creating a site for binding of oxygen. In the case of TauD, oxidative decarboxylation of αKG has been shown to produce an FeIV-oxo intermediate species that inserts oxygen into the unactivated C–H bond (Price et al. 2003a,b, 2005; Proshlyakov et al. 2004; Riggs-Gelasco et al. 2004; Grzyska et al. 2005).
Sequence alignments highlight several potential key residues of the (R)- and (S)-dichlorprop/αKG dioxygenases from S. herbicidovorans MH (Müller 2004). The His1-X-Asp/Glu-Xn-His2 motif of RdpA is comprised of residues His111, Asp113, and His270, while that of SdpA involves His102, Asp104, and His257. Fifteen residues beyond His2 are residues predicted to interact with the C-1 carboxylate of αKG, Arg285 and His272, respectively. Furthermore, Arg281 and Arg268 of the two proteins are predicted to form salt bridges to the αKG C-5 carboxylate, with additional interactions involving Thr138 of RdpA and Thr129 of SdpA. In contrast to these conserved residues, essentially nothing is known about the phenoxypropanoic acid-binding sites of these proteins, especially with regard to the structural basis of enantiospecificity.
Here, we describe the construction of homology models of SdpA and RdpA from S. herbicidovorans MH and the use of docking to identify residues likely to be involved in herbicide binding. Previous homology models have led to successes in elucidating or designing specificity-conferring interactions in ligands. For instance, homology modeling of a cercarial (human parasite) elastase led to the development of an effective elastase inhibitor (Cohen et al. 1991) and to understanding the specificity determinants for ligands binding to a parasite tRNA synthetase versus its human homolog (Sukuru et al. 2006). Here, we test by site-directed mutagenesis and kinetic analysis the residues predicted to be involved in substrate binding, enantiospecificity, or catalysis. The activity experiments are consistent with the key residues identified by modeling being involved in substrate binding. We provide additional evidence that several amino acids are responsible for the enantiospecificity of RdpA and SdpA, demonstrate that the active site of SdpA is less specific than RdpA for its substrate, and discuss the structural implications of these results.
Results
SdpA and RdpA homology models
RdpA and SdpA were aligned with TauD (Supplemental Fig. S1), and homology models were created as described in Materials and Methods (Supplemental Fig. S2) using the TauD structure as a structural template (O'Brien et al. 2003). The two phenoxypropionate-degrading proteins are predicted to contain jelly roll or double-stranded β-helix folds comprised of eight β-strands with connecting loops, as is typical of this enzyme family (Hausinger 2004; Clifton et al. 2006). The homology models contain FeII-binding sites (His111, Asp113, and His270 in RdpA or His102, Asp104, and His257 in SdpA), as expected from former sequence alignments with other FeII/αKG-dependent dioxygenases (Müller 2004). The high degree of active-site sequence identity and strong orientation of key side chains by interactions with the FeII are supportive of the active site being the most conserved and structurally accurate part of the RdpA and SdpA models. Detailed analysis of favored αKG-binding motifs in other members of this enzyme family indicate the iron is chelated by αKG with its keto group positioned opposite Asp113 of RdpA or Asp104 of SdpA and its C-1 carboxylate located so as to interact with His111 and His102, respectively. The positively charged residues Arg285 in RdpA and His272 in SdpA are well positioned to provide additional stabilization of the αKG C-1 carboxylate, and, in each protein, the C-5 carboxylate of αKG forms a salt bridge with Arg268 and Arg281, respectively. Whereas the RdpA structure represents only one subunit of the predicted trimeric protein, SdpA is suggested to be monomeric, based on gel-filtration experiments (Müller 2004).
Docking of substrates into the RdpA and SdpA structures
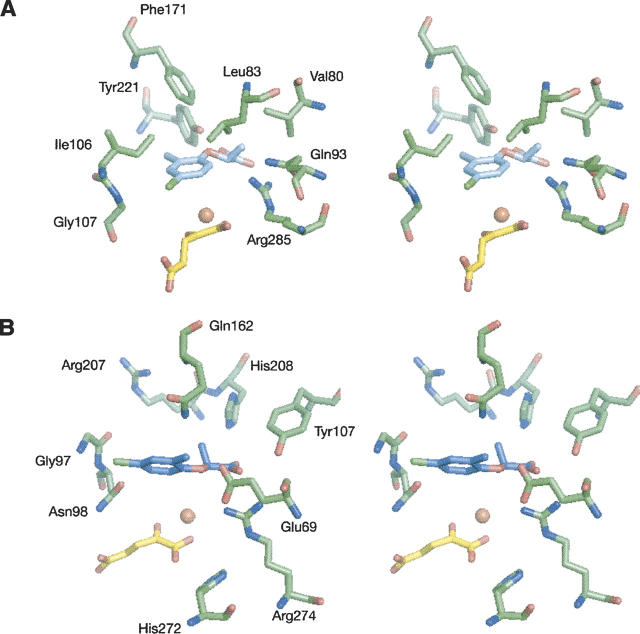
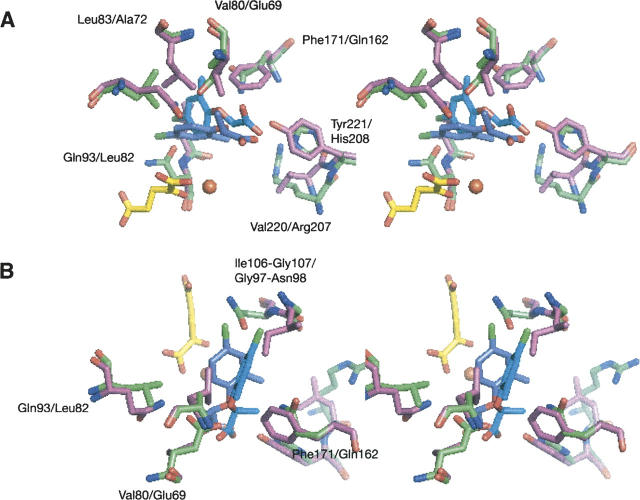
The natural substrates (R)- and (S)-mecoprop were docked into the active sites of RdpA and SdpA to gain insight into the basis of enzyme enantiospecificity. First, the cosubstrate αKG was modeled into the active sites of RdpA and SdpA using two distinct conformations, as found in the crystal structures of FeII/αKG-dependent dioxygenases (Clifton et al. 2006). The flat conformation has the five-member ring formed by the metal chelate coplanar with the C-5 carboxylate, whereas the twist conformation has the two planes forming a 90° angle. The resulting four models were energy-minimized and used as targets for substrate docking with the program SLIDE (Zavodszky et al. 2002) with the assumption that the substrate carbon undergoing hydroxylation would be located approximately at the same position relative to the iron center as the key carbon atom of taurine in TauD (Elkins et al. 2002; O'Brien et al. 2003). The mecoprop-docking interactions with RdpA and SdpA were analyzed in detail, and one model of each protein was selected based on the most favorable interactions between enzyme and substrate (see below). These models are illustrated in Figure 1, with the corresponding plots of mecoprop interactions shown in Figure 2.
Figure 1.
Stereo views of the most favorable models of the active sites of RdpA (A) and SdpA (B) with bound substrates. Shown are selected protein residues that are predicted to interact with the substrate. For clarity reasons, the FeII ligands are not depicted. (R)- and (S)-mecoprop are indicated in light blue and in turquoise, respectively, and αKG in yellow.
Figure 2.
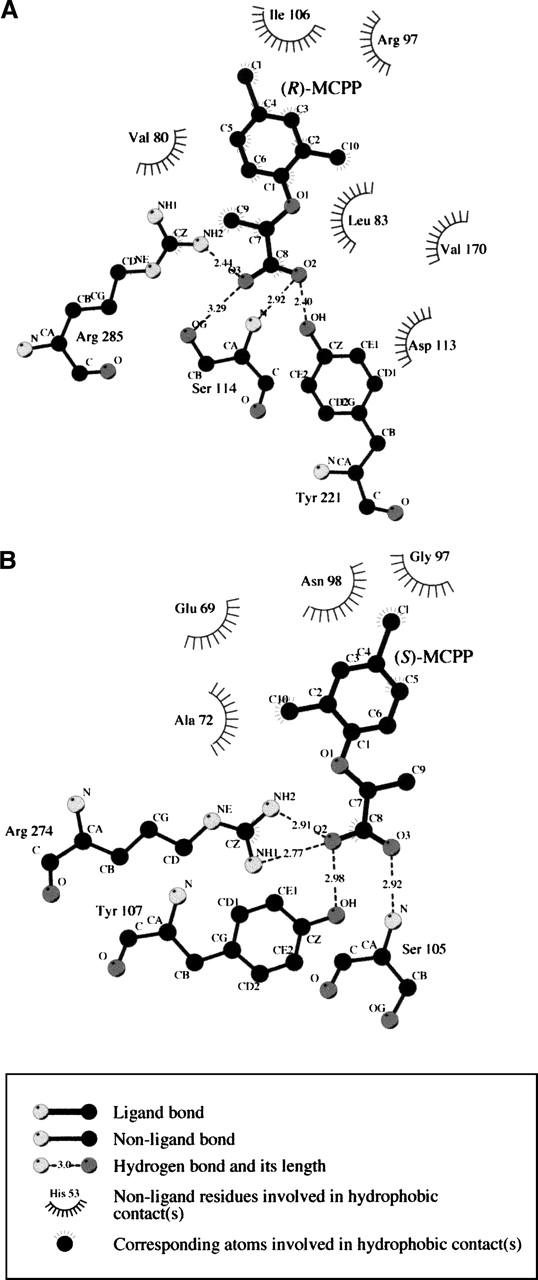
LIGPLOT diagrams of the active site models of RdpA (A) and SdpA (B). Shown are the most important interactions between the substrate and the active site according to the model.
Binding of (R)-mecoprop to RdpA
The substrate (R)-mecoprop consists of a hydrophobic phenoxy ring and a polar propanoic acid, with both components needing to be accommodated and bound by the active site. The αKG conformation leading to the most favorable interactions has αKG in the twist conformation and positions the phenoxy ring of (R)-mecoprop as illustrated in Figure 1A (with the corresponding interactions plotted in Fig. 2A). The mecoprop carboxylate interacts with the amide nitrogen of Ser114, the hydroxyl group of Tyr221, and a guanidino nitrogen of Arg285. The Tyr221 hydroxyl group also is predicted to lie near (3.5 Å) the substrate ether oxygen atom and could play a role in directing enantiospecificity. Residues lining the hydrophobic substrate-binding pocket include Val80, Leu83, Ile106, Gly107, and Phe171 (Figs. 1A, 2A), with Val80 and Leu83 being well positioned to interact with the propanoic acid methyl group. The terminal carbon atom (CZ) of the Phe171 side chain is 4.1 Å from the phenoxy group of (R)-mecoprop; since LigPlot has a 4.0 Å threshold for hydrophobic interactions, this interaction is missed in Figure 2A.
To directly test the importance of potential substrate-binding residues of RdpA identified by the homology modeling and substrate docking procedures, variant forms of the enzymes were created by site-directed mutagenesis. To eliminate the bulky and polar Tyr221 and Arg285 residues, Y221A and R285A mutants were generated. In the presumed “hydrophobic pocket,” Val80, Leu83, Ile106, and Phe171 each were changed to alanine to reduce hydrophobic interactions and thereby decrease the binding affinity of (R)-mecoprop to the active site. Replacing Gly107—positioned near and coplanar with the phenoxy ring—with a bulkier hydrophobic residue is expected to hinder substrate binding, so this residue was mutated to Ile and Asn in the double mutants I106G/G107I and I106G/G107N, respectively.
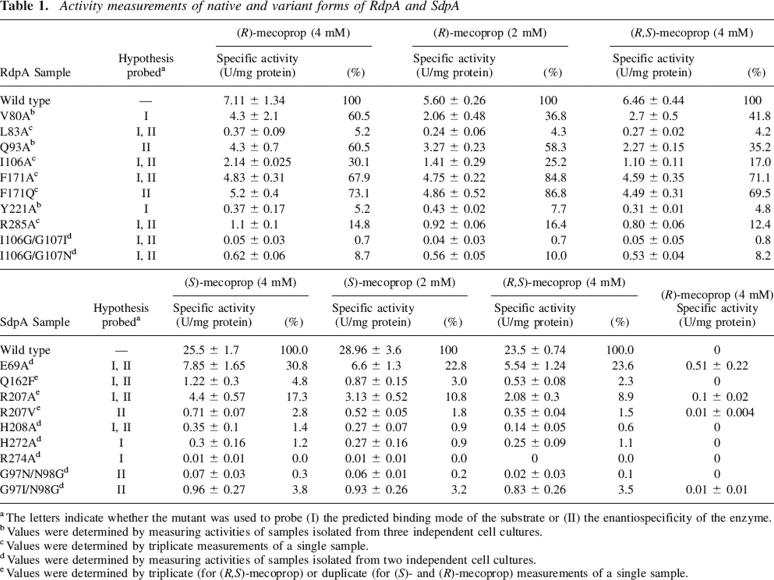
The specific activities of the RdpA variants were tested using (R)-mecoprop and the racemic mixture (Table 1). The (S) enantiomer is not sold commercially and was available in very limited supply, so the RdpA variants were not tested with this compound. When the RdpA variants were examined using 4 mM (R)-mecoprop, the V80A and F171A variants exhibited ∼60% of wild-type enzyme activity; I106A had ∼30% of that activity and L83A, Y221A, and R285A were ∼10% active. The activities of the double mutants I106G/G107I and I106G/G107N were further reduced compared with that of I106A, consistent with a bulkier side chain at position 107 presenting steric hindrance to substrate binding in a reactive conformation.
Table 1.
Activity measurements of native and variant forms of RdpA and SdpA
The RdpA variants retaining at least 10% of wild-type enzyme activity were subjected to more detailed kinetic characterization (Table 2). The maximal concentration of substrates that could be tested was 4 mM due to solubility limitations; therefore, Km values higher than 800 μM are only approximations and possess large errors. The Km values of V80A and R285A RdpA variants were at least fivefold increased over that of the wild-type enzyme, whereas that of the I106A RdpA variant was more than 20-fold greater, supporting the described docking orientation of the model. The effect on the R285A variant can be understood in terms of decreased interaction with the substrate carboxylate, while the changes observed for the V80A and I106A variants are likely to arise from loss of hydrophobic interactions. The F171A protein had a threefold lower Km, indicating either that this residue does not interact specifically with the phenoxy ring of (R)-mecoprop or that F171 is actually slightly hindering (R)-mecoprop binding. The calculated kcat of all mutant proteins was similar to the wild-type value with the exception of the R285A mutant enzyme. Arg285 is postulated to interact with both the mecoprop carboxylate and the C-1 carboxylate of αKG, so it could directly influence catalysis.
Table 2.
Kinetic parameters of selected variant forms of RdpA and SdpA
Binding of (S)-mecoprop to SdpA
The most favorable orientation of (S)-mecoprop was obtained with αKG in the twist conformation in SdpA and (S)-mecoprop bound as illustrated in Figure 1B (with the corresponding ligand interactions shown in Fig. 2B). In this model, the substrate carboxylate interacts with the amide nitrogen of Ser105, the hydroxyl group of Tyr107, and the guanidino nitrogens of Arg274. Additional active site residues near the polar carboxylate include His272 and His208. Also of interest, the substrate ether oxygen atom is predicted to lie within 3.4 Å of the two carboxyl oxygens of Glu69. If protonated or bridged by the proton of a bound water molecule, the Glu69 carboxyl group could confer specificity to the (S) enantiomer by making a hydrogen bond with the ether oxygen (a prediction not borne out by experimental results, vide infra). Residues predicted to be in contact with the hydrophobic ring of (S)-mecoprop include Ala71, Ala72, Leu82, Val84, Gly97, Asn98, Gln162, and Arg207 (with the latter four residues depicted in Fig. 1B). π-Cation interactions between positively charged side chains like Arg207 and aromatic side groups like that in mecoprop can contribute very favorably to ligand binding (Mitchell et al. 1994).
To experimentally test the importance of several of these residues for the binding of (S)-mecoprop in the active site of SdpA, a series of mutant proteins was constructed. Glu69, His208, His272, and Arg274 each were changed to alanine to eliminate the bulky and charged side chains proposed to interact with the substrate carboxylate. The extended side chain of Arg207 was eliminated in the R207A SdpA variant, and Gln162 was substituted with the corresponding hydrophobic residue found in RdpA to generate the Q162F variant.
The specific activities of the SdpA variants were tested using the (S) enantiomer, the racemic mixture, and the (R) enantiomer of mecoprop (Table 1). The activity of all SdpA mutants was strongly reduced. The most active SdpA variants were the E69A and R207A proteins, while all other variants exhibited <5% of the wild-type enzyme activity. No activity was detected in the R274A sample. E69A, R207A, and H208A were subjected to detailed kinetic characterization (Table 2). When analyzed with (S)-mecoprop, the R207A and H208A samples had 15- and 18-fold increases, respectively, in their Km values compared with the wild-type enzyme. The H208A result is compatible with an ionic interaction between the mecoprop carboxylate group and His208. A similar interaction might be possible with the highly flexible Arg207 residue, or this side chain may participate in a π-cation interaction with the aromatic group of the substrate, in either case accounting for the large Km change in the R207A variant. In contrast to these results, the E69A variant exhibited a lower Km than the native SdpA, arguing against the ether oxygen having a significant role in substrate binding. With (S)-mecoprop, the kcat was reduced fourfold for the E69A and R207A SdpA mutants compared with wild-type enzyme, and even greater reductions were noted for the H208A variant.
Residues determining enantiospecificity in RdpA and SdpA
A direct comparison of the active sites in the RdpA and SdpA models is shown in Figure 3. The different substrate enantiomers are predicted to bind with remarkably similar geometries in the two predicted structures, with the carboxyl groups and aromatic rings nearly overlapping but at distinct angles. RdpA residues Leu83, Gln93, Ile95 (omitted for clarity), Ile106, Phe171, and Arg285 (shown in Fig. 1A) could hinder binding of (S)-mecoprop; the corresponding residues in SdpA are Ala72, Leu82, Val84, Gly97, Gln162, and His272, respectively. Similarly, SdpA residues Glu69, Asn98, Ser161, His208, and Arg207 could restrict binding of (R)-mecoprop; Val80, Gly107, Val170, Tyr221, and Val220 occupy these positions in RdpA. Ala72, Gly97, and Gln162 are predicted to allow access by the (S) enantiomer. In general, side-chain interactions suggest that SdpA is less specific than RdpA for its substrate. This finding is compatible with activity measurements (Table 1), such as the activity with 2 mM (S)-mecoprop versus that with 4 mM of the racemate, that show inhibition of SdpA activity by (R)-mecoprop. However, RdpA remains completely active toward the (R) enantiomer in the presence of (S)-mecoprop (Müller 2004).
Figure 3.
Stereo views of the superimposition of the substrate-bound active site models of RdpA and SdpA. Shown are selected residues predicted to be involved in dictating enantiospecificity. Amino acid residues of RdpA are depicted in pink; residues of SdpA, in green. (R)-mecoprop is shown in purple; (S)-mecoprop, in turquoise. (A) View with the focus on the propanoic acid moiety and its binding to the active sites. (B) Same as in A but rotated to focus on the phenoxy ring and its interaction with the amino acid residues Ile106/Gly107 in RdpA and Gly97/Asn98 in SdpA.
Site-directed variants of RdpA and SdpA, created and purified as described earlier, were used to test a subset of the above-mentioned residues for their importance in controlling enantioselectivity. Single substitutions included the L83A, Q93A, I106A, F171A, F171Q, and R285A variants of RdpA along with the E69A, Q162F, R207A, R207V, and H208A variants of SdpA. In addition, to test the possibility that Ile106/Gly107 (large residue/small residue) in RdpA versus Gly97/Asn98 (small residue/large residue) in SdpA confers some measure of enantiospecificity to the enzymes, the I106G/G107I and I106G/G107N double variants of RdpA along with G97N/N98G and G97I/N98G double variants of SdpA were generated. Each enzyme was tested for activity using its enantiomer, the racemate, and in the case of the SdpA mutants, with (R)-mecoprop, too (Tables 1, 2).
Among the RdpA variants were several that show evidence of inhibition by the (S) enantiomer. For example, the Q93A variant retains 60% of wild-type enzyme activity, but this dropped to 35% when the racemate was provided, suggesting that Gln93 prevents the incorrect enantiomer from binding to and inhibiting the enzyme. Significant decreases in activity also were apparent when comparing rates using the racemic mixture of mecoprop versus the (R)-enantiomer with the Y221A and R285A variants; these results suggest that Tyr221 and Arg285 contribute to enantiospecificity. These variant proteins exhibited ∼5% to ∼15% of the wild-type enzyme activity. Less dramatic differences were observed using the I106A, F171A, F171Q, and I106G/G107N variants, but in each case the activity was reduced in the presence of the incorrect enantiomer. The I106A, F171A, F171Q, and R285A proteins were subjected to detailed kinetic analysis with (R)-mecoprop (Table 2). With the exception of the R285A variant (which exhibited ∼20% of the wild-type enzyme kcat), the proteins retained >70% of the wild-type enzyme kcat. The I106A and R285A variants exhibited 23- and fourfold increases in Km; the F171Q form exhibited a Km similar to the wild-type enzyme, and the F171A mutant had a threefold lower Km. The latter result is consistent with Phe171 hindering binding of the correct (R) enantiomer while helping to exclude the (S) enantiomer from the RdpA active site, so that the expanded active site in the F171A variant more readily binds its substrate.
Significantly, four SdpA variants (E69A, R207A, R207V, and G97I/N98G) showed slight activity with the opposite enantiomer, (R)-mecoprop (Table 1). These results suggest that Glu69, Arg207, and Gly97/Asn98 contribute to determining the enantiomeric specificity, as predicted by docking. They are compatible both with the postulated interaction of Glu69 with the (S)-mecoprop ether oxygen and with steric constraints on (R)-mecoprop binding imposed by Glu69, Arg207, and Asn98. Kinetic analyses of the E69A variant (Table 2) argue against a positive, stabilizing interaction between Glu69 and the substrate ether atom, because the Km of this variant is less than that of the wild-type enzyme. These results are most consistent with Glu69 discriminating against binding by the opposite enantiomer while slightly hindering binding of the correct enantiomer. The apparent Km of wild-type SdpA was approximately doubled when examined using the racemic mixture, as expected from earlier work (Müller 2004). Similar results were obtained for the E69A variant, while the Km was essentially unchanged for the R207A protein when comparing the single enantiomer versus the racemate. The R207A Km results are consistent with both substrate enantiomers binding to the active site with similar affinities. This point was directly demonstrated by assaying the R207A enzyme with (R)-mecoprop, yielding a Km of 4.0 ± 1.3 mM and a kcat of ∼0.16 min−1. For comparison, the Km of E69A SdpA for (R)-mecoprop was determined to be 5.4 ± 1.7 mM with a kcat of 65 ± 15 min−1. The kcat was further reduced for the E69A and H208A variants when using the racemic mixture, consistent with binding and inhibition by the (R) enantiomer. The low activities of the H208A and G97I/N98G proteins precluded a similar analysis.
Discussion
On the basis of the modeled RdpA and SdpA structures, we identified several amino acid residues that are likely to interact with (R)- and (S)-mecoprop, including several that are likely to contribute to the enantiospecificities of the two enzymes. The structural modeling performed here incorporated small-scale conformational changes in optimizing the protein–ligand complex through flexible docking using SLIDE and energy minimization of the structures. However, many enzymes, including selected representatives of the FeII/αKG dioxygenases (Clifton et al. 2006), are known to undergo larger-scale conformational changes upon ligand binding. The problem then becomes one of predicting protein conformational change as well as the orientation of ligand binding, a prodigious task for which no generally proven method exists. Success has been shown on subproblems for some complexes, e.g., predicting relative degrees of flexibility within a protein and how this flexibility redistributes upon complex formation (Gohlke et al. 2004), predicting large-scale conformational change between two protein states (Tama et al. 2004), evaluating conformational ensembles of the ligand and protein structures that are consistent with binding (Zavodszky et al. 2004), and predicting ligand binding by docking into a series of experimentally known enzyme conformations that approximate larger-scale protein flexibility (Lorber and Shoichet 1998). Molecular dynamics simulation of the protein–ligand complex is another option. However, it is generally difficult to access substantially different ligand orientations and protein conformations in molecular dynamics simulations. In our case, the situation is further complicated given the trimeric state of RdpA and the possible influence of subunit interfaces on active-site conformations. Therefore, the accuracy of homology modeling and prediction of (R)- and (S)-mecoprop interactions with the RdpA and SdpA enzymes may be limited by not anticipating medium- to large-scale protein conformational change. The predictive value of this modeling—identifying residues likely to be responsible for recognizing the (R) versus (S) enantiomer of mecoprop—can be assessed by consistency with the binding effects of mutations of these residues. Site-specific variants of RdpA and SdpA were created, their activities measured, and, in selected cases, detailed kinetic experiments were performed. The results confirm the residues involved in substrate or cosubstrate binding and provide useful insights into the structural basis of enantiomeric specificity.
Basis of the substrate-binding specificity of RdpA and SdpA
(R)- and (S)-mecoprop docked to the predicted structures of RdpA and SdpA, respectively, indicate that the mecoprop carboxylates and phenoxy rings are similarly positioned in the two proteins, with the substrate methyl group oriented in opposite configurations. The activity and kinetic data for R285A and Y221A variants of RdpA are compatible with the (R)-mecoprop carboxylate interacting with both the guanidino group of Arg285 and the hydroxyl group of Tyr221, where Tyr221 also forms a potential hydrogen bond to the ether oxygen atom. Similarly, activity and kinetic measurements of the R274A and H208A proteins are consistent with SdpA using the corresponding residues Arg274 and His208 to bind the carboxylate of (S)-mecoprop. Comparable Arg residues are conserved in many FeII/αKG-dependent dioxygenases, where they play a dual role of binding the primary substrate and the C1-carboxyl group of αKG (Valegård et al. 1998; Elkins et al. 2002; O'Brien et al. 2003; Hausinger 2004; Müller et al. 2004a). Such a dual role is predicted for Arg285 of RdpA, whereas Arg274 and His272 split these functions in SdpA.
Examination of the mutant forms of RdpA and SdpA also provided insights into the specificity for the distinct mecoprop enantiomers. For RdpA variants, Gln93, Ile106, and Phe171 are proposed to sterically exclude binding by the incorrect enantiomer, whereas the Q93A variant yields results consistent with inhibition by (S)-mecoprop. Similarly, the results with the SdpA mutants are consistent with Glu69, Asn98, and Arg207 and His208 acting to prevent binding of the (R) enantiomer. In particular, the E69A, R207A, and G97I/N98G variants exhibited activity with the incorrect substrate, (R)-mecoprop. Hydrophobic interactions are also likely to be critical for substrate discrimination. For example, Val80 and Leu83 in RdpA are predicted to be well positioned to interact with the propanoic acid methyl group, whereas Glu69 and Ala72 occupy these positions in SdpA. We conclude that several residues in each protein play a role in dictating the substrate enantiospecificity.
Comparison of the proposed RdpA- and SdpA-substrate interactions with 2,4-D binding by TfdA
RdpA and SdpA are 30% and 37% identical to TfdA, for which a homology model was previously constructed and its 2,4-D substrate docked (Elkins et al. 2002). Predicted interactions in theTfdA model previously were tested by site-directed mutagenesis and kinetic analyses (Hogan et al. 2000; Dunning Hotopp and Hausinger 2002), similar to what is described here for RdpA and SdpA. In the case of TfdA, the substrate carboxylate was suggested to interact with Arg278, His214, and Lys71. While the TfdA Arg and His residues have direct counterparts in the mecoprop-degrading enzymes (Arg285 and Tyr221 in RdpA; Arg274 and His208 in SdpA), there is no indication of a carboxylate-stabilizing Lys in RdpA or SdpA. Also of interest, Lys71 and/or Lys95 of TfdA was suggested to interact with the ether oxygen atom of 2,4-D. Again, a Lys does not appear to function in this manner in the mecoprop-degrading enzymes; however, Tyr221 of RdpA is reasonably well positioned for this role.
TfdA is slightly active with (S)-dichlorprop, but not with (R)-dichlorprop (Saari et al. 1999). Based on secondary structure alignment, TfdA residues Ala109 and Asn110 correspond to the Gly97/Asn98 (small residue/large residue) pair in SdpA, versus Ile106/Gly107 in RdpA. Evidence described earlier is consistent with this pair of RdpA and SdpA residues contributing to the enantiospecificity of these enzymes. The presence of (S)-dichlorprop-degrading activity in TfdA supports this proposal. 2,4-D converting TfdAs from other strains also show slight activities with phenoxypropanoic acids. Like TfdA from C. necator JMP134(pJP4), TfdA from Burkholderia cepacia strain RASC cleaves the ether bond of (S)-dichlorprop (Saari et al. 1999), whereas TfdA from Alcaligenes denitrificans exclusively converts the (R) enantiomer (Tett et al. 1997). Interestingly, PCR-derived DNA fragments reveal that these enzymes share 86% amino acid sequence identity in the corresponding region of the proteins (Saari et al. 1999); if the entire proteins exhibit this level of identity, the enantiospecificity is likely to be conferred by a few key amino acid residues.
Conclusion and relationship to other enantiospecific enzymes
The stereocenter-recognition-model of Sundaresan and Abrol (2002, 2005), much like the simpler three-point interaction model (Davankov 1997), generalizes the interactions between a chiral substrate and an enzyme or a receptor by postulating that at least three points of interaction are needed to distinguish between enantiomers with one chiral center. For the enantiospecific binding of mecoprop to RdpA and SdpA, the active site could be expected to interact with the carboxylic acid, either the ether oxygen atom or the phenoxy ring, and the methyl group. The predicted substrate interactions and kinetic results for RdpA and its variants are consistent with such mecoprop interactions. That is, several protein side chains are predicted to position the carboxylate, and Tyr221 likely forms a hydrogen bond to the ether oxygen, while several residues, notably the Ile106/Gly107 pair, position the phenoxy ring, and Val80 and Leu83 interact with the methyl group. In contrast, the SdpA model displays only two clear interactions with (S)-mecoprop; Tyr107 and Arg274 side chains position the carboxylate, and several, including the Gly97Asn98 pair, position the phenoxy ring, but the methyl group exhibits little interaction with the active site. Therefore, the active site of SdpA appears to be inherently less enantiospecific for (S)-mecoprop and more easily binds the opposite (R) enantiomer. The observed inhibition of SdpA by the (R) enantiomer and the reactivity of certain SdpA variants with (R)-mecoprop support this hypothesis.
In general, enantioselectivity is known to arise from both stabilization of the preferred substrate and relative destabilization of binding by the other enantiomer (Sundaresan and Abrol 2002, 2005). For example, tropinone reductase (TR)-1 and TR-II reduce tropinone in a stereoselective manner to produce tropine and Ψ-tropine. Tropinone is differently oriented in the active sites of the two enzymes, with both proteins providing stabilizing interactions and TR-I acting to prevent the inappropriate binding mode by a repulsion interaction involving the positively charged nitrogen of tropinone and a positively charged histidine residue (Nakajima et al. 1998). RdpA and SdpA are proposed to conform to this general pattern by using both positive and negative interactions to enhance binding by the appropriate enantiomer.
Materials and methods
Recombinant plasmids
Mutagenesis of rdpA and sdpA within pMec15 and pMec19 was carried out by using the Stratagene QuikChange System (Stratagene) and the mutagenic primers listed in Supplemental Table S1. Each mutation was confirmed by sequence analysis (Davis Sequencing). These constructs produce “wild-type” and variant enzymes as His6-tagged fusion proteins.
Enzyme purification
Escherichia coli C41(DE3) (Miroux and Walker 1996) cells harboring pMec15, pMec151, pMec152, pMec153, pMec154, pMec155, pMec156, pMec157, pMec158, pMec160, and pMec161 were used to purify His6-tagged wild-type RdpA and its V80A, Q93A, Y221A, R285A, I106G/G107I, I106G/G107N, L83A, I106A, F171A, and F171Q variants, respectively. Similarly, E. coli BL21(DE3) (Novagen) cells harboring pMec19, pMec191, pMec192, pMec193, pMec194, pMec195, pMec196, pMec197, and pMec198 were used to purify His6-tagged wild-type SdpA and its E69A, H208A, H272A, H274A, G97N/N98G, G98I/N98G, Q162F, R207A, and R207V variants, respectively. The cells were grown with constant shaking in Erlenmeyer flasks containing terrific broth supplemented with 200 μg/mL of ampicillin. Starter cultures grown overnight at 37°C were used to inoculate (0.5% [v/v]) medium for growth at room temperature. Upon reaching an OD600 between 0.3 and 0.7, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 1 mM, and the cultures were grown for an additional 16–18 h. To overexpress wild-type and variant forms of His6-tagged RdpA in E. coli C41(DE3) cells, it was important to fill flasks to 50% of their volume with medium and that the shaking be kept at moderate levels to minimize aeration and decrease growth rate. The cells were harvested by centrifugation at 6000g for 10 min at 4°C. To prepare cell extracts, each 1 g of the cell pellet was resuspended in 2.5 mL of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole at pH 8), and the cells were disrupted by sonication with cooling on ice between pulses. The disrupted cells were centrifuged at 130,000g for 60 min at 4°C, and the clarified cell extracts were stored in aliquots at –80°C. RdpA and SdpA accounted for up to 23% and 33% of the protein in the respective soluble cell extracts.
His6-RdpA and its variants were purified by a one-step procedure carried out at room temperature. Up to 100 mg of protein was loaded onto a Ni-bound NTA-agarose column (5 × 2.5 cm; Invitrogen) in lysis buffer. Unbound proteins were removed from the column with five column volumes of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole at pH 8). His6-RdpA and its variants were released from the resin with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 200 mM imidazole at a pH adjusted to 8) containing 20% glycerol to increase stability. His6-SdpA and its mutants were purified as described for the His6-RdpA, except that no glycerol was included in the elution buffer (see representative purifications in Supplemental Fig. S3). Purified proteins were stored on ice for up to 4 d.
Protein concentrations were determined by using the Bio-Rad protein assay (Bio-Rad) with bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis was carried out according to established procedures (Laemmli 1970).
Assays
Typical assays contained 100 μM (NH4)2Fe(SO4)2, 1 mM ascorbic acid, 1 mM αKG, and the indicated amount of substrate in 100 mM imidazole buffer (pH 6.75). All assays were carried out at 30°C. To determine kinetic parameters for phenoxypropanoic acids, the coupled continuous enzyme assay was used as previously described (Müller 2004). Units of activity were defined as micromoles of product produced per min per mg protein. Values of kcat were calculated using Mr 34,950 for the His6-RdpA variants and 33,460 for the His6-SdpA variants.
Structural modeling
Homology models of RdpA and SdpA were generated on the basis of the structure of the related taurine/αKG dioxygenase (TauD) from E. coli (O'Brien et al. 2003). With 33% and 29% identity to RdpA and SdpA and no significant gaps in the alignment, TauD (PDB code 1OS7, chain A) meets the criterion of at least 25% identity over at least 80 residues (Sander and Schneider 1991) to be an accurate structural template for modeling. Multiple alignments for the dichlorprop hydroxylase structures were obtained with the Bioinfo Meta Server (http://bioinfo.pl/Meta), and one was selected based on optimal agreement with predicted secondary structures. Side chains of RdpA and SdpA were reconstructed with the MMTSB Tool Set (Feig et al. 2000, 2004). Nonconserved loops and missing residues were added with Modeller (Fiser et al. 2000, 2002). The iron was placed into the active site according to the TauD structure, and the side chains in the predicted binding sites were adjusted using Insight II (Accelrys) to have similar conformations to the corresponding side chains from TauD, since conserved residues in catalytic sites tend to be highly structurally conserved. Catalytic sites are even more strongly conserved in structure when they involve a metal center, which strongly orients its ligating side chains. In particular, residues His111, Asp113, and His270 in RdpA and His102, Asp104, and His257 in SdpA were repositioned to bind the FeII metallocenter as they do in TauD, and αKG was positioned with its C-1 carboxylate plus C-2 carbonyl group chelating the metal and its C-5 carboxylate forming an ion pair with Arg268 or Arg281, respectively. The conformation of the free αKG (ID code COTPAC) was obtained from the Cambridge Structural Database (CSD).
To model αKG interactions with RdpA and SdpA as accurately as possible, multiple bound αKG conformations were retrieved from crystal structures of other αKG-dependent dioxygenases (PDB codes 1OS7, 1GY9, 1GQW, 1OII, 1OIJ, 1OIK, 1NX4, and 1NX8). This analysis revealed two distinct conformations: The flat conformation has the five-member ring formed by chelating the FeII nearly coplanar with the C-5 carboxylate, whereas the twist conformation has the two planes at a 90° angle. Both conformations could be accommodated by RdpA and SdpA, and four models (RdpA and SdpA with the flat and the twist αKG conformation) were built and energy-minimized in the context of the protein structure, with an implicit aqueous solvent (Generalized Born with solvent accessibility correction) using the MMTSB tool set and the CHARMM22 force field (Brooks et al. 1983; Feig et al. 2004).
Docking of substrates
The energy-minimized structures with each of the αKG conformations were used as targets for docking with the program SLIDE (Schnecke and Kuhn 2000; Zavodszky et al. 2002), which models the small-scale protein and ligand side-chain rotations that are ubiquitous in induced fit between proteins and their ligands (Zavodszky and Kuhn 2005). The free (R)-mecoprop conformation was obtained from CSD. Alternative low-energy conformations for both the (R) and the (S) enantiomer, reflecting the range of conformations likely to be energetically accessible, were generated from connectivity information using Omega v1.8.b3 (OpenEye Scientific Software). The energy window allowed for the conformers was 20 kcal/mol, ensuring a very thorough search of the ligands' conformations. Nine conformers each of (R)- and (S)-mecoprop and of the CSD structure were generated, and all 27 conformers were docked into each of the four models (RdpA with flat and twisted αKG conformations, and likewise for SdpA) with SLIDE using the default parameter set (Schnecke and Kuhn 2000; Zavodszky et al. 2002). To identify the most likely binding orientation, the dockings of mecoprop into RdpA and SdpA were filtered using the following geometric criteria for catalysis: The distance between the FeII and the C7 carbon atom of mecoprop should be ∼4 Å, and the C7-H4 bond should orient toward the FeII. In addition, based on the interactions observed between enzymes of this class and their substrates, the carboxylate group of mecoprop is expected to bind in the same region and make similar interactions as the taurine sulfate group bound to TauD, anchoring the substrate for catalysis. Among the (R)- mecoprop dockings satisfying these criteria, the one with the most favorable interactions positioned the mecoprop phenoxy ring into the enlarged hydrophobic pocket formed by the replacement of bulky Tyr73 in TauD by Leu83 in RdpA, and the propanoic acid methyl group in the hydrophobic pocket created by replacing His70 of TauD with Val80. This comprised the favored RdpA-(R)-mecoprop model. The (S)-mecoprop dockings satisfying the above geometric criteria were very similar to each other, and the one with optimal protein-substrate hydrogen bonds was selected.
Figures 1, and 3, and Supplemental Figure S2 of RdpA and SdpA structures and active sites were produced using the program PyMOL (DeLano Scientific LLC). Ligand interactions were depicted using LIGPLOT v.4.4.2 (Wallace et al. 1995).
Electronic supplemental material
Coordinate files in PDB format containing the atomic coordinates of the modeled RdpA and SdpA structures in complex with (R)- and (S)-mecoprop and a document file containing a table, three figures, and their legends.
Acknowledgments
These studies were supported by a fellowship for prospective researchers from the Swiss National Science Foundation (to T.A.M.), by NSF CAREER grant 0447799 (to M.F.), and by the NIH (AI53877 to L.A.K and GM063584 to R.P.H.). We thank Professor Hans-Peter Kohler for generously providing (S)-mecoprop and OpenEye (Santa Fe, NM) for generously providing their Omega software for our use.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Robert P. Hausinger, Department of Microbiology and Molecular Genetics, 6193 Biomedical Physical Sciences, Michigan State University, East Lansing, MI 48824-4320, USA; e-mail: hausinge@msu.edu; fax: (517) 353-8957.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.052059406.
Abbreviations: 2,4-D, 2,4-dichlorophenoxyacetic acid; AlkB, alkylation-damaged DNA repair enzyme; AtsK, alkyl sulfatase; ANS, anthocyanidin synthase; CarC, carbapenam synthase; CAS, clavaminate synthase; CSD, Cambridge Structural Database; DAOCS, deacetoxycephalosporin C synthase; dichlorprop, 2-(2,4-dichlorophenoxy)propanoic acid; FIH, factor inhibiting hypoxia-inducible factor; αKG, α-ketoglutarate; mecoprop, 2-(4-chloro-2-methyl-phenoxy)propanoic acid; NTA, nitrilotriacetic acid; PAHX, phytanoyl-coenzyme A 2-hydroxylase; PDB, Protein Data Bank; RdpA, (R)-specific dichlorprop/αKG dioxygenase; SdpA, (S)-specific dichlorprop/αKG dioxygenase; TfdA, 2,4-D/αKG dioxygenase; TauD, taurine/αKG dioxygenase; TR, tropinone reductase; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
References
- Åberg B. 1973. Plant growth regulators. XXXI. Some monochloro-monomethylphenoxyacetic and related optically active propionic acids Swed. J. Agric. Res. 3 49–62. [Google Scholar]
- Ahrens W.H. 1994. Weed Science Society of America, Champaign, IL. In Herbicide handbook (WSSA) . 7th ed.
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations J. Comput. Chem. 4 187–217. [Google Scholar]
- Clifton I.J., Hsueh L.-C., Baldwin J.E., Harlos K., Schofield C.J. 2001. Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent dioxygenases Eur. J. Biochem. 268 6625–6636. [DOI] [PubMed] [Google Scholar]
- Clifton I.J., Doan L.X., Sleeman M.C., Topf M., Suzuki H., Wilmouth R.C., Schofield C.J. 2003. Crystal structure of carbapenem synthase (CarC) J. Biol. Chem. 278 20843–20850. [DOI] [PubMed] [Google Scholar]
- Clifton I.J., McDonough M.A., Ehrismann D., Kershaw N.J., Granatino N., Schofield C.J. 2006. Structural studies on 2-oxoglutarate oxygenases and related double-stranded β-helix fold proteins J. Inorg. Biochem. 100 644–669. [DOI] [PubMed] [Google Scholar]
- Cohen F.E., Gregoret L.M., Amiri P., Aldape K., Raily J., McKerrow J.H. 1991. Arresting tissue invasion of a parasite by protease inhibitors chosen with the aid of computer modeling Biochemistry 30 11221–11229. [DOI] [PubMed] [Google Scholar]
- Dann III C.E., Bruick R.K., Deisenhofer J. 2002. Structure of a factor-inhibiting hypoxia-inducible factor 1: An essential asparaginyl hydroxylase involved in the hypoxic response pathway Proc. Natl. Acad. Sci. 99 15351–15356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davankov V.A. 1997. The nature of chiral recognition: Is it a three-point interaction? Chirality 9 99–102. [Google Scholar]
- Donaldson D., Kiely T., Grube A. In Pesticides industry sales and usage. 1998 and 1999 market estimates pp. 1–32. 2002. U.S. Environmental Protection Agency, Washington, D.C.
- Dunning Hotopp J.C. and Hausinger R.P. 2002. Probing the 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase substrate binding site by site-directed mutagenesis and mechanism-based inactivation Biochemistry 41 9787–9794. [DOI] [PubMed] [Google Scholar]
- Elkins J.M., Ryle M.J., Clifton I.J., Dunning Hotopp J.C., Lloyd J.S., Burzlaff N.I., Baldwin J.E., Hausinger R.P., Roach P.L. 2002. X-ray crystal structure of Escherichia coli taurine/α-ketoglutarate dioxygenase complexed to ferrous iron and substrates Biochemistry 41 5185–5192. [DOI] [PubMed] [Google Scholar]
- Elkins J.M., Hewitson K.S., McNeill L.A., Seibel J.F., Schlemminger I., Pugh C.W., Ratcliffe P.J., Schofield C.J. 2003. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α J. Biol. Chem. 278 1802–1806. [DOI] [PubMed] [Google Scholar]
- Feig M., Rotkiewicz P., Kolinski A., Skolnick J., Brooks III C.L. 2000. Accurate reconstruction of all-atom protein representations from side-chain-based low-resolution models Proteins 41 86–97. [DOI] [PubMed] [Google Scholar]
- Feig M., Karanicolas J., Brooks III C.L. 2004. MMTSB tool set: Enhanced sampling and multiscale modeling methods for applications in structural biology J. Mol. Graph. Model. 22 377–395. [DOI] [PubMed] [Google Scholar]
- Fiser A., Do R.K., Sali A. 2000. Modeling of loops in protein structures Protein Sci. 9 1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A., Feig M., Brooks III C.L., Sali A. 2002. Evolution and physics in comparative protein structure modeling Acc. Chem. Res. 35 413–421. [DOI] [PubMed] [Google Scholar]
- Fukumori F. and Hausinger R.P. 1993a. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase J. Bacteriol. 175 2083–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori F. and Hausinger R.P. 1993b. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase J. Biol. Chem. 268 24311–24317. [PubMed] [Google Scholar]
- Gohlke H., Kuhn L.A., Case D.A. 2004. Change in protein flexibility upon complex formation: Analysis of Ras-Raf using molecular dynamics and a molecular framework approach Proteins 56 322–337. [DOI] [PubMed] [Google Scholar]
- Grzyska P.K., Ryle M.J., Monterosso G.R., Liu J., Ballou D.P., Hausinger R.P. 2005. Steady-state and transient kinetic analyses of taurine/α-ketoglutarate dioxygenase: Effects of oxygen concentration, alternative sulfonates, and active site variants on the Fe(IV) intermediate Biochemistry 44 3845–3855. [DOI] [PubMed] [Google Scholar]
- Hausinger R.P. 2004. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes Crit. Rev. Biochem. Mol. Biol. 39 21–68. [DOI] [PubMed] [Google Scholar]
- Hausinger R.P., Fukumori F., Hogan D.A., Sassanella T.M., Kamagata Y., Takami H., Saari R.E. 1997. Biochemistry of 2,4-D degradation: Evolutionary implications In Microbial diversity and genetics of biodegradation (eds. Horikoshi K.et al.) . pp. 35–51. Japan Scientific Press, Tokyo.
- Hegg E.L., Whiting A.K., Saari R.E., McCracken J., Hausinger R.P., Que Jr. L. 1999. Herbicide-degrading α-keto acid-dependent enzyme TfdA: Metal coordination environment and mechanistic insights Biochemistry 38 16714–16726. [DOI] [PubMed] [Google Scholar]
- Ho R.Y.N., Mehn M.P., Hegg E.L., Liu A., Ryle M.A., Hausinger R.P., Que Jr. L. 2001. Resonance Raman studies of the iron(II)-α-keto acid chromophore in model and enzyme complexes J. Am. Chem. Soc. 123 5022–5029. [DOI] [PubMed] [Google Scholar]
- Hogan D.A., Smith S.R., Saari E.A., McCracken J., Hausinger R.P. 2000. Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding J. Biol. Chem. 275 12400–12409. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4 Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Loos M.A. 1975. Phenoxyalkanoic acids In Herbicides chemistry: Degradation and mode of action (ed. Kaufmann D.D.) . pp. 1–128. Marcel Dekker, Inc, New York.
- Lorber D.M. and Shoichet B.K. 1998. Flexible ligand docking using conformational ensembles Protein Sci. 7 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matell M. 1953. Stereochemical studies on plant growth regulators. VII. Optically active α-(2-methyl-4-chlorophenoxy)-propionic acid and α-(2,4-dichlorophenoxy)-n-butyric acid and their steric relations Ark. Kemi 6 365–373. [Google Scholar]
- McDonough M.A., Kavanagh K.L., Butler D., Searls T., Oppermann U., Schofield C.J. 2005. Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease J. Biol. Chem. 280 41101–41110. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker J.E. 1996. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane protein and globular proteins at high levels J. Mol. Biol. 260 289–298. [DOI] [PubMed] [Google Scholar]
- Mitchell J.B.O., Nandi L., McDonald I.K., Thornton J.M., Price S.L. 1994. Amino/aromatic interactions in proteins: Is the evidence stacked against hydrogen bonding? J. Mol. Biol. 239 315–331. [DOI] [PubMed] [Google Scholar]
- Müller T.A. “Metabolism of phenoxyalkanoic acid herbicides in Sphingomonas herbicidovorans MH: Cloning and characterization of two enantiospecific α-ketoglutarate-dependent dioxygenases and degradation analysis,” pp. 167. Ph.D. thesis 2004. Swiss Federal Institute of Technology Zurich, Zurich.
- Müller R.H., Jorks S., Kleinsteuber S., Babel W. 1999. Comamonas acidovorans strain MC1: A new isolate capable of degrading the chiral herbicides dichlorprop and mecoprop and the herbicides 2,4-D and MCPA Microbiol. Res. 154 241–246. [DOI] [PubMed] [Google Scholar]
- Müller R.H., Kleinsteuber S., Babel W. 2001. Physiological and genetic characteristics of two bacterial strains utilizing phenoxypropionate and phenoxyacetate herbicides Microbiol. Res. 156 121–131. [DOI] [PubMed] [Google Scholar]
- Müller I., Kahnert A., Pape T., Sheldrick G.M., Meyer-Klaucke W., Dierks T., Kertesz M., Usòn I. 2004a. Crystal structure of the alkylsulfatase AtsK: Insights into the catalytic mechanism of the Fe(II) α-ketoglutarate-dependent dioxygenase superfamily Biochemistry 43 3075–3088. [DOI] [PubMed] [Google Scholar]
- Müller T.A., Byrde S.M., Werlen C., van der Meer J.R., Kohler H.-P.E. 2004b. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH Appl. Environ. Microbiol. 70 6066–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller I., Stückl C., Wakeley J., Kertesz M., Usòn I. 2005. Succinate complex crystal structures of α-ketoglutarate-dependent dioxygenase AtsK. Steric aspects of enzyme self-hydroxylation J. Biol. Chem. 280 5716–5723. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Yamashita A., Akama H., Nakatsu T., Kato H., Hashimoto T., Oda J., Yamada Y. 1998. Crystal structures of two tropinone reductases: Different reaction stereospecificities in the same protein fold Proc. Natl. Acad. Sci. 95 4876–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel K., Suter M.J.-F., Kohler H.-P.E. 1997. Involvement of two α-ketoglutarate-dependent dioxygenases in enantioselective degradation of (R)- and (S)-mecoprop by Sphingomonas herbicidovorans MH J. Bacteriol. 179 6674–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J.R., Schuller D.J., Yang V.S., Dillard B.D., Lanzilotta W.N. 2003. Substrate-induced conformational changes in Escherichia coli taurine/α-ketoglutarate dioxygenase and insight into the oligomeric structure Biochemistry 42 5547–5554. [DOI] [PubMed] [Google Scholar]
- Pavel E.G., Zhou J., Busby R.W., Gunsior M., Townsend C.A., Solomon E.I. 1998. Circular dichroism and magnetic circular dichroism spectroscopic studies of the non-heme ferrous active site in clavaminate synthase and its interaction with α-ketoglutarate cosubstrate J. Am. Chem. Soc. 120 743–753. [Google Scholar]
- Price J.C., Barr E.W., Glass T.E., Krebs C., Bollinger Jr. J.M. 2003a. Evidence for hydrogen abstraction from C1 of taurine by the high-spin Fe(IV) intermediate detected during oxygen activation by taurine:α-ketoglutarate dioxygenase (TauD) J. Am. Chem. Soc. 125 13008–13009. [DOI] [PubMed] [Google Scholar]
- Price J.C., Barr E.W., Tirupati B., Bollinger Jr. J.M., Krebs C. 2003b. The first direct characterization of a high-valent iron intermediate in the reaction of an α-ketoglutarate-dependent dioxygenase: A high-spin Fe(IV) complex in taurine/α-ketoglutarate dioxygenase (TauD) from Escherichia coli Biochemistry 42 7497–7508. [DOI] [PubMed] [Google Scholar]
- Price J.C., Barr E.W., Hoffart L.M., Krebs C., Bollinger Jr. J.M. 2005. Kinetic dissection of the catalytic mechanism of taurine:α-ketoglutarate dioxygenase (TauD) from Escherichia coli Biochemistry 44 8138–8147. [DOI] [PubMed] [Google Scholar]
- Proshlyakov D.A., Henshaw T.F., Monterosso G.R., Ryle M.J., Hausinger R.P. 2004. Direct detection of oxygen intermediates in the non-heme Fe enzyme taurine/α-ketoglutarate dioxygenase J. Am. Chem. Soc. 126 1022–1023. [DOI] [PubMed] [Google Scholar]
- Riggs-Gelasco P.J., Price J.C., Guyer R.B., Brehm J.H., Barr E.W., Bollinger Jr. J.M., Krebs C. 2004. EXAFS spectroscopic evidence for an Fe=O unit in the Fe(IV) intermediate observed during oxygen activation by taurine:α-ketoglutarate dioxygenase J. Am. Chem. Soc. 126 8108–8109. [DOI] [PubMed] [Google Scholar]
- Ryle M.J., Padmakumar R., Hausinger R.P. 1999. Stopped-flow kinetic analysis of Escherichia coli taurine/α-ketoglutarate dioxygenase: Interactions with α-ketoglutarate, taurine, and oxygen Biochemistry 38 15278–15286. [DOI] [PubMed] [Google Scholar]
- Saari R.E. and Hausinger R.P. 1998. Ascorbic acid-dependent turnover and reactivation of 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase using thiophenoxyacetic acid Biochemistry 37 3035–3042. [DOI] [PubMed] [Google Scholar]
- Saari R.E., Hogan D.A., Hausinger R.P. 1999. Stereospecific degradation of the phenoxypropionate herbicide dichlorprop J. Mol. Catal. B Enzym. 6 421–428. [Google Scholar]
- Sander C. and Schneider R. 1991. Database of homology-derived protein structures and the structural meaning of sequence alignment Proteins 9 56–68. [DOI] [PubMed] [Google Scholar]
- Schnecke V. and Kuhn L.A. 2000. Virtual screening with solvation and ligand-induced complementarity Perspect. Drug Discov. Des. 20 171–190. [Google Scholar]
- Sundaresan V. and Abrol R. 2002. Towards a general model for protein-substrate stereoselectivity Protein Sci. 11 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V. and Abrol R. 2005. Biological chiral recognition: The substrate's perspective Chirality 17(Suppl.), S30–S39. [DOI] [PubMed] [Google Scholar]
- Sukuru S.C.K., Crepin T., Milev Y., Marsh L.C., Hill J.B., Anderson R.J., Morris J.C., Rohatgi A., O'Mahony G., Grøtli M.et al. 2006. Discovering new classes of Brugia malayi asparaginyl tRNA synthetase inhibitors and relating specificity to conformational change J. Comput. Aided Mol. Des. (in press). [DOI] [PubMed]
- Tama F., Miyashita O., Brooks III C.L. 2004. Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis J. Mol. Biol. 337 985–999. [DOI] [PubMed] [Google Scholar]
- Tett V.A., Willetts A.J., Lappin-Scott H.M. 1997. Biodegradation of the chlorophenoxy herbicide (R)-(+)-mecoprop by Alcaligenes denitrificans Biodegradation 8 43–52. [Google Scholar]
- Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage Nature 419 174–178. [DOI] [PubMed] [Google Scholar]
- Vaillancourt F.H., Yeh E., Vosburg D.A., O'Connor S.E., Walsh C.T. 2005a. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis Nature 436 1191–1194. [DOI] [PubMed] [Google Scholar]
- Vaillancourt F.H., Yin J., Walsh C.T. 2005b. SyrB2 in syringomycin E biosynthesis is a nonheme FeII α-ketoglutarate- and O2-dependent halogenase Proc. Natl. Acad. Sci. 102 10111–10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K., Terwisscha van Scheltinga A.C., Lloyd M.D., Hara T., Ramaswamy S., Perrakis A., Thompson A., Lee W.-J., Baldwin J.E., Schofield C.J.et al. 1998. Structure of a cephalosporin synthase Nature 394 805–809. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thornton J.M. 1995. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions Protein Eng. 8 127–134. [DOI] [PubMed] [Google Scholar]
- Wilmouth R.C., Turnbull J.J., Welford R.W.D., Clifton I.J., Prescott A.G., Schofield C.J. 2002. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana Structure 10 93–103. [DOI] [PubMed] [Google Scholar]
- Worthing C.R. and Hance R.J. In The pesticide manual—A world compendium . 1991. 9th ed. The British Crop Protection Council, Farnham, UK.
- Yu B., Edstrom W.C., Benach J., Hamuro Y., Weber P.C., Gibney B.R., Hunt J.F. 2006. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB Nature 439 879–884. [DOI] [PubMed] [Google Scholar]
- Zavodszky M.I. and Kuhn L.A. 2005. Side-chain flexibility in protein-ligand binding: The minimal rotation hypothesis Protein Sci. 14 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky M.I., Sanschagrin P.C., Korde R.S., Kuhn L.A. 2002. Distilling the essential features of a protein surface for improving protein-ligand docking, scoring, and virtual screening J. Comput. Aided Mol. Des. 16 883–902. [DOI] [PubMed] [Google Scholar]
- Zavodszky M.I., Lei M., Day A.R., Thorpe M.F., Kuhn L.A. 2004. Modeling correlated main-chain motions in proteins for flexible molecular recognition Proteins 57 243–261. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ren J., Stammers D.K., Baldwin J.E., Harlos K., Schofield C.J. 2000. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase Nat. Struct. Biol. 7 127–133. [DOI] [PubMed] [Google Scholar]
- Zhou J., Kelly W.L., Bachmann B.O., Gunsior M., Townsend C.A., Solomon E.I. 2001. Spectroscopic studies of substrate interactions with clavaminate synthase 2, a multifunctional α-KG-dependent non-heme iron enzyme: Correlation with mechanisms and reactivities J. Am. Chem. Soc. 123 7388–7398. [DOI] [PubMed] [Google Scholar]
- Zipper C., Nickel K., Angst W., Kohler H.-P.E. 1996. Complete microbial degradation of both enantiomers of the chiral herbicide mecoprop [(RS)-2-(4-chloro-2-methylphenoxy)propionic acid] in an enantioselective manner by Sphingomonas herbicidovorans sp. nov Appl. Environ. Microbiol. 62 4318–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]