Abstract
The NADP-reducing hydrogenase complex from Desulfovibrio fructosovorans is a heterotetramer encoded by the hndABCD operon. Sequence analysis indicates that the HndC subunit (52 kDa) corresponds to the NADP-reducing unit, and the HndD subunit (63.5 kDa) is homologous to Clostridium pasteurianum hydrogenase. The role of HndA and HndB subunits (18.8 kDa and 13.8 kDa, respectively) in the complex remains unknown. The HndA subunit belongs to the [2Fe-2S] ferredoxin family typified by C. pasteurianum ferredoxin. HndA is organized into two independent structural domains, and we report in the present work the NMR structure of its C-terminal domain, HndAc. HndAc has a thioredoxin-like fold consisting in four β-strands and two relatively long helices. The [2Fe-2S] cluster is located near the surface of the protein and bound to four cysteine residues particularly well conserved in this class of proteins. Electron exchange between the HndD N-terminal [2Fe-2S] domain (HndDN) and HndAc has been previously evidenced, and in the present studies we have mapped the binding site of the HndDN domain on HndAc. A structural analysis of HndB indicates that it is a FeS subunit with 41% similarity with HndAc and it contains a possible thioredoxin-like fold. Our data let us propose that HndAc and HndB can form a heterodimeric intermediate in the electron transfer between the hydrogenase (HndD) active site and the NADP reduction site in HndC.
Keywords: NADP-reducing hydrogenase, thioredoxin-like domain, [2Fe-2S] ferredoxin, nuclear magnetic resonance (NMR), heteronuclear single quantum correlation (HSQC)
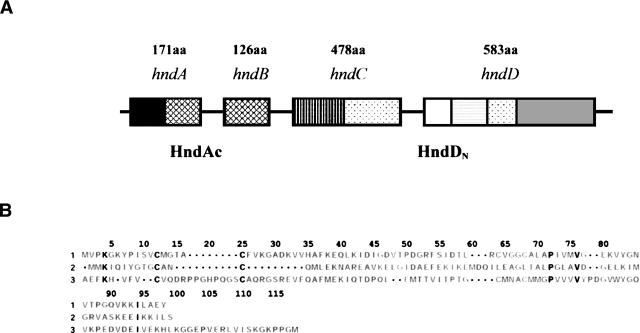
Hydrogenases are nonheme iron–sulfur proteins that catalyze the reversible oxidation of molecular hydrogen. They play a central role in the metabolism of sulfate-reducing bacteria belonging to the genus Desulfovibrio that use hydrogen as an electron donor and sulfate as a terminal electron acceptor. The Desulfovibrio fructosovorans (Df) strain differs from all other Desulfovibrio species by its ability to use fructose as a carbon source allowing to gain ATP by substrate level phosphorylation. This organism harbors a well-characterized [NiFe]-hydrogenase (Hatchikian et al. 1990) and a partially purified [Fe]-hydrogenase (Casalot et al. 1998). Both are periplasmic enzymes. In addition, a cytoplasmic enzyme encoded by an operon made up of four genes named hndA, hndB, hndC, and hndD (Fig. 1A), has been detected in this organism (Malki et al. 1995). HndD presents strong identity with Clostridium pasteurianum hydrogenase (Cp Hyd I) and constitutes the monomeric [Fe]-hydrogenase subunit. Indeed, HndD contains three domains in addition to the H-cluster domain: starting from the N terminus, a [2Fe-2S] plant ferredoxin-like domain, a [4Fe-4S] domain, and a domain homologous to the 2[4Fe-4S] bacterial ferredoxins. HndC is able to bind a flavin mononucleotide or dinucleotide, a NAD(P), and three [4Fe-4S] centers, and is designated as the NADP-reducing subunit. HndA contains a [2Fe-2S] cluster and might be involved in the intramolecular electron exchange. HndB was proposed to bind a prosthetic group (Dermoun et al. 2002). It was shown that the four subunits are organized in a cytoplasmic heterotetrameric complex that can perform H2-driven NADP-reduction and H2-driven methylviologen reduction. This tetrameric NADP reducing hydrogenase is closely related to the hexameric hydrogenase (HoxS) from Ralstonia eutropha (Re) encoded by the SH gene cluster hoxFUYHWI (Tran-Betcke et al. 1990; Bugdorf et al. 2005) and to the trimeric (HydABC) hydrogenase from Thermotoga maritima (Tm) (Verhagen et al. 1999). Nevertheless, the hydrogenase subunit is a [NiFe]-hydrogenase in R. eutropha and [Fe]-hydrogenases of the clostridial type are found in D. fructosovorans and T. maritima. The additional small subunits that are present in these enzymes increase the modular aspect of the hydrogenases (Vignais et al. 2001). HndA (from Df) and HydC (from Tm), which contain a [2Fe2S] cluster, are homologous to the NuoE subunit of complex I and to the N- terminal part of HoxF (from Re). These two small proteins are homologous to Escherichia coli [2Fe-2S] ferredoxin and to Aquifex aeolicus thioredoxin-like ferredoxin (Meyer et al. 1986; Yeh et al. 2000). HndB (from Df) is homologous to the N-terminal domain of HydB (from Tm) (Vignais et al. 2001; Dermoun et al. 2002).
Figure 1.
(A) Organization of the structural modules in the operon of the Hydrogenase NADP reductase from Desulfovibrio fructosovorans. (B) Sequence alignment of Desulfovibrio fructosovorans HndAc (1), Methanobacterium thermoautotrophicum thioredoxin (2), and Aquifex aeolicus ferredoxin (3) obtained by MOE software (http://www.chemcomp.com).
Df NADP-reducing hydrogenase is unique among sulfate reducing bacteria. The low level of expression of the HndABCD complex was a limit for a complete purification and thus a structural study of the whole enzyme. Overexpression of the different subunits has been attempted to perform reconstitution of the complex (De Luca et al. 1998; Dermoun et al. 2002). The HndA subunit was successfully overproduced in E. coli (De Luca et al. 1998). The UV-visible and EPR spectra of HndA indicated that a [2Fe-2S] cluster was correctly matured. However, the protein aggregated in the absence of its natural partners and was spontaneously hydrolyzed in two structural domains (De Luca et al. 1998). The C-terminal [2Fe-2S] domain of HndA (HndAc) and the truncated N-terminal [2Fe-2S] domain of HndD (HndDN) were expressed as soluble proteins in E. coli (Dermoun et al. 2002). An electron exchange between HndAc and HndDN redox centers was observed by EPR experiments, and a complex formation involving these two domains was evidenced by far-Western experiments (Dermoun et al. 2002). The folding of these structural domains and the maturation of their prosthetic groups proved the modular aspect of the NADP reducing hydrogenase (Fig. 1A). In this work, we report the structure of the HndAc domain and the mapping of the HndDN interacting site obtained by heteronuclear NMR spectroscopy, and we analyze the role of HndB within this NADP-reducing hydrogenase complex.
Results
HndAc structure calculation
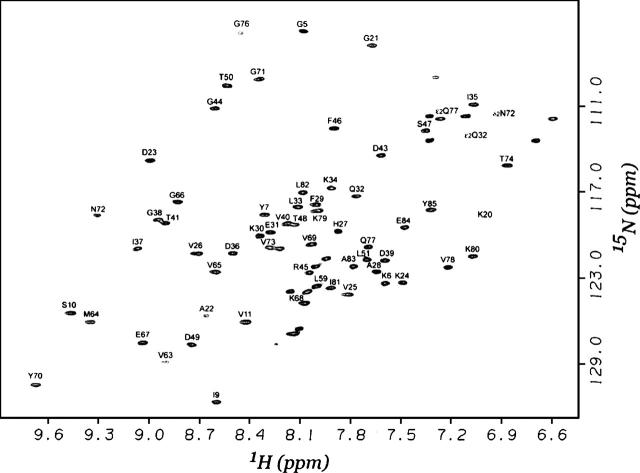
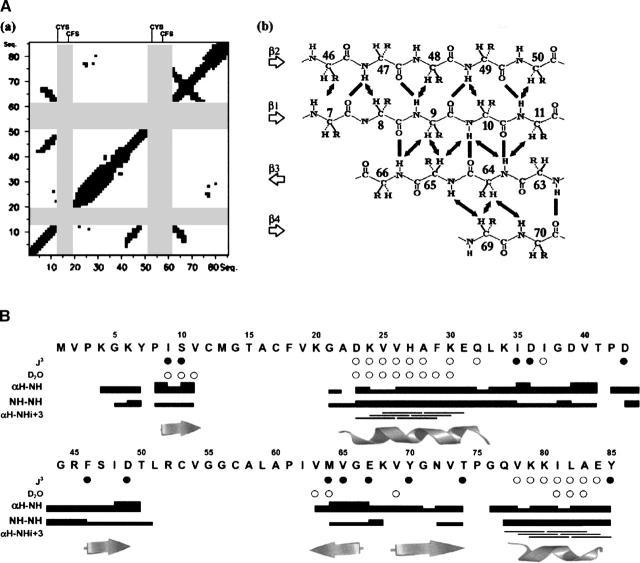
Backbone resonances of HndAc were assigned combining 3D NOESY-HSQC, TOCSY-HSQC, HNCA, HN(CO)CA, CBCA(CO)NH, HNCO, and HN(CA)CO experiments. The complete set of 1H, 13C, and 15N shifts has been deposited with the BioMagResBank (BMRB accession no. 6836). Due to the paramagnetic effect of the [2Fe-2S] cluster, the resonances of the residues C12–F18 and R52–I62 were not observed in the HSQC spectrum. As a consequence, among the 85 residues of the protein, the 15N-1H HSQC spectrum contains only 72 backbone amide resonances, and 61 were sequentially assigned (Fig. 2). Short- and medium-range nOes obtained from NOESY experiments and 13C chemical shift indices classify the protein in Class 3 (Mixed Alpha/Beta secondary elements) consisting of four β-strands (Fig. 3A) and two helices (Fig. 3B).
Figure 2.
2D 1H-15N HSQC spectrum of 1.6 mM 15N-labeled C-terminal domain of the HndA protein recorded at 293 K on a Bruker DRX 500 spectrometer. Sequential assignment is indicated. The NH2 of asparagine and glutamine side chains are connected by horizontal bars.
Figure 3.
(A) (a) Interresidue nOes identified in HndAc. One square was generated for each (i,j) pair of residues displaying nOe connectivities with each other (limits fixed to 6 Å). (b) The four β-strands are defined by interstrand main-chain nOes (arrows). Solid lines indicate predicted hydrogen bonds. (B) Characterization of HndAc secondary elements. 3JHNHA constants obtained from the HNHA experiment are represented by bold circles (coupling constants >7 Hz) and open circles (coupling constants <4 Hz). Nonexchanged amide protons in D2O spectra are represented by open circles. Interresidue nOes are represented by bars; their thickness indicates the intensity of the nOes. In gray are indicated the secondary elements.
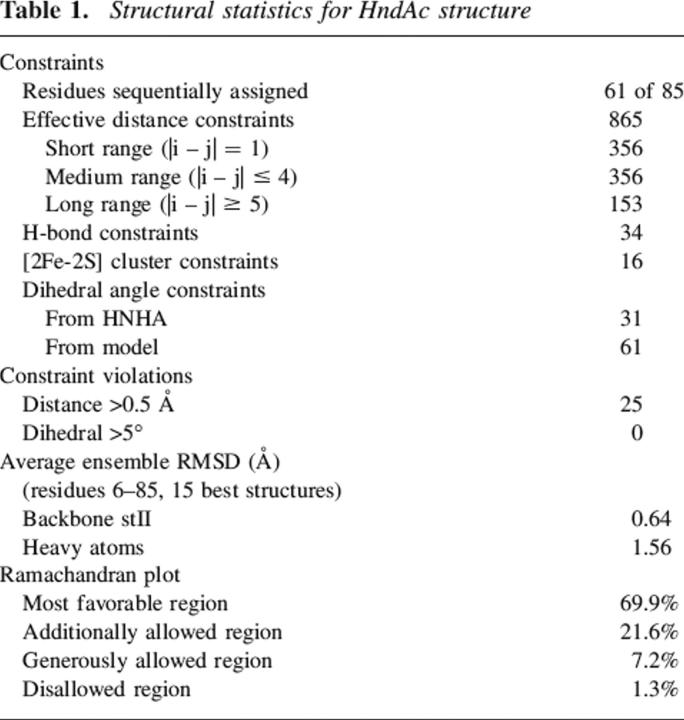
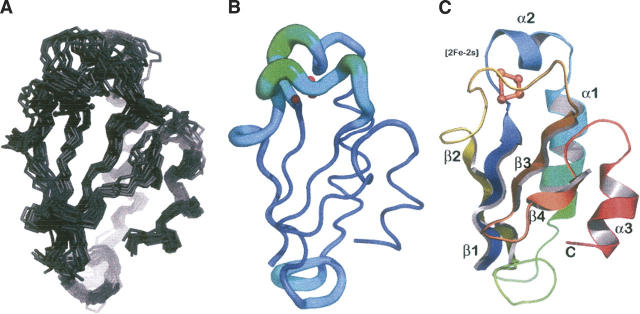
A total of 1007 constraints were used for HndAc structure calculation (Table 1). These comprise 356 short-range, 356 medium-range, and 153 long-range nOes, together with 31 dihedral constraints, as well as 34 H-bonds. We used the X-ray structure of A. aeolicus ferredoxin to model the [2Fe-2S] cluster and the unobservable part of HndAc. Sixteen [2Fe-2S] cluster distances and 61 dihedral angle constraints from the model were added to the previous experimental constraints. A superimposition of the final ensemble of 15 simulating annealed structures refined in an explicit water box is presented in Figure 4A. Among these structures, the representative structure (lowest energy and closest to average) is illustrated in Figure 4B. The conformers within the ensemble do not exhibit any nOe, dihedral angle, or scalar coupling violations >0.5 Å, 5°, and 2 Hz, respectively. Residues K6–Y85 are well structured in solution, and the coordinates of their backbone and heavy atoms can be superimposed to the average structure with a root mean square deviation (RMSD) of 0.64 (±0.19) Å and 1.56 (±0.60) Å, respectively. The structural statistics are summarized in Table 1. The ensemble of structures has been deposited with the Protein Data Bank (PDB code 2AUV). The τc measurements and nOe experiments are in agreement with a monomeric form of HndA.
Table 1.
Structural statistics for HndAc structure
Figure 4.
(A) Ensemble of 15 minimum energy structures of HndAc. (B) Representative structure of HndAc: The radius of the tube is proportional to the backbone RMSD of each residue. (C) Ribbon diagram of the representative structure of HndAc. The [2Fe-2S] cluster is shown in ball-and-stick form. The structural elements are labeled.
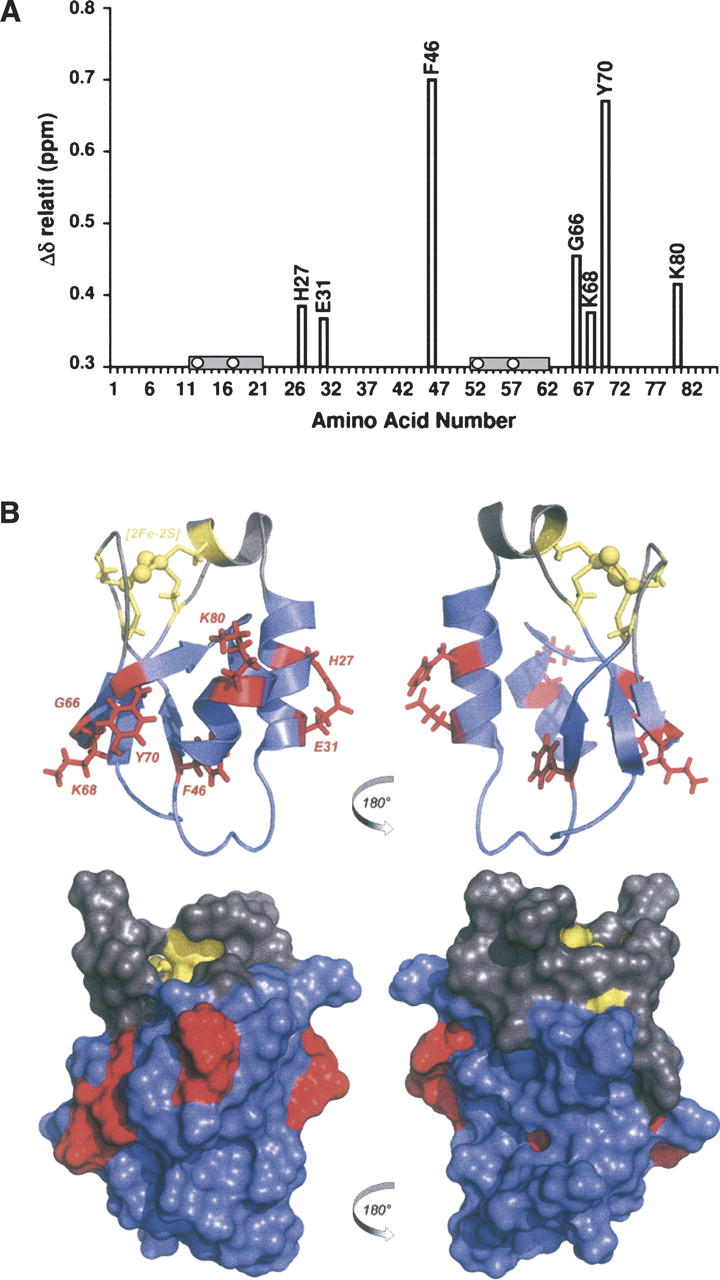
The HndAc protein adopts an overall structure classified as “three-layer αβα sandwich” or “thioredoxin-like domain,” which is characterized as having an N-terminal βαβ motif and a C-terminal ββα motif connected to each other by a third helix that does not exist in HndAc. The overall β-strand order is 2–1–3–4, with strand 3 antiparallel to the three other strands (Fig. 4C). The four β-strands in HndAc include residues I9–V11 (strand β1) and F46–T50 (strand β2), which run parallel to each other and antiparallel to the β-hairpin formed by residues V63–V65 (strand β3) and residues Y70–T74 (strand β4). The first helix (helix α1), from residues A16 to K20, is unusual in the thioredoxin-like motif. This helix is perpendicular to the helix α2, the longest helix, which runs from residues A22 to L33. Helix α3 is on the same side of the central β-sheet as helix α2 and is composed of residues Q77 to E84. Helix α3 is 30° tilted compared to helix α2. This spatial position stretches strand β4 bending the edge of the β-sheet. The main difference with Aa ferredoxin structure is the absence of the strand β5, which is involved in the interacting domain of the two subunits in Aa ferredoxin dimer.
Mapping of the HndDN interacting site on HndAc
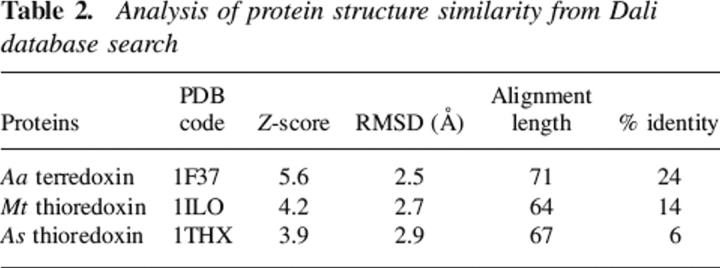
15N-1H HSQC spectra of HndAc domain were recorded in the presence and the absence of HndDN domain. Seven resonances (H27, E31, F46, G66, K68, Y70, and K80) were affected by the complex formation (Fig. 5A). The shift of these resonances during the titration indicates a fast exchange at the NMR timescale between the free and the bound forms of the two proteins. The mapping of the interacting surface is shown in Figure 5B. The contact interface is surrounded by charged residues (E31, K68, and K80) probably involved in electrostatic interactions with the HndDN subunit.
Figure 5.
Mapping of HndDN interacting site on HndA using NMR chemical shift variations. (A) Chemical shift variations observed for HndAc amide groups. In gray are the unobserved residues. Open circles indicate the position of the cysteine residues. (B) (Top) Ribbon diagram of HndAc structure; (bottom) molecular representation of NMR mapping of the HndDN interacting surface. In gray are the unobserved residues; in red, the affected residues; in blue, the unaffected residues; and in yellow, the FeS cluster. Two orientations with a 180° rotation of the molecule are presented.
Discussion
On the basis of the HndAc NMR structure, the search for homologous structures using the DALI (http://www.ebi.ac.uk/dali/) and SSM (http://www.ebi.ac.uk/msd-srv/ssm) databases (Table 2) identifies the thioredoxin-like [2Fe-2S] ferredoxin from A. aeolicus (PDB code 1F37) as the highest homologous protein of HndAc (0.35 Qscore and 2.13 Å RMSD from the SSM server). The thioredoxin from Methanobacterium thermoautotrophicum (PDB code 1ILO) is the second closely related protein (0.32 Qscore and 2.67 Å RMSD from the SSM server). Interestingly, the top hits from the two databases belong to known thioredoxins such as M. thermoautotrophicum (Mt) and Anabaena sp. (As) proteins. However, Dali server provides other related proteins belonging to multimeric enzymes, namely the thioredoxin-like domain of Hsco1 (Balatri et al. 2003; Williams et al. 2005) and the thioredoxin-like domain of SasA (Vakonakis et al. 2004). It was previously reported from sequence analysis that thioredoxin-like fold occurs in numerous proteins (Meyer 2001) and complexes such as hydrogenases (Albracht et al. 1997; Verhagen et al. 1999) or in complex I (NADH-ubiquinone oxidoreductase) of the aerobic respiratory chains (Yano et al. 1999).
Table 2.
Analysis of protein structure similarity from Dali database search
The secondary structure organization is highly conserved among the thioredoxin family. HndAc differs from this canonical motif βαβ(α)ββα by not having the central helix (Fig. 6). Instead, a supplemental helix is found after the first β-strand so that the overall topology from the N to the C terminus is β(α)αβββα. The four β-strands forming the core β-sheet are well conserved in Aa ferredoxin and in HndAc (Fig. 6). One can notice that an angle of 100° is formed by the β2 and β4 strands in HndAc. On the contrary, in Aa ferredoxin and thioredoxin structures this angle is ∼55°. The modification of the core β-sheet in HndAc is due to the increase of the length of the loop between β4 and α3. The orientation of the helices is conserved in the three proteins. Aa ferredoxin is a homodimer including an extended loop between β1 and α1, around the cluster. However, this loop, longer in Aa ferredoxin, is apparently not necessary for the redox properties or the stability of the [2Fe-2S] cluster in the thioredoxin-like domains (Yeh et al. 2000).
Figure 6.
Structural comparison of HndAc (A) and the two closest structural homologs: Aa ferredoxin (B) and Mt thioredoxin (C). The orientation of the α2 and α3 helices (HndAc numbering) is indicated. In orange are the cysteine residues of the active site of the thioredoxin (C) and the corresponding residues around the FeS cluster in Aa ferredoxin and HndAc. The FeS cluster is also in orange balls and sticks.
HndAc structure reveals that the [2Fe-2S] cluster is located near the surface of the protein, like it was already reported in the structure of the thioredoxin-like ferredoxin from A. aeolicus (Yeh et al. 2000): The [2Fe-2S] cluster is at the position of the disulfide bridge active site in thioredoxin (Fig. 1B). A thioredoxin-like fold appears to be relatively common and adopted as a protein design platform for incorporating metallocenters into proteins or essential residues for the activity of the protein (Coldren et al. 1997; Pinto et al. 1997; Wisz et al. 1998). Sco1 protein, which is required for the assembly of cytochrome oxidase, binds the copper atom through a highly conserved CxxxCP motif located at the thioredoxin active site and possibly histidine ligand (Balatri et al. 2003; Williams et al. 2005). Moreover, the particular placement of the cysteine residues at the active site of thioredoxin appears to be conserved for catalytically important residues located at the same position. This is the case of the N-terminal sensory domain of circadian clock-associated histidine kinase SasA, which lacks the catalytic cysteine residues of the thioredoxin fold. A patch of conserved solvent-exposed residues found near the canonical thioredoxin active site is supposed to be involved in SasA protein–protein interaction (Vakonakis et al. 2004).
To better understand the architecture of the NADP reducing hydrogenase complex from Desulfovibrio fructosovorans, we have investigated the interaction involving HndAc and HndDN domains. Electron transfer reaction within the complex was previously studied by EPR, and the complex formation was shown by far-Western experiments (Dermoun et al. 2002). In the present study we have mapped the interacting surface on HndAc using NMR chemical shifts. Interestingly the residues found affected by the complex formation are located on one side of the molecule and distributed into two patches of amino acids corresponding to the interacting site. An acidic patch including E31 and H27 is located on helix 1, and a basic patch containing G66, K68, Y70, and K80 is located in the strand β4 and the helix 2 (Fig. 7). Site-directed mutagenesis performed on the homologous ferredoxin from C. pasteurianum, pinpoints that three residues conserved in Aa ferredoxin, namely E29, Q32, and E36, are involved in the interaction with nitrogenase MoFe protein (Golinelli et al. 1997). Two of these residues located on the external face of helix α1 are structurally equivalent to H27 and E31 in HndAc. These data confirm the involvement of the helix α1 in the interacting surface of the thioredoxin-like ferredoxins. While the tyrosine residue of the basic patch in HndA is conserved in Aa ferredoxin, the basic residues correspond to acidic residues in Aa ferredoxin. These differences may be correlated to variations of the surface charges of the redox partners of these two proteins.
Figure 7.
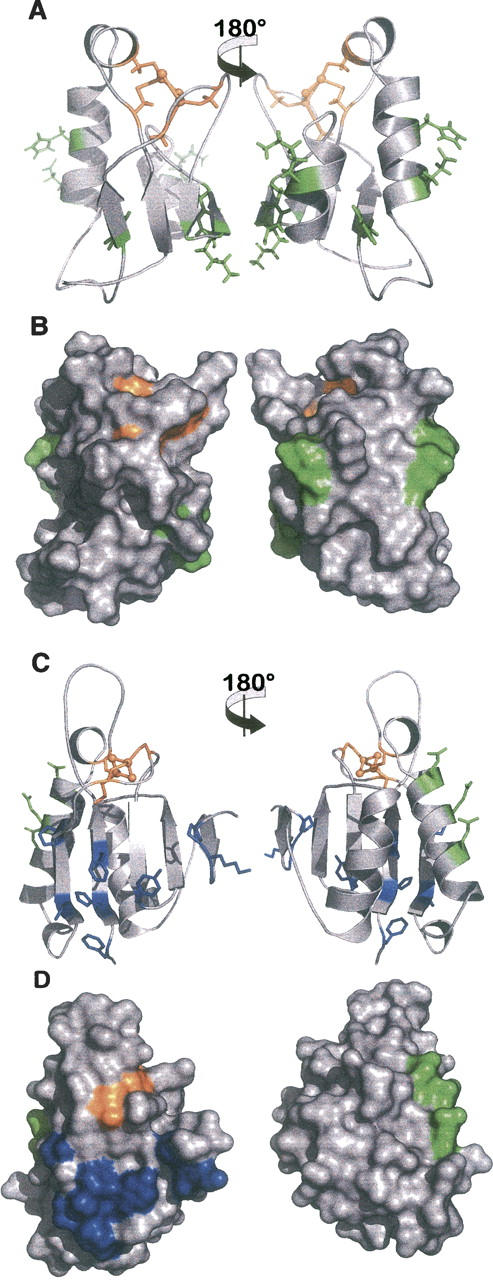
Stuctural comparison of the interacting surfaces in HndAc and Aa ferredoxin. (A) Stick and (B) surface representations of HndAc structure. (C) Stick and (D) surface representations of Aa ferredoxin. Two orientations of the molecules are presented with a 180° rotation. In orange is the FeS cluster; in green, the residues involved in the interaction sites of HndDN on HndAc (A,B) and of nitrogenase on Aa ferredoxin (C,D). In blue are indicated the residues involved in the dimer interface in Aa ferredoxin.
In the HndABCD complex, HndD corresponds to the hydrogenase subunit. From our study it is clear that the thioredoxin-like module present in the HndA subunit interacts with HndD in a first step of the intermolecular electron transfer within the enzymatic complex. The reduction of NADP+ final reaction is conducted by HndC. The role of HndB is still an open question. It has been outlined that the HydB subunit of NADPH hydrogenase from T. maritima is a fusion of HndB and HndC (Malki et al. 1995; Verhagen et al. 1999; Vignais et al. 2001), indicating that HndB can be an intermediate between HndA and HndC subunits. Sequence analysis of HndB indicates 41% similarity with HndA. A structural analysis of HndB using 3D PSSM server (http://www.sbg.bio.ic.ac.uk/~3dpssm/index2.html) reveals a potential thioredoxin-like fold for HndB (data not shown). The HndAc/HndB complex may be formed by the two subunits with a similar organization found in the homodimer of Aa ferredoxin. In the present work, we clearly established that the HndDN interacting site is not localized at the interaction site of the two subunits of Aa ferredoxin (Fig. 7). Further structural analysis of HndDN and HndB subunits will be carried out with the aim to generate structural models of the two intermediate complexes involving HndAc. One can expect that a ternary complex formed by the three electron transfer domains, namely HndDN, HndAc, and HndB, is necessary for the electron transfer pathway from the hydrogenase active site to the terminal NADP reducing site within the HndABCD complex. Another question is still open: What is the folding and the role of the N-terminal domain of HndA? We could not express this domain as a soluble protein; however, the spontaneous cleavage of HndA subunit in two separate domains suggests a distinct structural behavior of these two domains. A structure prediction analysis of the HndA N-terminal domain using the 3D PSSM server does not reveal a typical fold or homologous protein. It can be proposed that this highly hydrophobic domain may act in the metalloenzyme assembly.
Materials and methods
Expression of HndAc and HndDN
The plasmid constructions of pET-HndAc and pET-HndDN were previously described (Dermoun et al. 2002). In the two plasmids, the polyHis-tag codons were transcribed in phase with the end of the truncated hndAc and hndDN gene, respectively. E. coli BL21 (DE3) strains transformed with pET-hndAc were grown at 37°C in 30 mL of Luria–Bertani (LB), including ampicillin (100 μg/mL) to the late exponential growth phase. This suspension was mixed at a 1:1 ratio with 60% glycerol and frozen at −70°C. Two hundred microliters of this suspension were partially thawed on ice and inoculated in 2 × 500 mL of LB medium containing ampicillin (100 μg/mL) and glycerol (12 mg/mL). Cells were grown at 37°C to OD600nm = 2.0 and then stored at 4°C for 30 min. Ferric ammonium citrate (10 μg/mL), sodium sulfide (10 μM), and isopropyl β-D-thiogalactopyranoside (IPTG) were subsequently added (0.25 mM), and the cells were grown for 15 h at 15°C. For uniform 15N-labeling of HndAc, cells were grown on M9 medium supplemented with 15NH4Cl (1 g/L) and glycerol (12 g/L). For the 15N 13C labeling, glycerol was replaced by 13C glycerol (HO-13CH2-13CHOH-13CH2OH) (2 g/L). The same procedure was used for production of the truncated HndD (HndDN) subunit, but cells were grown for 15 h at 37°C after addition of IPTG.
The purification of HndAc and HndDN was performed using an Ni-NTA resin (Qiagen) as previously reported (Dermoun et al. 2002). The proteins were dialyzed against 50 mM potassium phosphate buffer (pH 5.9) in 100 mM NaCl for structure determination and in 25mM potassium phosphate buffer (pH 5.9) for protein interaction studies. H2O/D2O exchange was performed on an Amicon microconcentrator.
NMR experiments
NMR experiments were performed at 293 K, on the oxidized HndAc protein at 1.6 mM concentration in 50 mM potassium phosphate buffer (pH 5.9), 100 mM NaCl, and 10% D2O. The NMR tubes were filled under an argon atmosphere and flame-sealed. Spectra were acquired on a Bruker Avance DRX 500 spectrometer and on a Varian Inova 800-MHz spectrometer equipped with triple resonance (1H, 13C, 15N) probes. The spectra were analyzed with FELIX (Accelrys). Backbone resonance assignments were obtained by using a combination of three-dimensional heteronuclear experiments recorded on the 500-MHz spectrometer (HNHA, HNCA, HN[CO]CA, CBCA[CO]NH, HN[CA]CO, and HNCO). Side-chain assignments were obtained by using 3D heteronuclear 15N-TOCSY-HSQC, HCCH-TOCSY, 15N-NOESY-HSQC, and homonuclear 2D-TOCSY experiments (Table S1 of Supplemental Material). Distance constraints were collected from manual peak-picking using Felix 2002 (Accelrys) from 2D-NOESY and 3D-15N-NOESY-HSQC experiments recorded on the 800-MHz spectrometer. Dihedral angle constraints for φ and ψ were obtained from TALOS (Cornilescu et al. 1999).
Binding studies
Two sets of NMR titrations of the complex formation were performed recording 15N-1H HSQC spectra. Titration was monitored using a 15N-labeled HndAc sample in 25 mM phosphate buffer (pH 5.9). NMR spectra were recorded on an HndAc solution at 0.13-mM concentration in the presence of unlabeled HndDN at 0, 0.5, 1, 1.5, and 4 equivalents.
HndAc structure calculation
Distance constraints were collected from 15N NOESY-HSQC (80 msec mixing time) and homonuclear NOESY (150 msec mixing time) experiments performed on the 800-MHz spectrometer. In addition, 3JNH-Hα scalar coupling measured in a 3D HNHA spectrum yielded Φ-angle constraints. Backbone dihedral angle constraints were derived from chemical shifts with TALOS for residues located in regular secondary structure elements. Hydrogen bond constraints were deduced from the exchange rate of the amide proton in homonuclear TOCSY, NOESY, and COSY experiments recorded in 100% D2O. Due to the paramagnetism of the [2Fe-2S] cluster, signals from protons within a spherical distance of ∼7.8 Å from the iron ions are broadened and difficult to detect under standard NMR conditions (Oh and Markley 1990). As a consequence, the amide resonances of residues C12–F18 and R52–I62 are missing in the HSQC spectrum. In order to circumvent this difficulty, an accepted approach is to model the missing part based on structural homology (Lelong et al. 1995; Baumann et al. 1996; Im et al. 1998; Mo et al. 1999; Pochapsky et al. 1999; Müller et al. 2002). The backbone dihedral angle constraints from the X-ray structure of the thioredoxin from A. aeolicus (PDB code 1F37) were selected as a template to model the residues V11–A22 and D49–V65 of HndAc (Fig. 1B), resulting in 57 dihedral angle constraints. The [2Fe-2S] cluster was built using two modified residues, named CFS, comprising a FeS group bound to Sγ of a cysteine residue. These new residues were implemented at positions CFS17 and CFS57, which define the pole of the cluster. Fe1 was bound to C12 and CFS17, and Fe2 was bound to C53 and CFS57. On the basis of the structural homology with the thioredoxin-like ferredoxin from A. aeolicus, 16 distances and four dihedral χ constraints (provided as Table S2 of Supplemental Material) were applied to fix the [2Fe-2S] cluster geometry (Yeh et al. 2000).
For structure calculations we used the force field and the water refinement protocol of ARIA1.2 software (Nilges et al. 1997) with modified parameters to increase convergence (Molecular Dynamic step parameters *12) (Fossi et al. 2005). However, the software was not operated in an automatic way. Calculations were performed on eight 2.4-GHz bi-processor cluster operating under Linux with Hyper Threading Technology. The ARIA routine for automatic calibration could not be applied because of the paramagnetic effect of the [2Fe-2S] cluster. The volumes of the assigned cross-peaks were obtained using the integration routines in FELIX (Accelrys). Cross-peak intensities were converted into interatomic distances using known proton–proton distances in identified secondary structure elements. We used 2.8 Å for sequential NHi-NHi+1, 3.5 Å for Hαi-HNi+1 and 3.4 Å for Hαi-NHi+3 in α-helices not affected by the paramagnetic effect of the [2Fe-2S] cluster. The first step of the calculation was run from an extended structure containing CFS residues using the assigned intraresidue restraints considered as ambiguous nOes and the 16 restraints of the [2Fe-2S] cluster. At this point, ARIA standard parameters were used, with 100 structures calculated. The 10 best structures regarding nOe violations were refined and analyzed. Distance restraint violations were inspected and cross-peak assignment corrected when necessary. Successive runs were done until no nOe violation exceeded 0.3 Å. The 356 intraresidue nOes were then considered as unambiguous. The next stages were completed using a similar approach as described in the first step. The 205 sequential restraints were added first. The 151 medium range restraints were then injected to determine the secondary structures. These secondary structure elements were strengthened by the angle restraints deduced from the TALOS software and HNHA experiment. Twenty-seven hydrogen bonds deduced from the secondary elements and confirmed by D2O TOCSY spectra were added to the restraint list. At this stage, 160 long-range restraints were applied. The final step concerned the introduction of the backbone dihedral angle constraints of the non observed Val11–Ala22 and Asp49–Val65 residues as described here above.
The final 15 best structures were analyzed with MOLMOL (Koradi et al. 1996), PROCHECK, and PROCHECK_NMR (Laskowski et al. 1993, 1996), and the molecular representations were displayed with the program PYMOL v0.97 (DeLano Scientific).
Electronic supplemental material
The supplementary material consists of Table S1, acquisition parameters for NMR experiments performed on HndAc, and Table S2, distance constraints in the [2Fe-2S] cluster used in the ARIA program.
Acknowledgments
We thank Dominique Marion for NMR experiments recorded on the 800MHz NMR spectrometer (IBS-Grenoble) and Nicolas Rachlin for helpful experimental analysis of NMR data.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Françoise Guerlesquin, Unité de Bioénergétique et Ingénierie des Protéines, IBSM-CNRS, 31 Chemin Joseph Aiguier, 13402 Marseille Cedex 20, France; e-mail: guerlesq@ibsm.cnrs-mrs.fr; fax: +33-4-91-16-45-40.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051916606.
References
- Albracht S.P., Mariette A., de Jong P. 1997. Bovine-heart NADH: Ubiquinone oxidoreductase is a monomer with 8 Fe-S clusters and 2 FMN groups Biochim. Biophys. Acta 318 92–106. [DOI] [PubMed] [Google Scholar]
- Balatri E., Banci L., Bertini I., Cantini F., Ciofi-Baffoni S. 2003. Solution structure of Sco1: A thioredoxin-like protein involved in cytochrome c oxidase assembly Structure 11 1431–1443. [DOI] [PubMed] [Google Scholar]
- Baumann B., Sticht H., Scharpf M., Sutter M., Haehnel W., Rosch P. 1996. Structure of Synechococcus elongatus [Fe2S2] ferredoxin in solution Biochemistry 35 12831–12841. [DOI] [PubMed] [Google Scholar]
- Bugdorf T., van der Linden E., Bernhard M., Yin Q.Y., Back J.W., Hartog A.F., Muijsers A.O., de Koster C.G., Albracht S.P.J., Friedrich B. 2005. The soluble NAD+-reducing [NiFe]-hydrogenase from Ralstonia eutropha H16 consists of six subunits and can be specifically activated by NADPH J. Bacteriol. 187 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalot L., Hatchikian E.C., Forget N., de Philip P., Dermoun Z., Bélaïch J.P., Rousset M. 1998. Molecular study and partial characterization of iron-only hydrogenase in Desulfovibrio fructosovorans Anaerobe 4 45–55. [DOI] [PubMed] [Google Scholar]
- Coldren C.D., Hellinga H.W., Caradonna J.P. 1997. The rational design and construction of a cuboidal iron–sulfur protein Proc. Natl. Acad. Sci. 94 6635–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G., Delaglio F., Bax A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- De Luca G., Asso M., Bélaïch J.P., Dermoun Z. 1998. Purification and characterization of the HndA subunit of NADP-reducing hydrogenase from Desulfovibrio fructosovorans overproduced in Escherichia coli Biochemistry 37 2660–2665. [DOI] [PubMed] [Google Scholar]
- Dermoun Z., De Luca G., Asso M., Bertrand P., Guerlesquin F., Guigliarelli B. 2002. The NADP-reducing hydrogenase from Desulfovibrio fructosovorans: Functional interaction between the C-terminal region of HndA and the N-terminal region of HndD subunits Biochim. Biophys. Acta 1556 217–225. [DOI] [PubMed] [Google Scholar]
- Fossi M., Oschkinat H., Nilges M., Ball L.J. 2005. Quantitative study of the effects of chemical shift tolerances and rates of SA cooling on structure calculation from automatically assigned NOE data J. Magn. Reson. 175 92–102. [DOI] [PubMed] [Google Scholar]
- Golinelli M.P., Gagnon J., Meyer J. 1997. Specific interaction of the [2Fe-2S] ferredoxin from Clostridium pasteurianum with the nitrogenase MoFe protein Biochemistry 36 11797–11803. [DOI] [PubMed] [Google Scholar]
- Hatchikian C.E., Traore A.S., Fernandez V.M., Cammack R. 1990. Characterization of the nickel–iron periplasmic hydrogenase from Desulfovibrio fructosovorans Eur. J. Biochem. 187 635–643. [DOI] [PubMed] [Google Scholar]
- Im S.C., Liu G., Luchinat C., Sykes A.G., Bertini I. 1998. The solution structure of parsley [2Fe-2S] ferredoxin Eur. J. Biochem. 258 465–477. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter M., Wüthrich K. 1996. MOLMOL: A program for display and analysis of macromolecular structures J. Mol. Graph. 14 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur W., Moss D., Thornton J. 1993. PROCHECK: A program to check the stereochemical quality of protein structures J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Laskowski R.A., Rullmann J.A.C., MacArthur W., Kaptein R., Thornton J. 1996. AQUA and PROCHECK-NR: Programs for checking the quality of protein structures solved by NMR J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Lelong C., Setif P., Bottin H., Andre F., Neumann J.M. 1995. 1H and 15N NMR sequential assignment, secondary structure, and tertiary fold of [2Fe-2S] ferredoxin from Synechocystis sp. PCC 6803 Biochemistry 34 14462–14473. [DOI] [PubMed] [Google Scholar]
- Malki S., Saimmaime I., De Luca G., Rousset M., Dermoun Z., Bélaïch J.P. 1995. Characterization of an operon encoding an NADP-reducing hydrogenase in Desulfovibrio fructosovorans J. Bacteriol. 177 2628–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. 2001. Ferredoxins of the third kind FEBS Lett. 509 1–5. [DOI] [PubMed] [Google Scholar]
- Meyer J., Bruschi M.H., Bonicel J.J., Bovier-Lapierre G.E. 1986. Amino acid sequence of [2Fe-2S] ferredoxin from Clostridium pasteurianum Biochemistry 25 6054–6061. [DOI] [PubMed] [Google Scholar]
- Mo H., Pochapsky S.S., Pochapsky T.C. 1999. A model for the solution structure of oxidized terpredoxin, a Fe2S2 ferredoxin from Pseudomonas Biochemistry 38 5666–5675. [DOI] [PubMed] [Google Scholar]
- Müller J., Lugovskoy A.A., Wagner G., Lippard S. 2002. NMR structure of the [2Fe-2S] ferredoxin domain from soluble methane monooxygenase reductase and interaction with its hydroxylase Biochemistry 41 42–51. [DOI] [PubMed] [Google Scholar]
- Nilges M., Macias M.J., O'Donoghue S.I., Oschkinat H. 1997. Automated NOESY interpretation with ambiguous distance restraints: The refined NMR solution structure of the pleckstrin homology domain from β-spectrin J. Mol. Biol. 269 408–422. [DOI] [PubMed] [Google Scholar]
- Oh B.H. and Markley J.L. 1990. Multinuclear magnetic resonance studies of the 2Fe-2S ferredoxin from Anabaena species strain PCC 7120. 3. Detection and characterization of hyperfine-shifted nitrogen-15 and hydrogen-1 resonances of the oxidized form Biochemistry 29 4012–4017. [DOI] [PubMed] [Google Scholar]
- Pinto A.L., Hellinga H.W., Caradonna J.P. 1997. Construction of a catalytically active iron superoxide dismutase by rational protein design Proc. Natl. Acad. Sci. 94 5562–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochapsky T.C., Jain N.U., Kuti M., Lyons T.A., Heymont J. 1999. A refined model for the solution structure of oxidized putidaredoxin Biochemistry 38 4681–4690. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Warnecke U., Böcker C., Zaborosch C., Friedrich B. 1990. Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase of Alcaligenes eutrophus H16 J. Bacteriol. 172 2920–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakonakis I., Klewer D.A., Williams S.B., Golden S.S., LiWang A.C. 2004. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA J. Mol. Biol. 342 9–17. [DOI] [PubMed] [Google Scholar]
- Verhagen M.F., O'Rourke T., Adams M.W. 1999. The hyperthermophilic bacterium, Thermotoga maritima, contains an unusually complex iron-hydrogenase: Amino acid sequence analyses versus biochemical characterization Biochim. Biophys. Acta 1412 212–229. [DOI] [PubMed] [Google Scholar]
- Vignais P., Billoud B., Meyer J. 2001. Classification and phylogeny of hydrogenases FEMS Microbiol. Rev. 25 455–501. [DOI] [PubMed] [Google Scholar]
- Williams J.C., Sue C., Banting G.S., Yang H., Glerum D.M., Hendrickson W.A., Schon E.A. 2005. Crystal structure of human SCO1: Implications for redox signaling by a mitochondrial cytochrome c oxidase “assembly” protein J. Biol. Chem. 280 15202–15211. [DOI] [PubMed] [Google Scholar]
- Wisz M.S., Garrett C.Z., Hellinga H.W. 1998. Construction of a family of Cys2His2 zinc binding sites in the hydrophobic core of thioredoxin by structure-based design Biochemistry 37 8269–8277. [DOI] [PubMed] [Google Scholar]
- Yano T., Magnitsky S., Sled' V.D., Ohnishi T., Yagi T. 1999. Characterization of the putative 2x[4Fe-4S]-binding NQO9 subunit of the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Expression, reconstitution, and EPR characterization J. Biol. Chem. 274 28598–28605. [DOI] [PubMed] [Google Scholar]
- Yeh A.P., Chatelet C., Soltis S.M., Kuhn P., Meyer J., Rees D.C. 2000. Structure of a thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus J. Mol. Biol. 300 587–595. [DOI] [PubMed] [Google Scholar]