Abstract
Outer surface protein A (OspA) from Borrelia burgdorferi has an unusual dumbbell-shaped structure in which two globular domains are connected with a “single-layer” β-sheet (SLB). The protein is highly soluble, and it has been recalcitrant to crystallization. Only OspA complexes with Fab fragments have been successfully crystallized. OspA contains a large number of Lys and Glu residues, and these “high entropy” residues may disfavor crystal packing because some of them would need to be immobilized in forming a crystal lattice. We rationally designed a total of 13 surface mutations in which Lys and Glu residues were replaced with Ala or Ser. We successfully crystallized the mutant OspA without a bound Fab fragment and extended structure analysis to a 1.15 Å resolution. The new high-resolution structure revealed a unique backbone hydration pattern of the SLB segment in which water molecules fill the “weak spots” on both faces of the antiparallel β-sheet. These well-defined water molecules provide additional structural links between adjacent β-strands, and thus they may be important for maintaining the rigidity of the SLB that inherently lacks tight packing afforded by a hydrophobic core. The structure also revealed new information on the side-chain dynamics and on a solvent-accessible cavity in the core of the C-terminal globular domain. This work demonstrates the utility of extensive surface mutation in crystallizing recalcitrant proteins and dramatically improving the resolution of crystal structures, and provides new insights into the stabilization mechanism of OspA.
Keywords: crystallization, crystal packing, hydration, antiparallel β-sheet, conformational entropy, Lyme disease
Outer surface protein A (OspA) from Borrelia burgdorferi has been a focus of vaccine development for Lyme disease (Luft et al. 2002; Koide et al. 2005) and an excellent model system for studying β-sheet folding and peptide self-assembly (Pham et al. 1998; Koide et al. 1999, 2000; Ohnishi et al. 2000; Yan et al. 2002, 2004). The natural form of OspA is 273 residues long and lipidated at an N-terminal Cys. Most of biophysical studies to date have been performed on a truncated form in which the lipidation signal is removed (Dunn et al. 1990). This truncated form (256 residues; for brevity, we will refer to the truncated form as “wild type”) is highly water soluble, presumably because it contains disproportionately large numbers of Glu (a total of 22) and Lys (a total of 40) residues. The wild-type protein by itself has been recalcitrant to crystallization in initial trials by Li and Lawson (1995) and in more recent trials in our laboratory. However, its crystal structures have been determined as the complexes with two different antibody Fab fragments (Protein Data Bank [PDB] IDs 1OSP and 1FJ1) with resolutions of 1.95 and 2.68 Å, respectively (Li et al. 1997; Ding et al. 2000).
These structures revealed that OspA has an unusual “dumbbell” shape composed of sequential 21 antiparallel β-strands followed by a single α-helix at the C terminus (Fig. 1; Li et al. 1997). In the middle of the molecule is a single-layer β-sheet (SLB) segment. Both faces of this SLB segment are exposed to the solvent, and consequently the segment is not associated with a hydrophobic core. The amino acid sequences of the SLB segment are dominated by polar and charged amino acids, and they do not exhibit an alternating binary pattern of polar and nonpolar amino acid residues typical of an amphipathic β-sheet. Solution NMR spectroscopy and small-angle X-ray scattering studies have confirmed that free OspA is monomeric and that the crystal structure of OspA in the Fab complexes represents its predominant form in solution (Bu et al. 1998; Pham and Koide 1998). Furthermore, we have demonstrated that the SLB segment is highly stable (Pham et al. 1998; Yan et al. 2002). It can be extended by inserting copies of a β-hairpin unit, which mimics peptide self-assembly (Koide et al. 2000). To gain a fundamental understanding of the mechanism of SLB formation, we have characterized thermodynamic effects of a large number of OspA mutants (Yan et al. 2004), and we would now like to determine their high-resolution structures in order to fully understand the structural basis for the mutation effects (Matthews 1993). Although Li et al. (Li and Lawson 1995; Li et al. 1997) successfully determined the 1.95 Å crystal structure of the OspA-Fab complex (1OSP), this method of using a monoclonal antibody is expensive and poorly reproducible, possibly due to sample heterogeneity of the Fab fragment. Thus, it is not an ideal method for crystallizing a large number of OspA mutants. Also, we need to improve the resolution of the crystal structure in order to characterize details of structural perturbations by the mutations. Here, we aimed to develop a reproducible method for producing crystals of OspA by itself that diffract at a much higher resolution.
Figure 1.
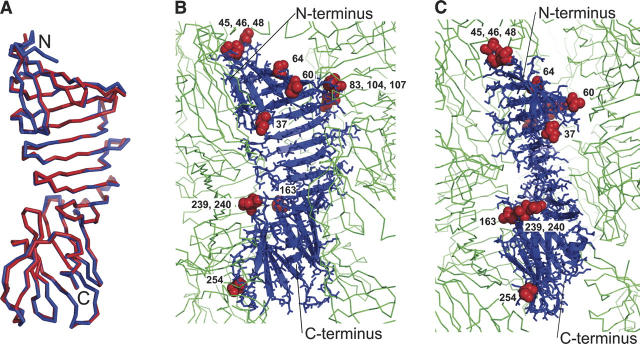
(A) Superposition of the OspA structure in the 184.1 Fab complex (1OSP; blue) and that of OspAsm1 (red). Only the backbone traces are shown. Note that the N terminus of OspAsm1 is truncated. (B,C) Crystal packing of OspAsm1 and the locations of mutated residues in two orthogonal views. The central monomer is shown as a ribbon model, and its adjacent molecules are shown as backbone line traces. For clarity, the symmetry-related molecule directly in front of the central molecule has been removed. The residues mutated in OspAsm1 are shown as red spheres and labeled.
Recently, Derewenda et al. (for review, see Derewenda 2004) pioneered the “surface entropy reduction” method for protein crystallization. This method is based on the insightful observation that side chains must be immobilized at crystal contacts, and thus large flexible amino acids such as Glu and Lys are expected to disfavor crystallization due to a large entropy loss upon immobilization. The charges at their side-chain termini also need to be properly compensated at crystal contacts. Replacing them with a short, neutral side chain reduces such entropy loss and the requirement for electrostatic complementarity and thus promotes crystallization. The method has primarily been used to crystallize proteins of unknown structure. Based on the unusual prevalence of Lys and Glu in OspA, we hypothesized that the high conformational entropy of OspA surface residues was one of the origins of the difficulty in crystallizing OspA alone and that the surface entropy reduction method might overcome this difficulty.
In this work, we report successful crystallization of OspA using rational surface engineering, which resulted in a 1.15 Å resolution crystal structure. This high-resolution structure revealed several features of OspA that were obscured in previous, lower resolution structures. We will discuss the stabilization mechanism of OspA SLB based on the new structural information.
Results and Discussion
Mutation design and crystallization
Our previous NMR analysis of the OspA solution structure revealed that the N-terminal region (residues 16–27) in the original construct (residues 16–273) is highly dynamic (Pham and Koide 1998). Thus, we first truncated the N terminus so that the new construct contains residues 27–273 in the hope that the removal of the flexible tail might enhance the crystallizability of the protein. We performed extensive crystallization of this truncated protein in the absence of a bound Fab fragment including high-throughput microbatch screening (1536 conditions) at the Hauptman-Woodward Medical Research Institute (HWI) (Luft et al. 2003) and in-house sitting drop vapor diffusion screening (196 conditions). However, we could not obtain any kind of crystals in these conditions, indicating that factors other than the flexible N-terminal tail prevent the crystallization of OspA.
OspA contains a large number of Lys and Glu (62 residues of the total 251 residues [25%]) most of which are located on the surface (Li et al. 1997). This Glu/Lys content exceeds that of RhoGDI (20%), which was used as a test case for the surface entropy reduction approach by Derewenda et al. (2004). Thus, we suspected that these Glu/Lys residues serve as an “entropy shield” that prevents crystallization, as proposed by Derewenda and colleagues (Derewenda 2004). They and others have demonstrated that a replacement of flexible residues on the protein surface with a smaller amino acid leads to successful preparation of diffraction-quality crystals. Consequently, we decided to apply this strategy to OspA.
We first chose three positions (K48, E196, and K230) based on the crystal structures of OspA-Fab complexes (1OSP and 1FJ1) (Li et al. 1997; Ding et al. 2000). The two Fabs (184.1 and LA-2) bind to distinct epitopes of OspA, and thus the two crystal structures have totally different modes of packing. Nevertheless, these three residues are involved in crystal contacts (with Fab) in both crystal structures, suggesting that these positions have high likelihood of being located in crystal contacts, regardless of the mode of crystal packing. Thus, we hypothesized that these may be crystal contact “hot spots” and reducing the side-chain entropy at these positions might enhance the probability of crystallization. Note that this is a slightly different strategy from that of Derewenda (2004) in which a single cluster of Glu/Lys residues are replaced. Mutants containing K230A were often expressed as inclusion bodies in Escherichia coli, while the wild-type and the other mutants were expressed as a soluble protein. Thus, we decided not to mutate this position. Crystallization trials of a double mutant (K48A:E196A) were not successful. We then introduced two additional mutations, K60A and K83A, because these positions are also highly exposed on the surface, but this quadruple mutant (K48A:K60A:K83A:E196A) also failed to crystallize.
Because the first set of mutations did not promote crystallization, we then decided to simultaneously introduce an additional nine mutations (E37S, E45S, K46S, K64S, E104S, K107S, K239S, E240S, and K254S) to the above quadruple mutant, resulting a total of 13 mutations. We selected positions that were highly exposed and located in a turn or bulge in the OspA crystal structure (1OSP). This time we replaced these nine residues with a Ser rather than an Ala to prevent the surface from becoming too hydrophobic. Also, three pair of mutations (positions 45 and 46, 104 and 107, and 239 and 240) each replaced a Glu/Lys cluster. We deliberately aimed to select mutation positions within the N- and C-terminal globular domains so that the SLB segment was almost left unchanged. The SLB segment contains only two mutations, E104S and K107S, located in a turn. We will refer to this mutant as “OspAsm1.”
Despite the large number of surface mutations, OspAsm1 was expressed as a soluble protein, and we were able to purify it in a similar manner as the wild type. The free energy difference of unfolding for OspAsm1 was ∼11 kcal/mol, as determined by urea denaturation. Not unexpectedly from the large number of mutations, OspAsm1 was destabilized by 5.3 kcal/mol relative to the wild type (Yan et al. 2004), although it was still sufficiently stable for maintaining the native conformation.
OspAsm1 crystallized readily in a wide variety of solution conditions. It crystallized in 299 of the 1536 conditions (19.5%) in the WHI high-throughput screening and 24 out of the 196 conditions (12.5%) in the in-house screening. The best single crystals were obtained from solutions containing PEG400 as a precipitant.
Overall structure and molecular contacts
We determined the structure of OspAsm1 at a resolution of 1.15 Å by the molecular replacement method. The data collection and refinement statistics are presented in Table 1. The final model contains 246 amino acids (residues 28–273). The residues at the N terminus, Lys27, and an additional four residues (GSHM) originated from the expression vector were disordered in the crystal structure. The structure contains 403 water molecules. Two continuous arch-like electron densities around Lys80 and Tyr147 have been fitted with polyethylene glycol (PEG) molecules. The electron density near Tyr147 with the appearance of a five-member ring has been assigned to an imidazole molecule.
Table 1.
Data collection and refinement statistics

The overall structure of OspAsm1 is very close to that of the wild-type OspA in the Fab complex (1OSP) (Fig. 1A). The root-mean-squared (RMS) deviation of the Cα atoms common in these structures is 0.823 Å, with large deviations (>1.5 Å) found only in loop or turn regions. There are three regions exhibiting Cα rms deviations >2 Å (residues 48, 49, 81, and 117). Of these, only residue 117 is involved in antibody binding in 1OSP. These results indicate that the extensive surface mutations did not significantly affect the overall structure of OspA and that OspA undergoes little backbone conformational change upon antibody binding.
We then investigated the roles of the mutated residues in forming crystal contacts. Derewenda et al. (Derewenda 2004) have shown that mutated residues are often involved in direct crystal contact. We found that eight of the 13 mutated residues were involved in crystal contacts via hydrogen bonding and/or van der Waals contacts in the OspAsm1 crystal (Fig. 1B,C). The involvement of each of the 13 mutated residues in crystal packing is summarized in Supplemental Table 1.
Backbone hydration of OspA SLB
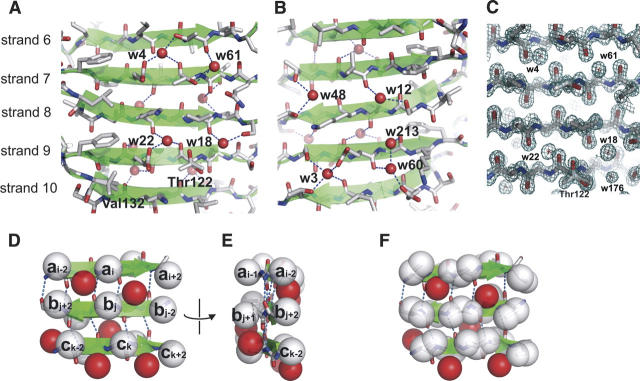
The 1.15 Å resolution structure determined in this work enabled us to unambiguously identify many water molecules. Of a total of 403 water molecules, 41 are located near the main-chain oxygen and/or nitrogen atoms in the SLB region. Particularly noteworthy are the nine water molecules that hydrate the backbone near the center of SLB region (Fig. 2A,B), for which clear electron density were observed (Fig. 2C).
Figure 2.
Backbone hydration and multiple side-chain conformers of the SLB. (A,B) The SLB region of OspAsm1 showing backbone hydration (red spheres). Panels A and B show the opposite faces of the β-sheet. Hydrogen bonds involving the water molecules are shown as dashed lines. The side chains are depicted as sticks. Two conformers for Thr122 and Val132 are shown. (C) The SigmaA-weighted electron density map contoured at 1.5σ for the region shown in A. (D,E) Schematic drawing of a section of the SLB. The Cβ atoms are shown as white spheres, and the rest of the side-chain atoms are omitted. The backbone atoms and the hydrogen bonds between them are shown as sticks and dashed lines, respectively. Water molecules are shown as red spheres. The cross-strand pairs, ai − 2/bj + 2, ai/bj, and ai + 2/bj − 2, represent hydrogen-bonded pairs because the two residues in each of these pairs form mutual backbone hydrogen bonds. The pairs bj + 2/ck − 2, bj/ck, and bj − 2/ck + 2 represent non-hydrogen-bonded pairs because they do not form mutual backbone hydrogen bonds. Note that between the β-strands a and b the side chains of hydrogen-bonded pairs point to the reader and the side chains of non-hydrogen-bonded pairs point away from the reader, while between the β-strands b and c the side-chain directions are reversed. The four Cβ atoms of two hydrogen-bonded pairs surround the water molecules. Thus, these water molecules are located over a non-hydrogen-bonded pair. The occupation by the water molecules of alternating faces of the β-strand interfaces is evident in the side view of the β-sheet (E). (F) Inability of nonaromatic side chains to occlude the water molecules hydrating the SLB backbone. This figure depicts the same region as in D but includes the Cγ atoms. Note that the Cγ atoms cannot reach the space occupied by the water molecules over non-hydrogen-bonded pairs, but they can occupy the equivalent space over hydrogen-bonded space, as pointed out by Finkelstein and Nakamura (1993).
Except for the water molecule “w213” (Fig. 2B), all of these water molecules that hydrate the SLB main chain occupy the “weak points” of an antiparallel β-sheet identified by Finkelstein and Nakamura (1993). A weak point is defined as a region at the surface of a β-sheet that cannot be filled by the surrounding nonaromatic side chains belonging to the same sheet. A weak point is located on the opposite face to the side chains of a non-hydrogen-bonded cross-strand pair, because the four side chains surrounding such a point are farther away than those surrounding an equivalent point for the hydrogen-bonded pair (Fig. 2D–F; Finkelstein and Nakamura 1993). The distinction between the hydrogen-bonded pair and the non-hydrogen-bonded pair is elaborated in Figure 2D and its caption (Wouters and Curmi 1995).
In a given strand–strand interface (e.g., that between strands 8 and 9) (Fig. 2A), the backbone hydration water molecules are located on one face, and ones in the adjacent strand–strand interface (e.g., that between strands 7 and 8) are on the other face. This arrangement creates an unusual, alternating backbone hydration pattern (Fig. 2E). This interesting pattern is due to the fundamental properties of the antiparallel β-sheet. In an antiparallel β-sheet such as the SLB, hydrogen-bonding pairs and non-hydrogen-bonding pairs alternate along the interface between two β-strands (Fig. 2D). The side-chain direction also alternates along the β-strand. Thus, for a given β-strand interface, all the non-hydrogen-bonded pairs have the side chains located on the same face of the β-sheet (e.g., bj + 2/ck − 2, bj/ck and bj − 2/ck + 2 in Fig. 2D), and those in the adjacent β-strand interface have the side chains on the opposite face (ai − 1/bj + 1 and ai + 1/bj − 1). Consequently, the weak points in a given β-strand interface and those in its adjacent β-strand interface are located on the opposite faces of the β-sheet (e.g., the interface between strands a and b and that between strands b and c in Fig. 2D), and thus the water molecules that hydrate the SLB backbone are found on alternating faces of the β-sheet as seen in Figure 2.
Finkelstein and Nakamura (1993) found that the weak points in antiparallel β-sheets are often filled by an aromatic side chain of the same β-sheet or side-chain atoms of another structural element through hydrophobic packing. The SLB does not interact with another structural element, and it contains few aromatic amino acids. Therefore, the SLB architecture leaves most of the weak points unoccupied by side-chain atoms. In comparison, we found 14 other water molecules that filled the weak spots of the N-terminal and C-terminal globular domains. These water-filled weak spots naturally exist only on the solvent-accessible face of these amphipathic β-sheets. These results indicate that the hydration of the weak spots is an inherent feature of antiparallel β-sheets and that the alternating backbone hydration pattern is unique to the SLB architecture.
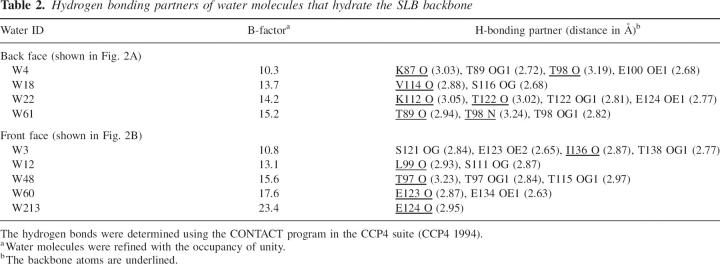
Although one might expect that the SLB structure may be vulnerable due to many unoccupied weak points on both faces, NMR studies have demonstrated that the OspA SLB is highly rigid and stable (Pham et al. 1998; Pham and Koide 1998; Pawley et al. 2002). The water molecules identified in the OspAsm1 structure fill the weak points and bridge adjacent β-strands. These water molecules are hydrogen-bonded to the backbone oxygen and/or nitrogen atoms and also to side-chain atoms (Table 2; Fig. 2A,B). There is a good correlation (r = –0.76) between the B-factor for these water molecules and the number of hydrogen bonds, while the B-factor and the shortest hydrogen-bonding distance show a weaker correlation (r = 0.58), suggesting the importance of multiple hydrogen bonds in pinning down the water molecules in these pockets. These observations suggest that these water molecules are an integral part of the SLB architecture and play an important role in maintaining the rigidity of this highly exposed β-sheet.
Table 2.
Hydrogen bonding partners of water molecules that hydrate the SLB backbone
Structural features of OspA revealed in the high-resolution structure
Multiple side-chain conformers were identified for 18 residues (Table 1). Immobilization of a side chain in the folded state results in an entropy loss. Thus, it is advantageous in terms of conformational stability to maintain multiple side-chain conformers in the native state, as long as such multiple conformers do not lead to a loss of enthalpic stabilization. In the SLB, Thr122 and Val132 exhibit clear signs of two distinct conformers (Fig. 2A,B). These multiple conformers may contribute to the stabilization of the SLB segment that must overcome the inherent lack of extensive hydrophobic packing.
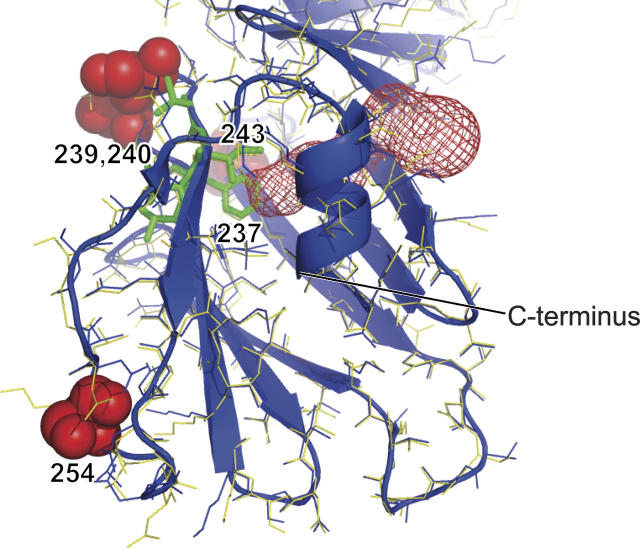
Li et al. (1997) identified a cavity in the C-terminal globular domain in the original OspA structure, and they proposed that it might be a ligand-binding site. Interestingly, a similar cavity is also found in the recently determined crystal structure of the OspB C-terminal domain (Becker et al. 2005). The cavity is preserved in the OspAsm1 structure. However, we found that the cavity has a channel that connects it to the bulk solvent (Fig. 3). Such a channel was not obvious in the original structure. We also found one water molecule in the cavity. No surface mutations are located in the close vicinity of this cavity, and thus it is highly unlikely that the channel formation is an artifact of surface mutations. While the ligand for this cavity is yet to be identified, previous study suggested that Phe237 and Ile243 that line the cavity are involved in receptor binding (Pal et al. 2000). Thus, the channel identified in the OspAsm1 structure implies that a potential ligand may be able to bind to this site without a large conformational change. Regardless of the mechanism of action, these observations strengthen the hypothesis that this cavity is important for the biological role of OspA, i.e., the attachment of the spirochete to the host (tick) gut cells (Yang et al. 2004).
Figure 3.
A solvent-accessible cavity in the C-terminal domain (shown in red mesh). The OspAsm1 structure is shown in blue lines and in ribbon. The OspA structure from 1OSP is shown in yellow. The orientation of the molecules is close to that in Figure 1A. The mutated residues in OspAsm1 (239, 240, and 254) are shown as red spheres. Residues 243 and 237 that are implicated in receptor binding are shown as green sticks.
Surface mutation as a means to enhance resolution
In this work, we demonstrated a dramatic effect of surface engineering on crystallization of OspA. Our results are in a good agreement with Derewenda's (2004) on RhoGDI. Replacements of Glu and Lys residues generally reduce solubility and stability but improve crystallizability. These surface mutations do not significantly alter the protein conformation. While most of surface engineering trials to date have been performed for de novo structure determination, Munshi et al. (2003) designed double and triple mutants based on a low-resolution structure of insulin-like growth factor-1 receptor kinase and successfully obtained a 1.5 Å resolution structure. OspAsm1 appears to be the most extensively surface engineered protein to date, with a total of 13 mutations. Also, this work seems to be the first to combine surface engineering of multiple, discrete patches rather than focusing on a single patch. Targeting multiple “high-entropy” patches may be a powerful strategy to obtain/improve crystals, although one might significantly reduce the solubility and/or stability of the protein of interest.
Recently, Eisenberg's and Derewenda's groups developed a Web-based tool for designing surface mutations for better crystallization (http://nihserver.mbi.ucla.edu/SER/). The program combines secondary structure prediction, side-chain entropy, and sequence conservation and suggests mutation positions. The program suggested eight “clusters” of potential surface mutation sites. These are (1) K27A (N terminus), (2) E37A, (3) K104A;D105A;K107A, (4) D118A;K119A, (5) D141A, (6) E148A, (7) K239A;E240A, and (8) K269A:K273A (C terminus). The clusters 2, 3, and 7 are mutated in OspAsm1. We avoided the SLB region in our mutation design, which eliminates clusters 4, 5, and 6. Thus, the successful prediction of residues that have been mutated in OspA and make crystal contacts suggests that this program will be a useful addition to the crystallographer's toolkit.
OspAsm1 as a “crystallization scaffold”
The SLB segment is a good model system for investigating the structure and stability of antiparallel β-sheets without complications from extensive long-range interactions due to hydrophobic core packing. We designed the surface mutations in OspAsm1 in such a way that most of them are located in the N- and C-terminal globular domains, i.e., outside the SLB segment. Therefore, OspAsm1 should serve as a “crystallization scaffold” for SLB variants. We are now crystallizing many SLB variants using OspAsm1, and our preliminary results suggest that the OspAsm1 scaffold generally produce high-quality crystals even when the SLB segment is highly mutated. Systematic analysis of the structure and energetics of these SLB mutants will advance our understanding of the molecular basis of β-sheet formation. More generally, initial surface engineering for the development of a powerful crystallization scaffold may be an attractive strategy when one has a large number of mutants for which high-resolution structures are desired.
Materials and methods
Mutagenesis and protein production
Mutagenesis was performed on the expression vector encoding residues 27–273 of OspA as described previously (Yan et al. 2004). Mutant proteins were expressed in E. coli BL21 (DE3) as described before (Yan et al. 2004), except that overnight autoinduction media of Studier was used (Studier 2005).
Cells were lysed with hen-egg lysozyme and sonication in 50 mM Tris-HCl (pH 8.0) buffer, NaCl was added at a final concentration of 500 mM, and cell debris was removed by centrifugation. The supernatant was loaded onto an immobilized metal affinity chromatography column (Histrap HP, Amersham Biosciences) equilibrated in 50 mM HEPES (pH 8.0) containing 500 mM NaCl. The column was washed with 50 mM HEPES (pH 8.0) containing 500 mM NaCl and 20 mM imidazole, and then the protein was eluted with 50 mM HEPES (pH 8.0) containing 500 mM imidazole. The protein was cleaved with thrombin (Novagen) to remove the N-terminal His-tag. The solution was dialyzed against 20 mM sodium acetate buffer (pH 4.6) and purified with cation-exchange chromatography (Resource S, Amersham Biosciences) using a linear gradient of NaCl.
Conformational stability was determined from urea-induced unfolding as described previously (Yan et al. 2004).
Crystallization
High-throughput crystallization screening of mutant proteins was performed at the HWI (Luft et al. 2003) with protein concentrations between 10 and 60 mg/mL.
The optimal concentration of OspAsm1 for crystallization trials was determined using PCT kit (Hampton Research). Crystal screening was performed by the sitting-drop vapor-diffusion method using Hampton Research Crystal Screens I and II and Emerald Biostructures Wizard 1 and 2. Optimization was performed using the hanging drop vapor-diffusion method, where a drop initially contained 1 μL each of the reservoir solution and purified OspAsm1 (18 mg/mL in 10 mM Tris-HCl at pH 8). Crystals for data collection were obtained with the use of a reservoir solution consisting of 0.1M imidazole (pH 7.7) and 30% PEG400.
X-ray data collection and structure determination
Crystals of OspAsm1 were flash-frozen in liquid nitrogen using the mother liquid as a cryo-protectant. The X-ray diffraction data were collected at the 17ID beam-line (Advanced Photon Source at the Argonne National Laboratory). Crystal data and data collection statistics are summarized in Table 1. X-ray data were processed with HKL2000 (Otwinowski and Minor 1997) and SCALA of the CCP4 program suite (CCP4 1994). The structure of the OspAsm1 was determined by molecular replacement with the program MOLREP in CCP4. The wild-type OspA structure (PDB ID 1OSP) was used as the search model. In the further refinement 5% randomly chosen reflections were set aside for calculating R-free. After rigid body refinement with program Refmac5 (Murshudov et al. 1997), further refinement was carried out with the simulated annealing and energy minimization protocols using the program CNS1.1 (Brünger et al. 1998). The model was rebuilt using the Coot program (Emsley and Cowtan 2004) against a composite omit map calculated in CNS1.1. Topology and parameter files for the imidazole molecule were obtained from the Heterocompound Information Center of Uppsala (HIC-Up) (Kleywegt and Jones 1998). Final positional and anisotropic temperature factor refinement was performed using Refmac5 software (Murshudov et al. 1997). The hydrogen atoms were included in the riding positions. The positional and temperature factor refinement was restrained with the target sigma values of 0.022 Å, 2.0°, and 3.0 Å2 for covalent bonds, bond angles, and anisotropic thermal factors, respectively. The van der Waals contacts and hydrogen bonds were analyzed by the CONTACT program in the CCP4 suite. Molecular graphics were generated using Pymol (http://www.pymol.org). Cavities were searched with the CAVER program (http://loschmidt.chemi.muni.cz/caver/). The coordinates have been deposited in the PDB with entry code 2G8C.
Acknowledgments
We are thankful to the staff of the Hauptman-Woodward Medical Research Institute for the initial screening of crystallization conditions. We are thankful to Drs. Yoshikazu Tanaka, Akiko Koide, and Anthony A. Kossiakoff for helpful discussions. This work was supported by NIH grants R01-GM57215 and U54-GM074946. Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Shoehi Koide, Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA; e-mail: skoide@uchicago.edu; fax: (773) 702-0439.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062246706.
Abbreviations: OspA, outer surface protein A; SLB, single-layer β-sheet; NMR, nuclear magnetic resonance; HWI, Hauptman-Woodward Medical Research Institute.
References
- Becker M., Bunikis J., Lade B.D., Dunn J.J., Barbour A.G., Lawson C.L. 2005. Structural investigation of Borrelia burgdorferi OspB, a bactericidal Fab target. J. Biol. Chem. 280: 17363–17370. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Bu Z., Koide S., Engelman D.M. 1998. A solution SAXS study of Borrelia burgdorferi OspA, a protein containing a single-layer β-sheet. Protein Sci. 7: 2681–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (CCP4). 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Derewenda Z.S. 2004. Rational protein crystallization by mutational surface engineering. Structure 12: 529–535. [DOI] [PubMed] [Google Scholar]
- Ding W., Huang X., Yang X., Dunn J.J., Luft B.J., Koide S., Lawson C.L. 2000. Structural identification of a key protective B-cell epitope in Lyme disease antigen OspA. J. Mol. Biol. 302: 1153–1164. [DOI] [PubMed] [Google Scholar]
- Dunn J.J., Lade B.N., Barbour A.G. 1990. Outer surface protein A (OspA) from the Lyme disease spirochete Borrelia burgdorferi: High level expression and purification of a soluble recombinant form of OspA. Protein Expr. Purif. 1: 159–168. [DOI] [PubMed] [Google Scholar]
- Emsley P. and Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Finkelstein A.V. and Nakamura H. 1993. Weak points of antiparallel β-sheets. How are they filled up in globular proteins? Protein Eng. 6: 367–372. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones T.A. 1998. Databases in protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 54: 1119–1131. [DOI] [PubMed] [Google Scholar]
- Koide S., Bu Z., Risal D., Pham T.-N., Nakagawa T., Tamura A., Engelman D.M. 1999. Multi-step denaturation of Borrelia burgdorferi OspA, a protein containing a single-layer β-sheet. Biochemistry 38: 4757–4767. [DOI] [PubMed] [Google Scholar]
- Koide S., Huang X., Link K., Koide A., Bu Z., Engelman D.M. 2000. Design of single-layer β-sheets without a hydrophobic core. Nature 403: 456–460. [DOI] [PubMed] [Google Scholar]
- Koide S., Yang X., Huang X., Dunn J.J., Luft B.J. 2005. Structure-based design of a second-generation Lyme disease vaccine based on a C-terminal fragment of Borrelia burgdorferi OspA. J. Mol. Biol. 350: 290–299. [DOI] [PubMed] [Google Scholar]
- Li H. and Lawson C.L. 1995. Crystallization and preliminary X-ray analysis of Borrelia burgdorferi outer surface protein A (OspA) complexed with a murine monoclonal antibody Fab fragment. J. Struct. Biol. 115: 335–337. [DOI] [PubMed] [Google Scholar]
- Li H., Dunn J.J., Luft B.J., Lawson C.L. 1997. Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab. Proc. Natl. Acad. Sci. 94: 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luft B.J., Dunn J.J., Lawson C.L. 2002. Approaches toward the directed design of a vaccine against Borrelia burgdorferi. J. Infect. Dis. 185:(Suppl. 1):, S46–S51. [DOI] [PubMed] [Google Scholar]
- Luft J.R., Collins R.J., Fehrman N.A., Lauricella A.M., Veatch C.K., DeTitta G.T. 2003. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J. Struct. Biol. 142: 170–179. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. 1993. Structural and genetic analysis of protein stability. Annu. Rev. Biochem. 62: 139–160. [DOI] [PubMed] [Google Scholar]
- Munshi S., Hall D.L., Kornienko M., Darke P.L., Kuo L.C. 2003. Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 59: 1725–1730. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Ohnishi S., Koide A., Koide S. 2000. Solution conformation and amyloid-like fibril formation of a polar peptide derived from a β-hairpin in the OspA single-layer β-sheet. J. Mol. Biol. 301: 477–489. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pal U., de Silva A.M., Montgomery R.R., Fish D., Anguita J., Anderson J.F., Lobet Y., Fikrig E. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Invest. 106: 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley N.H., Koide S., Nicholson L.K. 2002. Backbone dynamics and thermodynamics of Borrelia outer surface protein A. J. Mol. Biol. 324: 991–1002. [DOI] [PubMed] [Google Scholar]
- Pham T.-N. and Koide S. 1998. NMR studies of Borrelia burgdorferi OspA, a 28 kDa protein containing a single-layer β-sheet. J. Biomol. NMR 11: 407–414. [DOI] [PubMed] [Google Scholar]
- Pham T.-N., Koide A., Koide S. 1998. A stable single-layer β-sheet without a hydrophobic core. Nat. Struct. Biol. 5: 115–119. [DOI] [PubMed] [Google Scholar]
- Studier F.W. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41: 207–234. [DOI] [PubMed] [Google Scholar]
- Wouters M.A. and Curmi P.M. 1995. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins 22: 119–131. [DOI] [PubMed] [Google Scholar]
- Yan S., Gawlak G., Smith J., Silver L., Koide A., Koide S. 2004. Conformational heterogeneity of an equilibrium folding intermediate quantified and mapped by scanning mutagenesis. J. Mol. Biol. 338: 811–825. [DOI] [PubMed] [Google Scholar]
- Yan S., Kennedy S., Koide S. 2002. Thermodynamic and kinetic exploration of the energy landscape of Borrelia burgdorferi OspA by native-state hydrogen exchange. J. Mol. Biol. 323: 363–375. [DOI] [PubMed] [Google Scholar]
- Yang X.F., Pal U., Alani S.M., Fikrig E., Norgard M.V. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199: 641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]