Abstract
The rate of quinol oxidation by cytochrome bc1/b6f complex is in part associated with the redox potential (Em) of its Rieske [2Fe-2S] center, for which an approximate correlation with the number of hydrogen bonds to the cluster has been proposed. Here we report comparative resonance Raman (RR) characterization of bacterial and archaeal high-potential Rieske proteins and their site-directed variants with a modified hydrogen bond network around the cluster. Major differences among their RR spectra appear to be associated in part with the presence or absence of Tyr-156 (in the Rhodobacter sphaeroides numbering) near one of the Cys ligands to the cluster. Elimination of the hydrogen bond between the terminal cysteinyl sulfur ligand (St) and Tyr-Oη (as with the Y156W variant, which has a modified histidine Nɛ pKa,ox) induces a small structural bias of the geometry of the cluster and the surrounding protein in the normal coordinate system, and significantly affects some Fe-Sb/t stretching vibrations. This is not observed in the case of the hydrogen bond between the bridging sulfide ligand (Sb) and Ser-Oγ, which is weak and/or unfavorably oriented for extensive coupling with the Fe-Sb/t stretching vibrations.
Keywords: resonance Raman, Rieske protein, [2Fe-2S] cluster, hydrogen bond, cytochrome bc1 complex, Rhodobacter, archaea
Proteins containing Rieske-type [2Fe-2S] clusters play important roles in many biological electron transfer reactions such as aerobic respiration, photosynthesis, and biodegradation of various alkene and aromatic compounds (Mason and Cammack 1992; Trumpower and Gennis 1994; Link 1999; Berry et al. 2000). Their distinct biological function is in part associated with the cluster redox potential (Em), for which an approximate correlation with the number of hydrogen bonds to the cluster has been proposed (Iwata et al. 1996; Colbert et al. 2000; Hunsicker-Wang et al. 2003). Despite this fundamental importance, evaluation of the contribution of structure to function for each hydrogen bond in the immediate cluster environment is often difficult to address experimentally in many iron-sulfur proteins, because several hydrogen bonds are contributed by the peptide backbone (Iwata et al. 1996; Colbert et al. 2000; Hunsicker-Wang et al. 2003; Iwasaki et al. 2004b).
In the cytochrome (cyt) bc1/b6f family, a Rieske protein contributes to the high-potential electron transfer chain involved in the bifurcated mechanism at the quinol-oxidizing Qo site, catalyzing a crucial step in the coupled electron–proton transfer that is linked to generation of a transmembrane electrochemical proton gradient (Berry et al. 2000; Crofts 2004). All of its Sb/t atoms are involved in a complex hydrogen bonded network (through which the cluster is linked to the surrounding polypeptide chain), which includes contributions from the side-chain hydroxyl groups of highly conserved Ser-154 (to Sb) and Tyr-156 (to St of Cys-129) (in Rhodobacter sphaeroides numbering [Guergova-Kuras et al. 2000]; equivalent to Ser-163 and Tyr-165, respectively, in bovine numbering [Iwata et al. 1996]) (Fig. 1A). This peculiarity has provided an opportunity to explore the influence of these hydrogen bonds on the cluster Em of the cyt bc1-associated Rieske proteins by site-directed mutagenesis (Denke et al. 1998; Schröter et al. 1998; Guergova-Kuras et al. 2000). Elimination of each hydroxyl group lowers the cluster Em, changes the EPR spectrum of the reduced Rieske [2Fe-2S] center, and decreases the rate of ubiquinol oxidation by cyt bc1 complex (Denke et al. 1998; Schröter et al. 1998; Guergova-Kuras et al. 2000). In at least one case, it also influences the pKa,ox value, which was attributed to the Nɛ of the histidyl ligands of the cluster (Guergova-Kuras et al. 2000). Although these studies have provided the basis for understanding its particular mechanistic importance, the nature of each hydrogen bond and its contribution to the immediate cluster environment have not been investigated in detail.
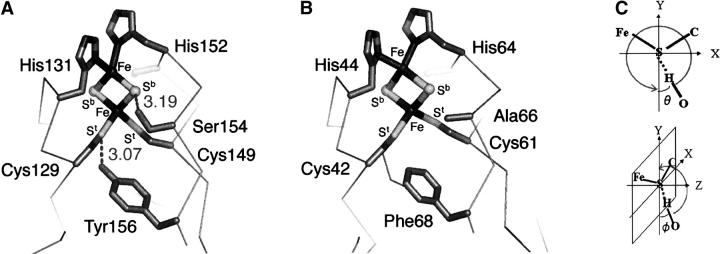
Figure 1.
The structures of the Rieske [2Fe-2S] cluster site in R. sphaeroides ISP (A) (taken from 1rie.pdb but in R. sphaeroides numbering [Guergova-Kuras et al. 2000]) and S. tokodaii SDX (B) (Uchiyama et al. 2004; T. Iwasaki, T. Uchiyama, A. Kounosu, and T. Kumasaka, unpubl.) showing the two targeted residues of interest and the four ligand residues. The proline-rich loop region was omitted for clarity. (C) The hydrogen bond orientation used for the preliminary normal mode calculation, as defined by ϕ between the Fe-S-C (or Fe-S-Fe) plane and S–HO bond; and θ of the S–HO bond within the Fe-S-C (or Fe-S-Fe) plane.
We report here a comparative study of variations in resonance Raman (RR) spectra of a repertoire of bacterial and archaeal high-potential Rieske proteins with different hydrogen-bond networks. These have been varied by site-directed mutagenesis in three well-characterized mutant proteins of the Rieske cluster binding domain solubilized by proteolytic cleavage from the hydrophobic N-terminal tail (ISP) of R. sphaeroides cyt bc1 complex (Em, acid pH = +315 mV; pKa,ox1, 7.6 ± 0.1) (Fig. 1A; Guergova-Kuras et al. 2000; Zu et al. 2003) and two mutant proteins of the high-potential, archaeal sulredoxin (SDX) from Sulfolobus tokodaii strain 7 (Em, acid pH = +188 mV; pKa,ox1 of the visible CD transition [corresponding to pKa,ox2 of the Em], 8.4 ± 0.2) (Iwasaki et al. 1995, 1996, 2004a; Kounosu et al. 2004; Uchiyama et al. 2004) with weak homology with the regular cyt bc1-associated Rieske proteins (DDBJ-EMBL accession no. AB023295) (Fig. 1B). In contrast with the wild-type ISP, there is no hydrogen-bonding interaction between the Sb/t atoms and side-chain groups in the wild-type SDX because of the presence of Ala-66 and Phe-68 in place of Ser-154 and Tyr-156, respectively, as confirmed by the preliminary crystal structure of the wild-type SDX being refined at 2.0 Å resolution (Fig. 1B; Uchiyama et al. 2004; T. Iwasaki, T. Uchiyama, A. Kounosu, and T. Kumasaka, unpubl.).
Results and Discussion
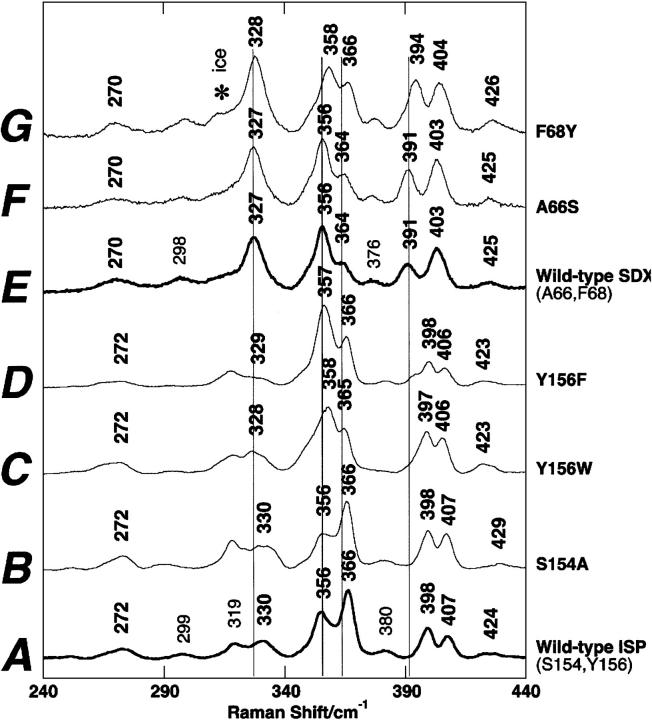
The RR spectra at 77 K exhibit similar low symmetry of the oxidized [2Fe-2S] clusters in bacterial ISP (Fig. 2A) and archaeal SDX (Fig. 2E) with distinguishable features in the 240–440 cm−1 region that are indicative of some structural differences in the surrounding protein (Iwasaki et al. 2004a). In the ISP mutants (Guergova-Kuras et al. 2000; S. Lhee, D.R. Kolling, C.J. Yoon, and A.R. Crofts, unpubl.), a negligible RR spectral change was detected for S154A (Fig. 2B) with reference to the wild-type ISP (Fig. 2A), whereas marked changes in the relative intensities of two dominant bands at 356 and 366 cm−1 with small band shifts (<2 cm−1) are observed for Y156W (Fig. 2C) and Y156F (Fig. 2D). In the SDX mutants, essentially no RR spectral change was detected between the wild-type (Fig. 2E) and A66S spectra (Fig. 2F), whereas the upshift (<4 cm−1) of the vibrational modes in the 300–410 cm−1 region was observed for F68Y spectra (Fig. 2G). Thus, the elimination (in ISP [Fig. 2C,D]) or import (into SDX [Fig. 2G]) of the hydrogen bond between St and Tyr-Oη causes marked effects on the RR bands in the 320–400 cm−1 region of these high-potential proteins, whereas the elimination (in ISP [Fig. 2B]) or import (into SDX [Fig. 2F]) of the hydrogen bond between Sb and Ser-Oγ group does not. Because the crystal structure (at 1.6 Å resolution) of the bovine mitochondrial ISP (Iwata et al. 1996) shows similar hydrogen bond distances between Sb and Ser-Oγ (3.19 Å) and between St and Tyr-Oη (3.07 Å), these variations cannot solely be ascribed to the differences in their hydrogen bond strengths. Indeed, the larger effect on Em (Denke et al. 1998; Schröter et al. 1998; Guergova-Kuras et al. 2000) would suggest a stronger effect from the serine.
Figure 2.
Comparative RR spectra of the wild-type and site-directed variant forms of R. sphaeroides ISP (A–D) and S. tokodaii SDX (E–G) in the oxidized state, obtained at 77K using 488.0-nm Ar+ laser excitation (Iwasaki et al. 2004a). An asterisk in the spectra indicates an ice made from buffer.
Preliminary normal mode analyses using simple hydrogen bonding models of the Rieske [2Fe-2S] cluster system (Supplemental Figs. S1, S2) suggest that the Fe-St/b stretching vibrations are affected by the hydrogen bond orientation (as defined by ϕb between the Fe-Sb-Fe plane and the Sb–HOγ bond, θb of the Sb–HOγ bond within the Fe-Sb-Fe plane, ϕt between the Fe-St-C plane and the St–HOη bond, and θt of the St–HOη bond within the Fe-St-C plane; see Fig. 1C). For instance, the Fe-St/b stretching vibrations are estimated to exhibit their upshift pattern around the 320–370 cm−1 region at ϕb∼0° or ∼180° (i.e., the Sb–HOγ bond is near-parallel with the Fe-Sb-Fe plane) and θb fulfilling the angular conditions where the Sb–HOγ bond is near perpendicular to the Fe–Fe axis, whereas the shift is negligible (<1 cm−1) at ϕb∼90° (i.e., the Sb–HOγ bond being near perpendicular to the Fe-Sb-Fe plane) (Fig. 1C; Supplemental Fig. S1). Thus, our experimental RR results, in conjunction with the preliminary normal mode analyses (Supplemental Figs. S1, S2A), suggest that (1) the hydrogen bond between Sb and Ser-Oγ is unfavorably oriented for extensive coupling of the normal modes of the (Fe-)Sb…HO type hydrogen bonding with the Fe-Sb/t stretching vibrations (Fig. 2A,B,E,F), i.e., the ϕb being close to 90° (like the mitochondrial Rieske protein (Fig. 1A; Iwata et al. 1996), and/or (2) it is simply weak. The absence of band shifts in the 390–440 cm−1 region of the experimental A66S spectrum (cf. Fig. 2E,F and Supplemental Fig. S2A), in which contributions of Fe-Sb stretching modes become dominant in this protein system (Rotsaert et al. 2003; Iwasaki et al. 2004a), supports this conclusion.
In contrast, formation of the hydrogen bond between St of the Cys ligand (Cys-129 in ISP and Cys-42 in SDX) and Tyr-Oη significantly affects the relative intensities of the pair of adjacent intense RR bands in the 350–370 cm−1 region, rather than slightly shifting some related (Fe-St) vibrational modes (Fig. 2A,C,D,E,G; Supplemental Fig. S2B). First, the effect of the extra hydrogen bonding with St on the upshift of the Fe-St/b stretching vibrations in this region (Fig. 2E,G) can be expected with ϕt∼0° or ∼180° (i.e., the St–HOη bond is near parallel with the Fe-St-C plane) and θt fulfilling the angular conditions where the St–HOη bond is near perpendicular to the St-C bond axis (or near-parallel with the Fe-St bond axis) (Fig. 1C; Supplemental Fig. S1). Second, in addition to this orientational effect, one would expect more significant RR band shifts with the protein variants having the modified (Fe-)St…HO type hydrogen bond in the 320–370 cm−1 region than those having the modified (Fe-)Sb…HO type hydrogen bond (in agreement with the spectra in Fig. 2), because of a higher probability for stronger coupling of the former hydrogen bond to the Fe-Sb/t stretching vibrations (dominating in the experimental spectra) than the latter hydrogen bond in the Rieske [2Fe-2S] cluster system (Supplemental Fig. S2). Third, small discrepancies of the band-shift patterns observed among the experimental spectra of the ISP and SDX variants (Fig. 2, cf. A,C,D and E,G), and the calculated normal vibrations having Fe-Sb/t stretching contribution in a simple model (Supplemental Fig. S2), can be explained by also considering that the hydrogen bonding between St and Tyr-Oη can induce a small structural bias affecting the geometry of the Rieske [2Fe-2S] cluster core and the conformation of the surrounding protein in the normal coordinate system, thereby promoting different resonance enhancements of some Fe-St/b vibrational modes and modifying relative (Sb-)Fe-St bond strengths that are primarily defined by extensive coupling to the surrounding protein. This is clearly seen in the spectra of the ISP variants in the 350–370 cm−1 region (Fig. 2A,C,D), which look more like the SDX spectrum (Fig. 2G). Thus, the upshift of the pKa,ox1 of the histidine Nɛ in the Y156W variant, which also affects the pH dependence of the rate of quinol oxidation in the cyt bc1 complex (Guergova-Kuras et al. 2000), can be attributed (in part) to the structural bias of the surrounding protein that increases the negative charge density around the sulfur atoms of the cluster. It will be interesting to see the atomic details of these structural biases when high-resolution X-ray crystal structures of the ISP and SDX variants become available.
In conclusion, our present results demonstrate that (1) individual hydrogen bonds (with Sb vs. St) affect the geometry and electronic properties of the Rieske [2Fe-2S] cluster environment differently, and (2) there is an orientation dependence for the extent of coupling of the internal coordinates contributed from hydrogen bonding with the Fe-Sb/t stretching vibrations. Some of these effects can be interpreted in terms of simple kinematic aspects (see Supplemental Figs. S1, S2) and may be applicable to other iron–sulfur protein systems. The modification of the hydrogen bond between St and the Tyr-Oη group induces a larger structural bias of the geometry of the Rieske [2Fe-2S] cluster and the surrounding protein in the normal coordinate system and significantly affects the Fe-Sb/t stretching vibrations mixed with many protein modes. This is not the case with modification of the hydrogen bond between Sb and Ser-Oγ, because the hydrogen bond is weak and/or unfavorably oriented for extensive coupling with the Fe-Sb/t stretching vibrations. Intriguingly, in agreement with the conclusions drawn herein, natural selection has exclusively utilized the latter hydrogen bond in the cyt bc1/b6f family for evolutionary tuning of the Rieske cluster Em with different quinol substrates (e.g., menaquinol vs. ubiquinol) (Mason and Cammack 1992; Trumpower and Gennis 1994; Link 1999; Berry et al. 2000).
Materials and methods
Escherichia coli strain HB101 (TaKaRa) used for cloning was grown in Luria-Bertani (LB) or Terrific broth (TB) medium, with 50 μg/mL kanamycin when required. The expression vector, pET28a, was purchased from Novagen. Water was purified by a Milli-Q purification system (Millipore). Other chemicals mentioned in this study were of analytical grade.
The nucleotide sequence determination was performed by the dideoxy chain termination method with an automatic DNA sequencer, ABI PRISM 310 Genetic Analyzer (PE Biosystems). The DNA sequence was processed with the DNASIS v3.6 software (Hitachi Software Engineering Co., Ltd). The homology search against databases was performed with the BEAUTY and BLAST network service (Worley et al. 1995). The structural alignments and figures were generated with PyMOL (DeLano Scientific).
The sdx gene coding for S. tokodaii SDX has been cloned and sequenced (Kounosu et al. 2004). Site-directed mutagenesis was performed by the PCR mutagenesis technique with a QuikChange Site-Directed Mutagenesis Kit (Stratagene), using a pET28aSDX vector harboring the sdx gene (Kounosu et al. 2004) as a long template. PCR mutagenesis was carried out with sets of the following PCR primers: 5′-TGCCATCTATCATTATTTGATCTAAGG-3′ and 5′-CCTTAGATCAAATAATGATAGATGGCA-3′ for the A66S mutant (replacement of Ala-66 by serine), and 5′-TGCCATCTAGCATTATATGATCTAAGG-3′ and 5′-CCTTAGATCATATAATGCTAGATGGCA-3′ for the F68Y mutant (replacement of Phe-68 by tyrosine). Each amplified PCR product was individually treated with DpnI and transformed into E. coli HB101 competent cells. In each case, the nucleotide sequences of the resultant vectors were confirmed for both strands.
The pET28aSDX vectors harboring the sdx gene and its variants were transformed separately into the host strain, E. coli BL21-CodonPlus(DE3)-RIL strain (Stratagene). The transformants were grown overnight at 25°C in LB medium containing 50 μg/mL kanamycin and 100–200 μM FeCl3, and the recombinant holoprotein was overproduced with 1 mM IPTG for 24 h at 25°C. The cells were pelleted by centrifugation and stored at −80°C until use. Purification of each recombinant holoprotein having a hexahistidine tag at the N terminus was performed as described previously (Iwasaki et al. 2004a; Kounosu et al. 2004). After proteolytic removal of the hexahistidine tag from the recombinant protein for 16–22 h at ∼25°C using a Thrombin Cleavage Capture Kit (Novagen) according to the manufacturer's instruction, the recombinant holoprotein was fully oxidized for 1–2 min with a small amount of potassium ferricyanide, which was then removed by using a Sephadex G-75 gel filtration column (Amersham Bioscience) equilibrated with 350 mM NaCl, 20 mM Tris-HCl (pH 7.5). The extra hydrogen bond introduced at the targeted position was indicated by the substantial upfield change of gx in the EPR spectra of the dithionite-reduced variants thus prepared (from 1.79–1.73 in A66S and to 1.77 in F68Y) (cf. Denke et al. 1998; Schröter et al. 1998; Guergova-Kuras et al. 2000). The purified recombinant holoproteins were stored at either 4°C or −80°C until use.
The sample preparations of the wild-type ISP of R. sphaeroides and its variants have been reported (Guergova-Kuras et al. 1999, 2000; Iwasaki et al. 2004a). For the S154A mutant, site-directed mutagenesis was performed by the PCR mutagenesis technique with a QuikChange Site-Directed Mutagenesis Kit with the modification of 6% (v/v) DMSO in the final reaction mixture and using the pGBH6 vector harboring the fbc operon (Guergova-Kuras et al. 1999), with the following PCR primer containing the SacI site: 5′-TCTGCCCCTGCCACGGAGCTCACTACGACAGTGCC-3′ (replacement of Ser-154 by alanine). Each amplified PCR product was individually treated with DpnI and transformed into E. coli DH5α competent cells. The nucleotide sequence of the resultant vector was confirmed.
The fragment from the pGBH6 vector harboring the S154A fbc operon restricted with EcoRI and HindIII was ligated into pRK415 (Keen et al. 1988; Guergova-Kuras et al. 1999) and transformed into the host strain, E. coli S17–1 (Simon et al. 1983). The modified pRK415 vector was introduced into R. sphaeroides BC17 (Guergova-Kuras et al. 1999) by facilitated conjugation with the transformed E. coli strain S17–1. The ISP from R. sphaeroides was purified as described previously (Iwasaki et al. 2004a), with the following modifications: In place of the DEAE-Sephacel column, the sample was applied to a 1-mL phenyl sepharose column (Amersham Biosciences) equilibrated with 1.5 M NaCl and 25 mM Na2HPO4 (pH 7.0). The column was washed with 10 column volumes of the equilibration buffer, and then the bound ISP was eluted by using a buffer of lower ionic strength (generally 25 mM Na2HPO4 at pH 7.0), concentrated with a Centriplus 7-kDa cutoff centrifugal concentrator (Amicon), buffer-exchanged into 400 mM NaCl, 50 mM K2HPO (pH 7.0), and 20% (v/v) glycerol, and stored at −80°C.
Absorption spectra were recorded with a Hitachi U3210 spectrophotometer or a Beckman DU-7400 spectrophotometer equipped with a thermoelectric cell holder. Electron paramagnetic resonance (EPR) measurements were performed at 8–15 K by using a JEX-RE3X spectrometer equipped with an ES-CT470 Heli-Tran cryostat system and a Scientific Instruments digital temperature indicator/controller Model 9650 (Iwasaki et al. 2000). Low-temperature RR spectra were recorded at 77 K using a Spex 750M Raman spectrometer fitted with a Spectrum-One 2048 × 512 CCD camera and a Spectra-Physics 2017 Ar+ laser (output 500 mW) by collecting 45° backscattering off the surface of a frozen sample in a quartz microcell (Iwasaki et al. 2004a). The slit width of the spectrometer is 80 μm, and a multiscan signal-averaging technique was employed to improve the signal-to-noise ratio. The spectral data were processed using KaleidaGraph v3.51 (Synergy Software). Preliminary normal coordinate analysis was performed by the Wilson's GF matrix method using a Urey-Bradley force field, as described previously (Saito et al. 1991). Some preliminary results are presented in the Supplemental Material (Figs. S1, S2), along with the force constant values for the Sb/t-H stretching modes and Fe-Sb/t-H bending motion of the Fe-Sb/t…HO type hydrogen bonding (PDF).
Electronic supplemental material
Influence of the hydrogen bond orientation on the Fe-S stretching vibration, by using simple five-atom models derived from the Cartesian coordinates taken from the PDB code 1rie.pdb (Iwata et al. 1996) is presented in Supplemental Fig. S1. Influence of a strong Sb/t-HO hydrogen bond within a Rieske [2Fe-2S] protein environment, by using simple “five-residue” models (Cys42, His44, Cys61, and His64, plus Ser66 or Tyr68) derived from the Cartesian coordinates taken from the X-ray crystal structure of S. tokodaii SDX (Uchiyama et al. 2004; T. Iwasaki, T. Uchiyama, A. Kounosu, and T. Kumasaka, unpubl.) is presented in Supplemental Fig. S2 (PDF).
Acknowledgments
This work was supported in part by the Japanese MEXT grants-in-aid 15770088 and 18608004 (T.Iw.), by JSPS Grant BSAR-507 (T.Iw.), by NSF Grant 9910113 (S.A.D.), by NIH grants GM35438 (A.R.C.) and GM62954 (S.A.D.), by the National Priority Research Program, Protein 3000 Project (T.K.), by the Frontier Project “Mechanism of Adaptation and Evolution of Extremophiles and Their Applications” to Rikkyo University, and by financial support to the Research Center for Measurement in Advanced Science of Rikkyo University.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Toshio Iwasaki, Department of Biochemistry and Molecular Biology, Nippon Medical School, Sendagi, Bunkyo-ku, Tokyo 113-8602, Japan; e-mail: tiwasaki@nms.ac.jp; fax: +81-3-5685-3054.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.052035406.
Abbreviations: cyt, cytochrome; Em, redox potential; ISP, the Rieske cluster binding domain solubilized from cytochrome bc1 complex by proteolytic cleavage; SDX, sulredoxin (a high-potential Rieske [2Fe-2S] protein from Sulfolobus tokodaii); RR, resonance Raman; Sb, bridging sulfide ligand; St, terminal cysteinyl sulfur ligand; Sb/t, Sb and/or St; ϕb, hydrogen bond orientation as defined by the angle between the Fe-Sb-Fe plane and the Sb–HOγ bond; θb, hydrogen bond orientation as defined by the angle of the Sb–HOγ bond within the Fe-Sb-Fe plane; ϕt, hydrogen bond orientation as defined by the angle between the Fe-St-C plane and the St–HOη bond; θt, hydrogen bond orientation as defined by the angle of the St–HOη bond within the Fe-St-C plane.
References
- Berry E.A., Guergova-Kuras M., Huang L.-S., Crofts A.R. 2000. Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 69: 1005–1075. [DOI] [PubMed] [Google Scholar]
- Colbert C.L., Couture M.M.-J., Eltis L.D., Bolin J. 2000. A cluster exposed: Structure of the Rieske ferredoxin from biphenyl dioxygenase and redox properties of Rieske Fe-S proteins. Structure 8: 1267–1278. [DOI] [PubMed] [Google Scholar]
- Crofts A.R. 2004. The cytochrome bc1 complex: Function in the context of structure. Annu. Rev. Physiol. 66: 689–733. [DOI] [PubMed] [Google Scholar]
- Denke E., Merbitz-Zahradnik T., Hatzfeld O.M., Snyder C.H., Link T.A., Trumpower B.L. 1998. Alteration of the midpoint potential and catalytic activity of the Rieske iron-sulfur protein by changes of amino acids forming hydrogen bonds to the iron-sulfur cluster. J. Biol. Chem. 273: 9085–9093. [DOI] [PubMed] [Google Scholar]
- Guergova-Kuras M., Salcedo-Hernandez R., Bechmann G., Kuras R., Gennis R.B., Crofts A.R. 1999. Expression and one-step purification of a fully active polyhistidine-tagged cytochrome bc1 complex from Rhodobacter sphaeroides. Protein Expr. Purif. 15: 370–380. [DOI] [PubMed] [Google Scholar]
- Guergova-Kuras M., Kuras R., Ugulava N., Hadad I., Crofts A.R. 2000. Specific mutagenesis of the Rieske iron-sulfur protein in Rhodobacter sphaeroides shows that both the thermodynamic gradient and the pK of the oxidized form determine the rate of quinol oxidation by the bc1 complex. Biochemistry 39: 7436–7444. [DOI] [PubMed] [Google Scholar]
- Hunsicker-Wang L.M., Heine A., Chen Y., Luna E.P., Todaro T., Zhang Y.M., Williams P.A., McRee D.E., Hirst J., Stout C.D. et al. 2003. High-resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and comparison. Biochemistry 42: 7303–7317. [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Isogai T., Iizuka T., Oshima T. 1995. Sulredoxin: A novel iron-sulfur protein of the thermoacidophilic archaeon Sulfolobus sp. strain 7 with a Rieske-type [2Fe-2S] cluster. J. Bacteriol. 177: 2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T., Imai T., Urushiyama A., Oshima T. 1996. Redox-linked ionization of sulredoxin of Sulfolobus sp. strain 7, an archaeal Rieske-type [2Fe-2S] protein. J. Biol. Chem. 271: 27659–27663. [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Watanabe E., Ohmori D., Imai T., Urushiyama A., Akiyama M., Hayashi-Iwasaki Y., Cosper N.J., Scott R.A. 2000. Spectroscopic investigation of selective cluster conversion of archaeal zinc-containing ferredoxin from Sulfolobus sp. strain 7. J. Biol. Chem. 275: 25391–25401. [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Kounosu A., Kolling D.R.J., Crofts A.R., Dikanov S.A., Jin A., Imai T., Urushiyama A. 2004a. Characterization of the pH-dependent resonance Raman transitions of archaeal and bacterial Rieske [2Fe-2S] proteins. J. Am. Chem. Soc. 126: 4788–4789. [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Kounosu A., Uzawa T., Samoilova R.I., Dikanov S.A. 2004b. Orientation-selected 15N-HYSCORE detection of weakly coupled nitrogens around the archaeal Rieske [2Fe-2S] center. J. Am. Chem. Soc. 126: 13902–13903. [DOI] [PubMed] [Google Scholar]
- Iwata S., Saynovits M., Link T.A., Michel H. 1996. Structure of a water soluble fragment of the ‘Rieske’ iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1complex determined by MAD phasing at 1.5 Å resolution. Structure 4: 567–579. [DOI] [PubMed] [Google Scholar]
- Keen N.T., Tamaki S., Kobayashi D., Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70: 191–197. [DOI] [PubMed] [Google Scholar]
- Kounosu A., Li Z., Cosper N.J., Shokes J.E., Scott R.A., Imai T., Urushiyama A., Iwasaki T. 2004. Engineering a three-cysteine, one-histidine ligand environment into a new hyperthermophilic archaeal Rieske-type [2Fe-2S] ferredoxin from Sulfolobus solfataricus. J. Biol. Chem. 279: 12519–12528. [DOI] [PubMed] [Google Scholar]
- Link T.A. 1999. The structures of Rieske and Rieske-type proteins. Adv. Inorg. Chem. 47: 83–157. [Google Scholar]
- Mason J.R. and Cammack R. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46: 277–305. [DOI] [PubMed] [Google Scholar]
- Rotsaert F.A.J., Pikus J.D., Fox B.G., Markley J.L., Sanders-Loehr J. 2003. N-isotope effects on the Raman spectra of Fe2S2 ferredoxin and Rieske ferredoxin: Evidence for structural rigidity of metal sites. J. Biol. Inorg. Chem. 8: 318–326. [DOI] [PubMed] [Google Scholar]
- Saito H., Imai T., Wakita K., Urushiyama A., Yagi T. 1991. Resonance Raman active vibrations of rubredoxin: Normal coordinate analysis of a 423-atom model. Bull. Chem. Soc. Jpn. 64: 829–836. [Google Scholar]
- Schröter T., Hatzfeld O.M., Gemeinhardt S., Korn M., Friedrich T., Ludwig B., Link T.A. 1998. Mutational analysis of residues forming hydrogen bonds in the Rieske [2Fe-2S] cluster of the cytochrome bc1 complex in Paracoccus denitrificans. Eur. J. Biochem. 255: 100–106. [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Biotechnology (N.Y.) 1: 784–791. [Google Scholar]
- Trumpower B.L. and Gennis R.B. 1994. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: The enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu. Rev. Biochem. 63: 675–716. [DOI] [PubMed] [Google Scholar]
- Uchiyama T., Kounosu A., Sato T., Tanaka N., Iwasaki T., Kumasaka T. 2004. Crystallization and preliminary X-ray diffraction studies of the hyperthermophilic archaeal sulredoxin having the unique Rieske [2Fe-2S] cluster environment. Acta Crystallogr. D Biol. Crystallogr. 60: 1487–1489. [DOI] [PubMed] [Google Scholar]
- Worley K.C., Wiese B.A., Smith R.F. 1995. BEAUTY: An enhanced BLAST-based search tool that integrates multiple biological information resources into sequence similarity search results. Genome Res. 5: 173–184. [DOI] [PubMed] [Google Scholar]
- Zu Y., Couture M.M.-J., Kolling D.R.J., Crofts A.R., Eltis L.D., Fee J.A., Hirst J. 2003. Reduction potentials of Rieske clusters: Importance of the coupling between oxidation state and histidine protonation state. Biochemistry 42: 12400–12408. [DOI] [PubMed] [Google Scholar]