Abstract
Electrostatic interactions are important for both protein stability and function, including binding and catalysis. As protein design moves into these areas, an accurate description of electrostatic energy becomes necessary. Here, we show that a simple distance-dependent Coulombic function parameterized by a comparison to Poisson-Boltzmann calculations is able to capture some of these electrostatic interactions. Specifically, all three helix N-capping interactions in the engrailed homeodomain fold are recovered using the newly parameterized model. The stability of this designed protein is similar to a protein forced by sequence restriction to have beneficial electrostatic interactions.
Keywords: protein design, electrostatics, engrailed, N-capping
The energy functions used in protein design must be rapidly evaluable due to the large size of protein design calculations. However, the physical interactions these energy functions are developed to represent are highly complex. Approximations introduced to increase the speed of calculations must also capture the intricate balance of the stabilizing and destabilizing interactions that lead to the observed marginal stability of proteins (Dill 1990).
Electrostatic interactions contribute significantly to protein stability and function. Not only must intraprotein interactions be considered (hydrogen bonding, salt bridges, etc.), but effects due to the aqueous environment such as polar solvation and solvent screening need to be evaluated as well. It is computationally intractable to consider all individual water molecules surrounding a protein during a protein design calculation. Continuum approaches that consider the solvent at a macroscopic level using various numerical solutions of the Poisson-Boltzmann equation (Honig and Nicholls 1995) have been used to predict side chain pKa's, the electrostatic component of binding, and other biologically important processes. However, a full Poisson-Boltzmann calculation is far too time-consuming to be used at each step of a protein design calculation. Various methods attempting to reproduce the accuracy of Poisson-Boltzmann calculations within the restrictions of protein design include the adaptation of a solvent exclusion method (Lazaridis and Karplus 1999) in designing a novel protein fold (Kuhlman et al. 2003), a modified Tanford-Kirkwood approach (Havranek and Harbury 1999) to design specific protein–protein interactions (Havranek and Harbury 2003), use of a Born method in a new protein design force field (Pokala and Handel 2004), and a highly parameterized set of simple terms (Wisz and Hellinga 2003) in designing enzymatic activity onto a previously catalytically inactive scaffold (Dwyer et al. 2004). Work in this lab led to the development of a two-body decomposable implementation of a Poisson-Boltzmann calculation useful in protein design (Marshall et al. 2005).
Previous design studies have shown both the importance of electrostatics and the need to improve the electrostatic component (Marshall et al. 2002) of our protein design algorithm, ORBIT (Dahiyat and Mayo 1996, 1997). Local interactions were shown to be underrepresented and hydrogen bonding was overrepresented relative to long-range Coulombic interactions.
Here we show that a comparison of ORBIT electrostatic energies and those calculated using the finite difference Poisson-Boltzmann implementation in DelPhi (Rocchia et al. 2001) allowed a parameterization of the simple Coulombic equation term used in ORBIT. By scaling the dielectric value it is possible to approximate the energies calculated using the more accurate Poisson-Boltzmann method. Local interactions (side chain–backbone) and longer range interactions (side chain–side chain) are parameterized separately. The polar solvation model used in this study penalizes the burial of non-hydrogen-bonded, non-backbone polar hydrogens.
Results
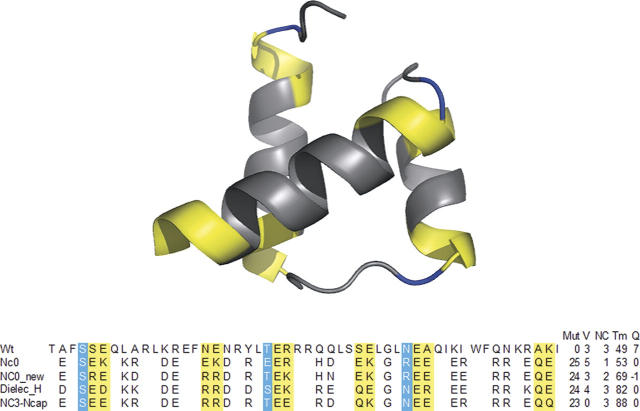
Electrostatic effects are studied in the background of the engrailed homeodomain, a small (51 amino acids) protein with three α helices (Fig. 1). The wild-type sequence is not optimized for stability with seven positive charges distributed across a small amount of surface. The protein sequence resulting from a design calculation with the unoptimized electrostatic terms of ORBIT is NC0 (Marshall et al. 2002). While eliminating the charge excess, this designed protein was shown to have incorporated a number of unfavorable electrostatic interactions relative to wild type: a reduced number of N-capping interactions and an increased number of potentially destabilizing interactions with the helix dipole. NC0 has a stability slightly greater than wild type and is used as the baseline for ORBIT's electrostatic performance in this study. An updated rotamer library that was shown to lead to similar designed sequences as a previous library was used in the study reported here. The sequence designed with this new library but with the unoptimized electrostatic term is NC0_new. This protein was designed as a control for the new electrostatic term.
Figure 1.
Engrailed homeodomain structure and designed sequences. N-capping positions are shown in blue. Designed positions that could interact with the helix dipoles are shown in yellow. Positions left blank were not varied in the design calculations. Also shown is Mut, the number of mutations from the wild-type sequence; V, the number of helix dipole violations at designed positions; NC, the number of N-capping interactions; Tm, the melting temperature; and Q, the theoretical charge of the sequence.
The analysis of a small set of proteins suggested that a distance-dependent dielectric of 5.1r for side chain–backbone interactions and 7.1r for side chain–side chain interactions more closely predicts the energy calculated by DelPhi (see Materials and Methods). Previous work in this lab used a distance-dependent dielectric of 40r. The new dielectric values led to a 7.8-fold increase in the strength of electrostatic interactions in the side chain–backbone case and a 5.6-fold increase in the side chain–side chain case. Thus, while the importance of electrostatics is increased significantly in the design calculations, it is further increased in the side chain–backbone case in order to address the concerns raised in Marshall et al. (2002). The design calculation using the optimized dielectric values and penalties based on the number of buried polar hydrogens is Dielec_H.
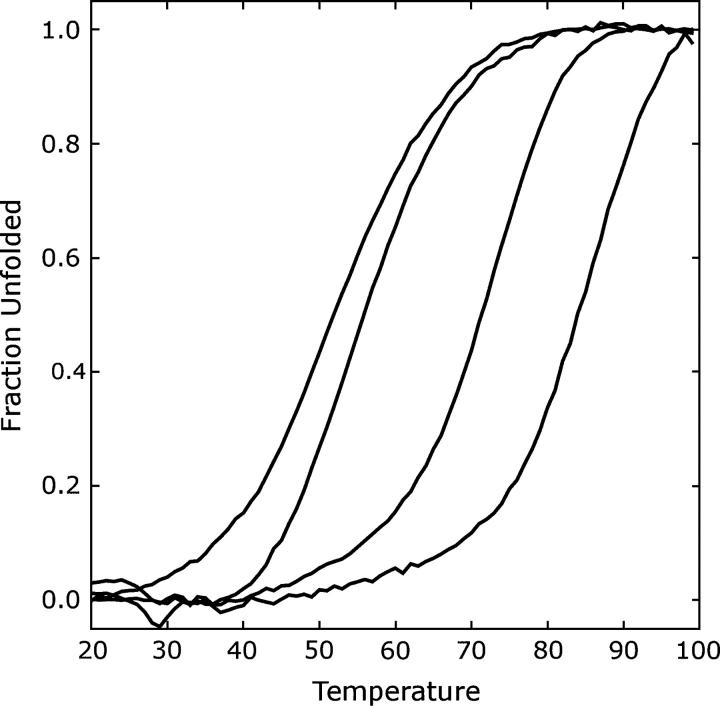
Circular dichroism wavelength scans indicated that the designed proteins were well folded and α-helical. Thermal denaturation studies were carried out on NC0, NC0_new, and Dielec_H (Fig. 2). All proteins unfolded completely and reversibly. For comparison, the thermal denaturation curve of wild-type engrailed is also included.
Figure 2.
Temperature denaturation curves of engrailed variants (from left to right): Wild type, NC0, NC0_new, Dielec_H.
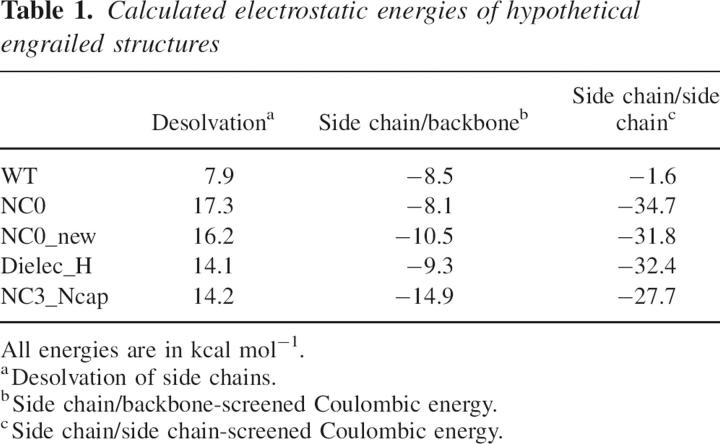
The dependence of protein design on the rotamer library used is shown clearly by the difference in sequence between NC0 and NC0_new (Fig. 1). While the rotamer libraries used have very similar numbers of rotamers and the calculations were otherwise identical, there are nine mutations between NC0 and NC0_new. The surface of a protein is much less constrained by sterics than the protein core, allowing a much greater choice of rotamers. A slight difference at one position leading to the choice of a different amino acid could propagate other changes across the protein surface during the design. The ORBIT calculated energies of these proteins are similar, but NC0_new is shown to have an unfolding transition temperature 21°C higher. By examining the electrostatic character of these sequences it is clear that NC0_new has both more beneficial N-capping interactions and less detrimental interactions with the helix dipole. There is some debate as to the importance of the helix dipole, especially at solvent-exposed positions (Gilson and Honig 1989; Sengupta et al. 2005). DelPhi analysis of the hypothetical structures of these sequences suggests that NC0_new experiences less of a desolvation penalty and better side chain–backbone interactions than NC0 (Table 1). Care must be taken with interpretation of this data due to the hypothetical nature of the structures and the strong conformation dependence of Poisson-Boltzmann calculations (Alexov 2003). Thus while NC0_new and NC0 were not designed to be significantly different, the differences that are observed both in sequence and measured stability can be explained, at least qualitatively, by electrostatic differences.
Table 1.
Calculated electrostatic energies of hypothetical engrailed structures
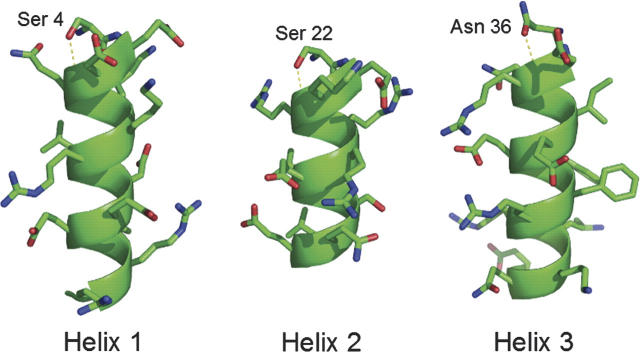
The sequences of Dielec_H and NC0_new can now be compared to determine the effects of simply modifying the electrostatic term to include lower dielectrics. Dielec_H is a very stable protein, approximately as stable as NC3_Ncap (Marshall et al. 2002), which was designed by preventing detrimental interactions with helix dipoles and forcing N-capping interactions by restricting amino acid composition. In this study, increasing the strength of side chain–backbone electrostatic interactions relative to side chain–side chain interactions appears to recover all three of the N-capping interactions in engrailed (Fig. 3). DelPhi calculations show that Dielec_H has more favorable desolvation and side chain–side chain energies than both NC0_new and NC3_Ncap (Table 1). However, these values are conformation-dependent, and the calculated difference in side chain–backbone energy between Dielec_H and NC3_Ncap is dependent primarily on the interactions of three glutamates that would likely assume more than one conformation in solution.
Figure 3.
Hypothetical structures of engrailed helices showing N-capping interactions in Dielec_H.
Discussion
Protein design requires the evaluation of a large number of functions for a complete calculation. These functions need to both be rapid and accurate. Unfortunately, the complexity of the protein energy surface does not lend itself to a simple representation. While a full Poisson-Boltzmann calculation at each step would lead to a more accurate view of the electrostatic environment of the protein, implementing such a procedure remains computationally intractable. The need for approximate functions necessitates the evaluation of both their accuracy and usefulness. A parameterization of the Coulombic term in ORBIT using the Poisson-Boltzmann equation implemented in DelPhi leads to an increase in the weight of electrostatics in the ORBIT force field as well as separate dielectric values for side chain–backbone and side chain–side chain interactions. While it is difficult to conclusively state the exact interactions that lead to differences in stability, in this work we show experimentally that simple modifications to the Coulombic term lead to a designed protein that is stabilized by electrostatic interactions as well as recovering helix N-capping interactions, a stabilizing feature of natural protein sequences.
Materials and methods
The new dielectric values were obtained by performing electrostatic calculations on a small set of proteins and determining the value that would lead to the best fit. The protein structures downloaded from the PDB are 1igh (β1 domain of protein G), 1rge (ribonuclease SA), 1rhe (Rhe VL), 1whi (L14 ribosomal protein), 1tta (transthyretin), 2rn2 (ribonuclease H), 3lzm (T4 lysozyme), and 1amm (γB-crystalin). The DelPhi (Rocchia et al. 2001) calculations used a grid spacing of 2.0 grids Å−1, an interior dielectric of 4.0, an exterior dielectric of 80.0, 0.050 M salt, and a probe radius of 1.4 Å. PARSE charges and radii were used (Sitkoff et al. 1994). In order to more directly compare with the terms in the ORBIT force field, the DelPhi results for both unfolded and folded states were separated into backbone and side chain desolvation and screened Coulombic interactions. The description of the unfolded and folded states of the backbone and the side chains can be found in Figures 1–3 in Marshall et al. (2005). The dielectric value used in the ORBIT Coulombic term is then scaled to more closely agree with the values calculated with DelPhi. Correlations coefficients of the fits between DelPhi electrostatic energies and scaled Coulombic energies (side chain/side chain and side chain/backbone) are >0.9 (data not shown). In order to facilitate comparison with previous work (Marshall et al. 2002), electrostatic calculations in Table 1 were performed with the same DelPhi parameters as above, with the exception of a probe radius of 0 Å.
The preparation of the engrailed homeodomain PDB (Berman et al. 2000) structure, 1enh, and the designed positions are the same surface positions as reported in Marshall et al. (2002). Residues allowed at the designed positions were Ala, Ser, Thr, Asp, Asn, His, Glu, Gln, Lys, and Arg. Rotamers are derived from the rotamer library of Dunbrack and Karplus (1994) with expansion of 1 standard deviation about angles χ1 and χ2 of aliphatic residues, expansion of 1 standard deviation around χ1 of hydrophobic residues, and no expansion of polar residue dihedral angles.
The force field in ORBIT contains van der Waals, Coulombic, hydrogen-bond, and solvation terms (Gordon et al. 1999). The hydrogen-bond term is geometry- and hybridization-dependent, as described in Dahiyat et al. (1997). The polar hydrogen burial term is calculated as 2.0 kcal/mol for each nonbackbone, non-hydrogen-bonded buried polar hydrogen, as described (Dahiyat et al. 1997). Sequence optimization was performed using DEE (Desmet et al. 1992; Goldstein 1994; Gordon and Mayo 1998) or HERO (Gordon et al. 2003). The one best sequence for each design was expressed and purified for biophysical analysis.
Genes for the engrailed variants were prepared by recursive PCR (Prodromou and Pearl 1992) and cloned into pET-11a (Novagen). Wild-type engrailed expresses poorly and was cloned into a plasmid that had been engineered to include N-terminal His-tags and a ubiquitin domain with a ubiquitin-specific cleavage site (Pilon et al. 1997). DNA sequencing confirmed the identity of all variants. Proteins were expressed in BL21(DE3) E. coli cells (Stratagene) and isolated with freeze-thaw (Johnson and Hecht 1994) or sonication. Proteins were purified by HPLC as in Marshall et al. (2002) or nickel exchange columns (Qiagen). Cleavage of the protein of interest from the fusion domain occurred by use of the protease UCH-L3 (Boston Biochem) at 37°C for 1–4 h. Proteins were confirmed with MALDI-TOF mass spectrometry. Temperature denaturation circular dichroism was carried out as described (Marshall et al. 2002).
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and the Ralph M. Parsons Foundation. E.S.Z. would like to thank the ARCS Foundation for funding.
Footnotes
Reprint requests to: Stephen L. Mayo, Biochemistry and Molecular Biophysics, California Institute of Technology, MC 114-96, 1200 E. California Blvd., Pasadena, CA 91125, USA; e-mail: steve@mayo.caltech.edu; fax: (626) 568-0934.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062105506.
References
- Alexov E. 2003. Role of the protein side-chain fluctuations on the strength of pair-wise electrostatic interactions: Comparing experimental with computed pKas. Proteins 50: 94–103. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiyat B.I. and Mayo S.L. 1996. Protein design automation. Protein Sci. 5: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiyat B.I. and Mayo S.L. 1997. De novo protein design: Fully automated sequence selection. Science 278: 82–87. [DOI] [PubMed] [Google Scholar]
- Dahiyat B.I., Gordon D.B., Mayo S.L. 1997. Automated design of the surface positions of protein helices. Protein Sci. 6: 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet J., De Maeyer M., Hazes B., Lasters I. 1992. The dead-end elimination theorem and its use in protein side-chain positioning. Nature 356: 539–542. [DOI] [PubMed] [Google Scholar]
- Dill K.A. 1990. Dominant forces in protein folding. Biochemistry 29: 7133–7155. [DOI] [PubMed] [Google Scholar]
- Dunbrack R.L. and Karplus M. 1994. Conformational analysis of the backbone-dependent rotamer preferences of protein side-chains. Nat. Struct. Biol. 1: 334–340. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A., Looger L.L., Hellinga H.W. 2004. Computational design of a biologically active enzyme. Science 304: 1967–1971. [DOI] [PubMed] [Google Scholar]
- Gilson M.K. and Honig B. 1989. Destabilization of an α-helix bundle protein by helix dipoles. Proc. Natl. Acad. Sci. 86: 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein R.F. 1994. Efficient rotamer elimination applied to protein side-chains and related spin-glasses. Biophys. J. 66: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.B. and Mayo S.L. 1998. Radical performance enhancements for combinatorial optimization algorithms based on the dead-end elimination theorem. J. Comput. Chem. 19: 1505–1514. [Google Scholar]
- Gordon D.B., Marshall S.A., Mayo S.L. 1999. Energy functions for protein design. Curr. Opin. Struct. Biol. 9: 509–513. [DOI] [PubMed] [Google Scholar]
- Gordon D.B., Hom G.K., Mayo S.L., Pierce N.A. 2003. Exact rotamer optimization for protein design. J. Comput. Chem. 24: 232–243. [DOI] [PubMed] [Google Scholar]
- Havranek J.J. and Harbury P.B. 1999. Tanford-Kirkwood electrostatics for protein modeling. Proc. Natl. Acad. Sci. 96: 11145–11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek J.J. and Harbury P.B. 2003. Automated design of specificity in molecular recognition. Nat. Struct. Biol. 10: 45–52. [DOI] [PubMed] [Google Scholar]
- Honig B. and Nicholls A. 1995. Classical electrostatics in biology and chemistry. Science 268: 1144–1149. [DOI] [PubMed] [Google Scholar]
- Johnson B.H. and Hecht M.H. 1994. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Biotechnology (N. Y.) 12: 1357–1360. [DOI] [PubMed] [Google Scholar]
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. 2003. Design of a novel globular protein fold with atomic-level accuracy. Science 302: 1364–1368. [DOI] [PubMed] [Google Scholar]
- Lazaridis T. and Karplus M. 1999. Effective energy function for proteins in solution. Proteins 35: 133–152. [DOI] [PubMed] [Google Scholar]
- Marshall S.A., Morgan C.S., Mayo S.L. 2002. Electrostatics significantly affect the stability of designed homeodomain variants. J. Mol. Biol. 316: 189–199. [DOI] [PubMed] [Google Scholar]
- Marshall S.A., Vizcarra C.M., Mayo S.L. 2005. One- and two-body decomposable Poisson-Boltzmann methods for protein design calculations. Proteins 14: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon A., Yost P., Chase T.E., Lohnas G., Burkett T., Roberts S., Bentley W.E. 1997. Ubiquitin fusion technology: Bioprocessing of peptides. Biotechnol. Prog. 13: 374–379. [DOI] [PubMed] [Google Scholar]
- Pokala N. and Handel T.M. 2004. Energy functions for protein design I: Efficient and accurate continuum electrostatics and solvation. Protein Sci. 13: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C. and Pearl L.H. 1992. Recursive PCR: A novel technique for total gene synthesis. Protein Eng. 5: 827–829. [DOI] [PubMed] [Google Scholar]
- Rocchia W., Alexov E., Honig B. 2001. Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent ions. J. Phys. Chem. B 105: 6507–6514. [Google Scholar]
- Sengupta D., Behera R.N., Smith J.C., Ullmann G.M. 2005. The α helix dipole: Screened out? Structure 13: 849–855. [DOI] [PubMed] [Google Scholar]
- Sitkoff D., Sharp K.A., Honig B. 1994. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 98: 1978–1988. [Google Scholar]
- Wisz M.S. and Hellinga H.W. 2003. An empirical model for electrostatic interactions in proteins incorporating multiple geometry-dependent dielectric constants. Proteins 51: 360–377. [DOI] [PubMed] [Google Scholar]